Hetionet in Neo4j: the tale of Project Rephetio
Seminar Group on Big and Scientific Data
University of Pennsylvania
February 10, 2017
DSL Conference Room
Moore 102
11:00 am – 12:00 pm
By Daniel Himmelstein
@dhimmel
Slides at slides.com/dhimmel/big-data-seminar
Greene Lab
I'm a data scientist
http://www.greenelab.com/

There are many graph databases. I'm most familiar with Neo4j which is:
- an ACID-compliant transactional database with native graph storage and processing
- the most popular graph database according to db-engines.com
- open source: Community (GPLv3 licensed) & Enterprise (AGPLv3 licensed) Editions
The Graph Mindset
how Susan Davidson's class was a microcosm of a larger academic network

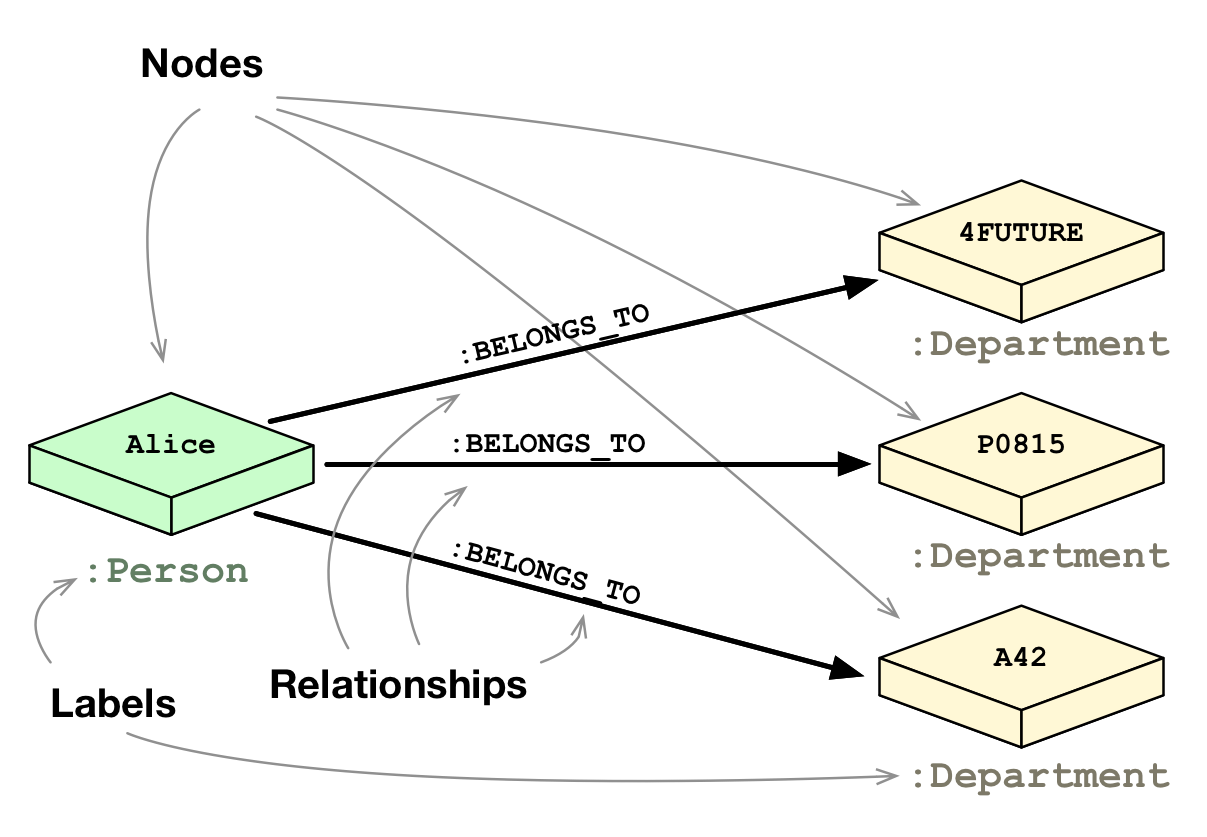
Graphs are composed of:
- Nodes
- Relationships
Nodes / relationships have type:
- node labels
(person, course, university) - relationship types
(lecturer, institution)

- first_name: Daniel
- last_name: Himmelstein
- twitter: @dhimmel
- SSN: 012-34-5678

- catalog: CIS550
- title: Database & Information Systems
- units: 5
- catalog: EPID600
- title: Data Science for Biomedical Informatics
- units: 1

- url: www.upenn.edu
- founded: 1740
- type: private
- league: ivy




- grade: A-
- grade: B
- grade: F

What can we do with this graph?
- Course statistics:
How many students are in CIS 550?
- Course recommendations:
What courses do other students in CIS 550 take?
- Course scheduling:
What room should a course be in so it's nearby other courses that its students are enrolled in?
Source: From Relational to Neo4j
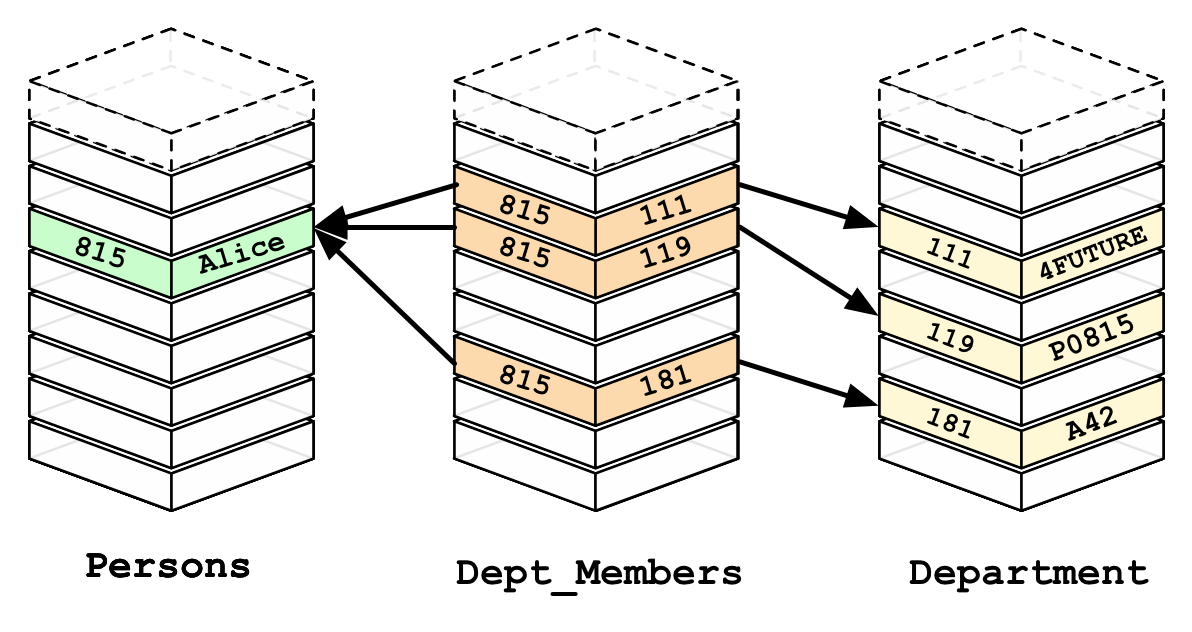
The Relational Database Model
Source: From Relational to Neo4j
The Relational Database Model
Limitations:
- Relationships require an intermediate table
- Schemas are cumbersome to create to maintain
Source: From Relational to Neo4j
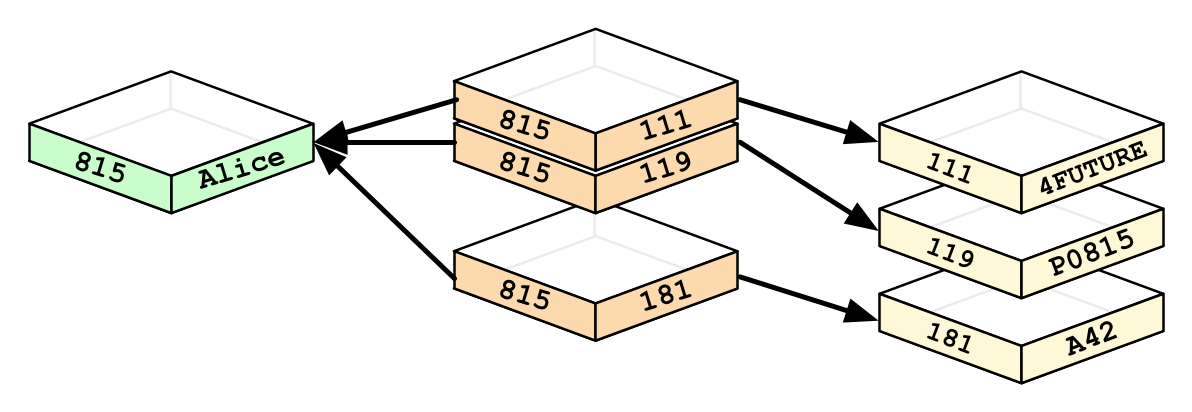
Relationships inherently form graphs
Source: From Relational to Neo4j
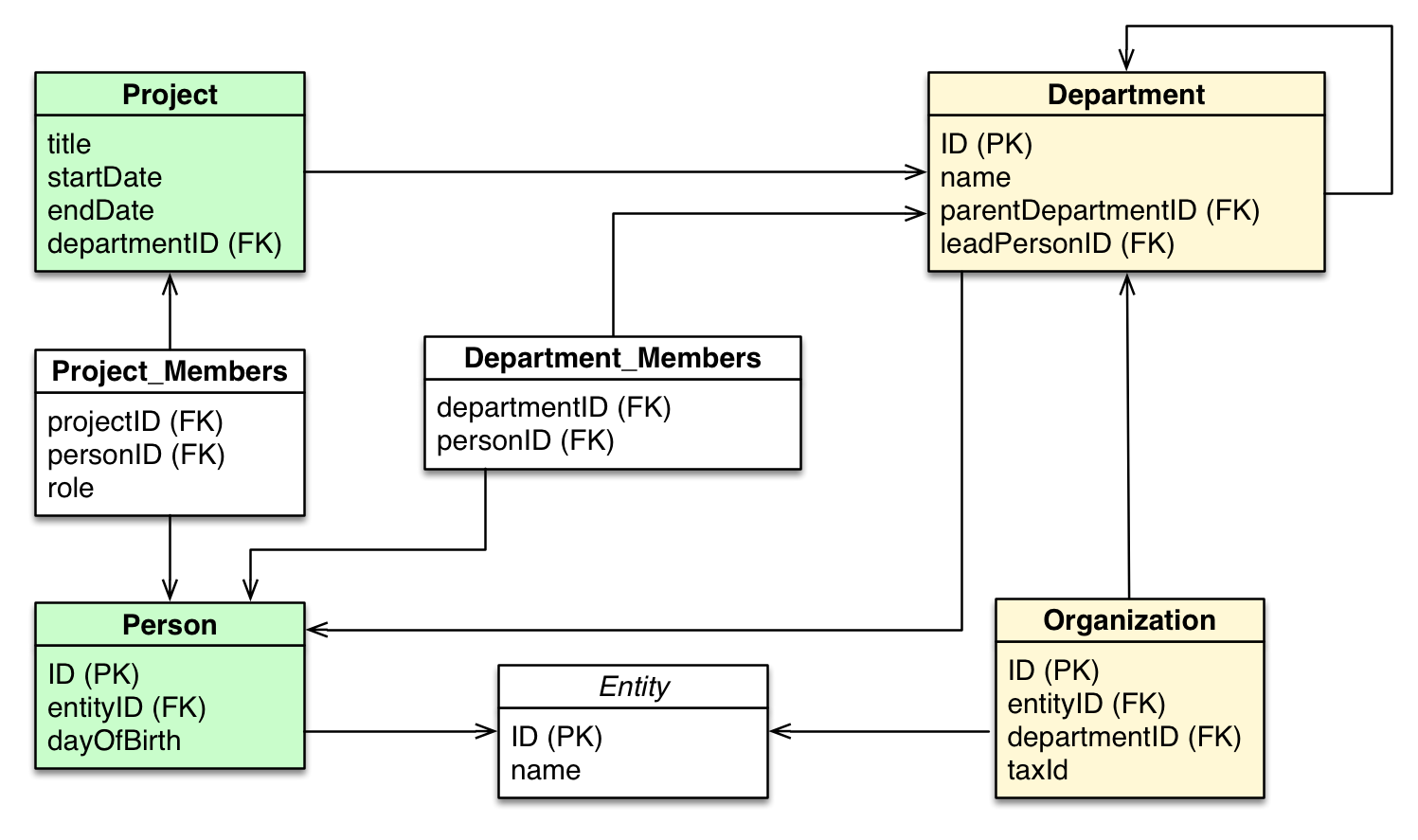
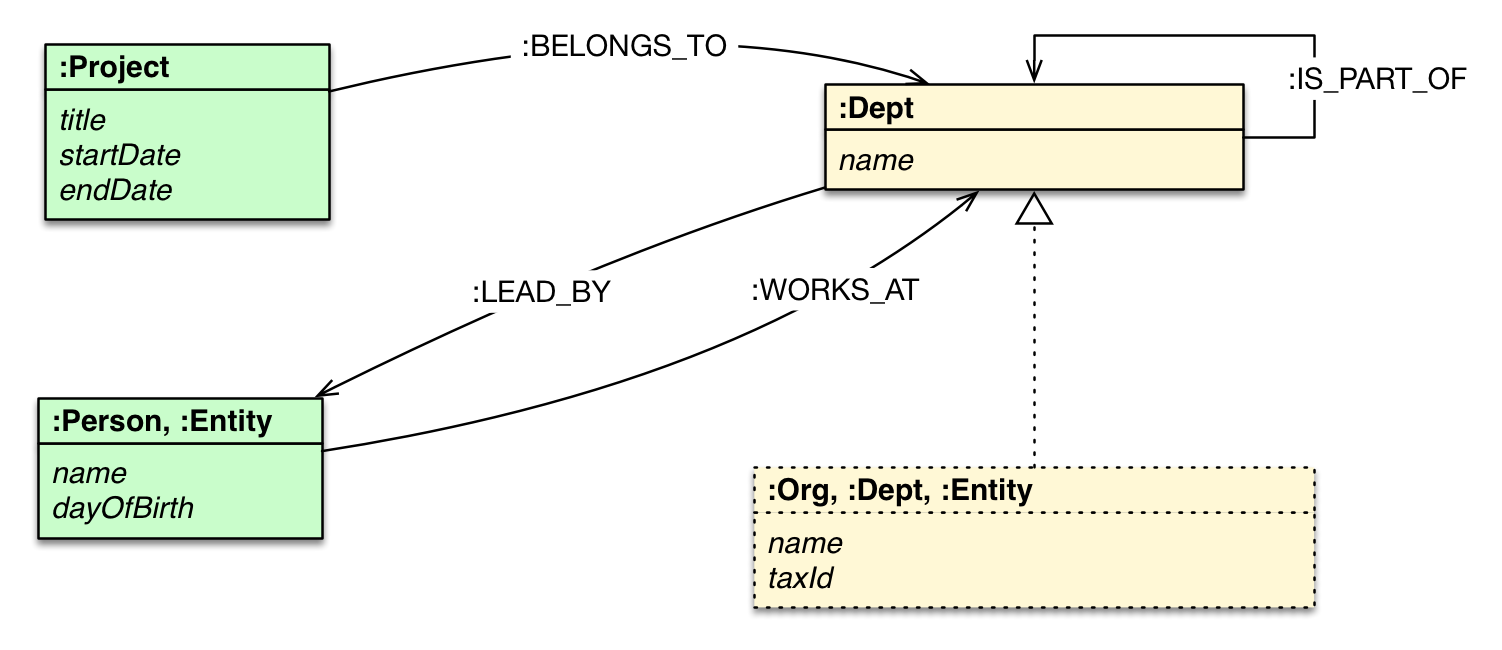
Relational database schema
Source: From Relational to Neo4j
Graph database schema (metagraph)
Emil Eifrem at GraphConnect 2015
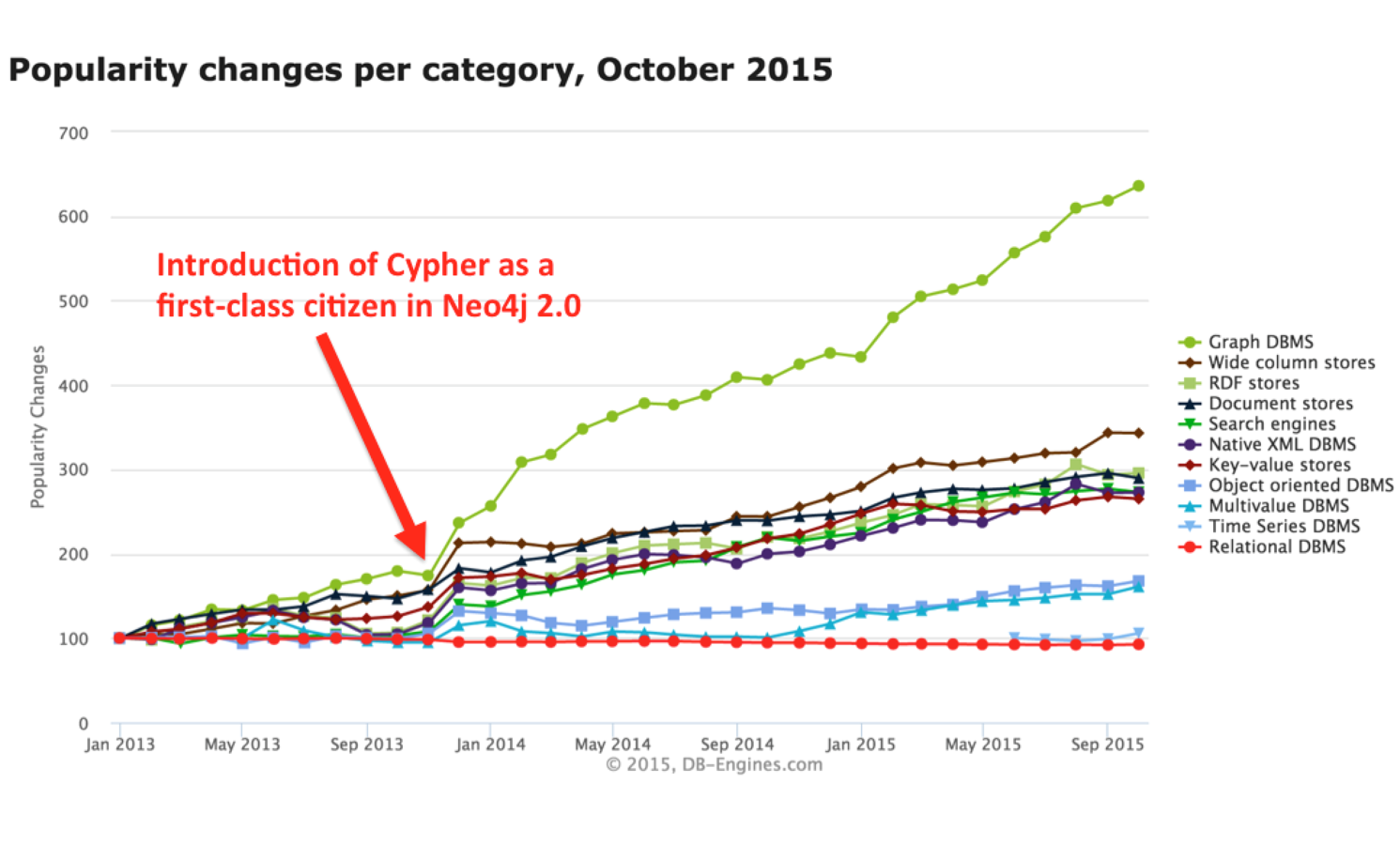
The rise of graph databases
Cypher accelerated graph database adoption
From Meet openCypher: The SQL for Graphs. Neo4j Blog
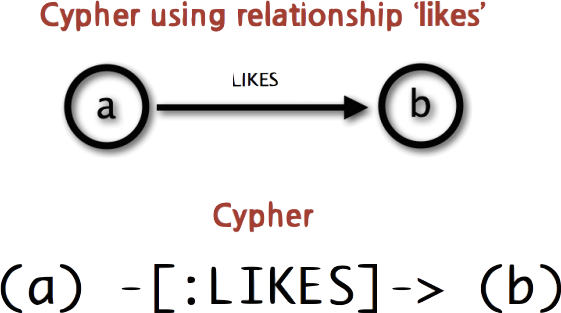
What Cypher looks like
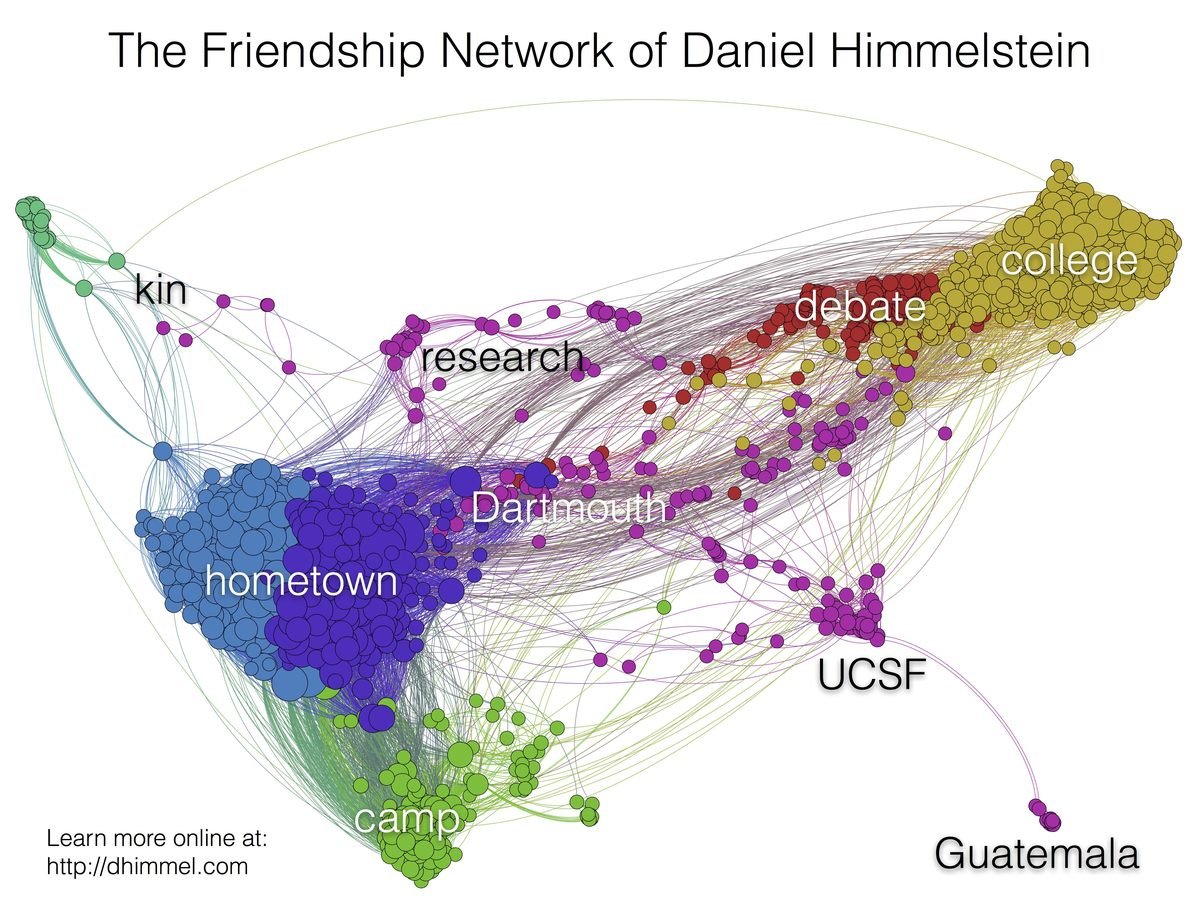
How I became intestested in graphs
http://blog.dhimmel.com/friendship-network/
How do you teach a computer biology?
multilayer network, multiplex network, multivariate network, multinetwork, multirelational network, multirelational data, multilayered network, multidimensional network, multislice network, multiplex of interdependent networks, hypernetwork, overlay network, composite network, multilevel network, multiweighted graph, heterogeneous network, multitype network, interconnected networks, interdependent networks, partially interdependent networks, network of networks, coupled networks, interconnecting networks, interacting networks, heterogenous information network
networks with multiple node or relationship types
A 2012 Study identified 26 different names for this type of network:
hetnet
What's the best software for storing and querying hetnets?
| dhimmel/hetio | |
|---|---|
| 86 | |
| 5 | |
| 2 |



| neo4j/neo4j |
|---|
| 42,498 |
| 3,071 |
| 1,007 |
GitHub stats from 2016-10-09
- Hetnet of biology designed for drug repurposing
- ~50 thousand nodes
11 types (labels)
- ~2.25 million relationships
24 types
- integrates 29 public resources
knowledge from millions of studies
- the hardest part:
licensing of publicly available data
Hetionet v1.0
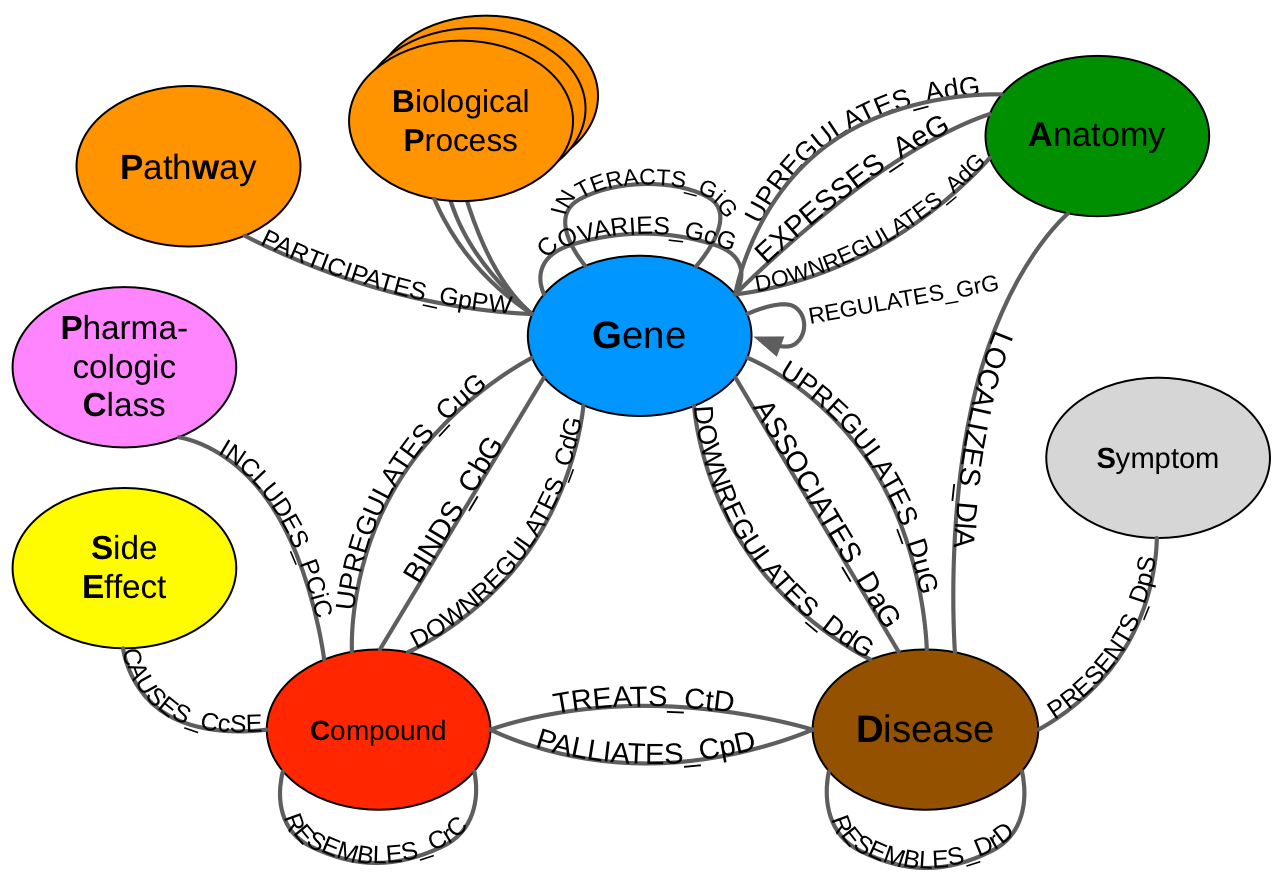
MetaGraph / Data Model / Schema
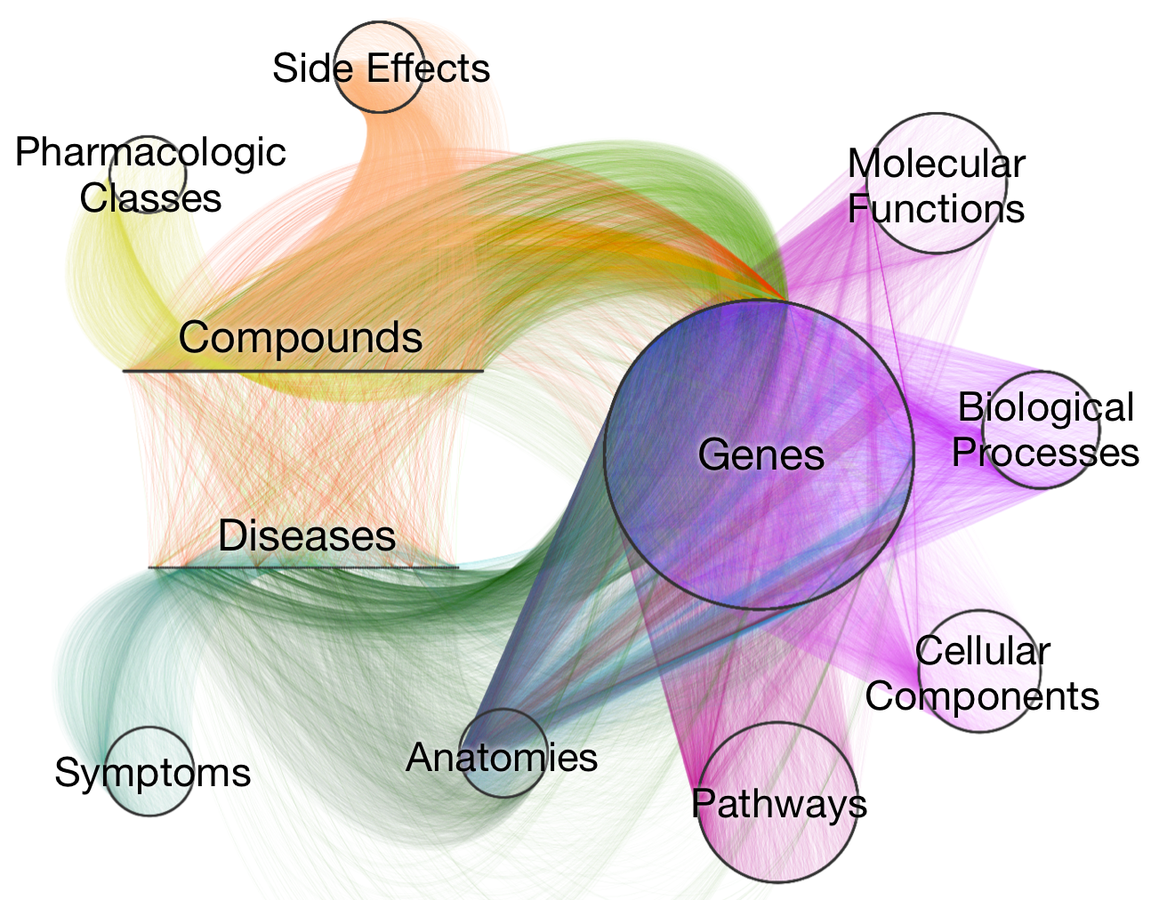
Visualizing Hetionet v1.0
Future: all biomedical knowledge in a single network
https://github.com/greenelab/snorkeling

- Teach computers how to read the literature and extract knowledge.
- Continuously and automatically refine and grow the hetnet.
- Free from any legal restrictions on reuse.
- Customized Docker image
- Digital Ocean droplet
- SSL from Let's Encrypt
- readonly mode with a query execution timeout
- Custom GRASS style
- Custom guides
Public Hetionet Neo4j Instance
Details at doi.org/brsc
MATCH path =
// Specify the type of path to match
(n0:Disease)-[e1:ASSOCIATES_DaG]-(n1:Gene)-[:INTERACTS_GiG]-
(n2:Gene)-[:PARTICIPATES_GpBP]-(n3:BiologicalProcess)
WHERE
// Specify the source and target nodes
n0.name = 'multiple sclerosis' AND
n3.name = 'retina layer formation'
// Require GWAS support for the
// Disease-associates-Gene relationship
AND 'GWAS Catalog' in e1.sources
// Require the interacting gene to be
// upregulated in a relevant tissue
AND exists(
(n0)-[:LOCALIZES_DlA]-(:Anatomy)-[:UPREGULATES_AuG]-(n2))
RETURN pathHow could multiple sclerosis could affect retina layer formation?
More queries at thinklab.com/d/220
Project Rephetio: drug repurposing predictions
-
Hetionet v1.0 contains:
- 1,538 connected compounds
- 136 connected diseases
- 209,168 compound–disease pairs
- 755 treatments
- 1,206 compound–disease metapaths with length ≤ 4
- machine learning classifier
- predict the probability of treatment for all 209,168 compound–disease pairs (het.io/repurpose)
- Project online at thinklab.com/p/rephetio
Systematic integration of biomedical knowledge prioritizes drugs for repurposing
Daniel S Himmelstein, Antoine Lizee, Christine Hessler, Leo Brueggeman, Sabrina L Chen, Dexter Hadley, Ari Green, Pouya Khankhanian, Sergio E Baranzini
bioRxiv. 2016. DOI: 10.1101/087619
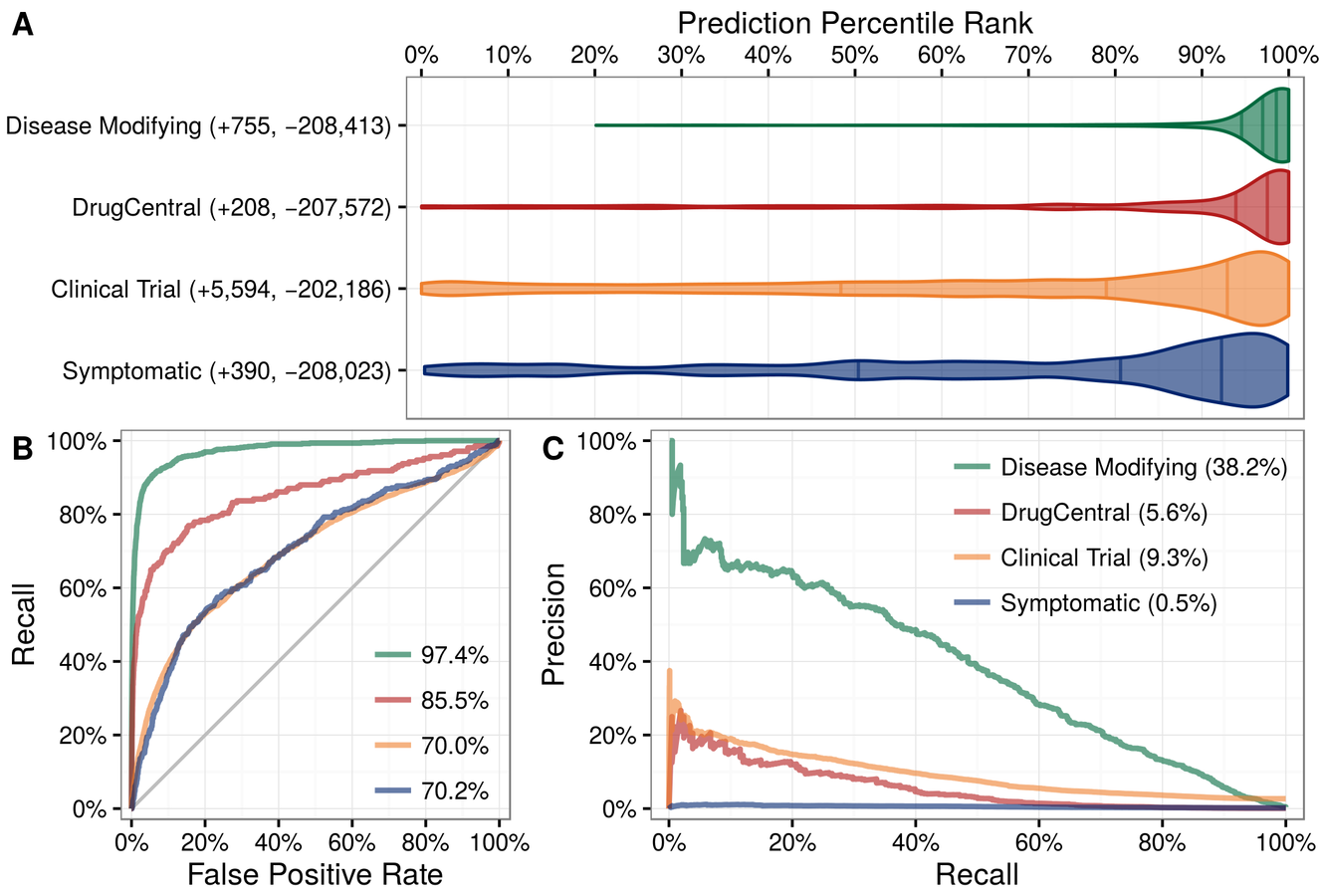
Predictions succeed at prioritizing known treatments

Project Rephetio: Does bupropion treat nicotine dependence?
- Bupropion was first approved for depression in 1985
-
In 1997, bupropion was approved for smoking cessation
- Can we predict this repurposing from Hetionet? The prediction was:
- 99.5th percentile for nicotine dependence
- probability 2.50-fold greater than null

Compound–causes–SideEffect–causes–Compound–treats–Disease

Compound–binds–Gene–binds–Compound–treats–Disease

Compound–binds–Gene–associates–Disease

Compound–binds–Gene–participates–Pathway–participates–Disease
MATCH path = (n0:Compound)-[:BINDS_CbG]-(n1)-[:PARTICIPATES_GpPW]-
(n2)-[:PARTICIPATES_GpPW]-(n3)-[:ASSOCIATES_DaG]-(n4:Disease)
USING JOIN ON n2
WHERE n0.name = 'Bupropion'
AND n4.name = 'nicotine dependence'
AND n1 <> n3
WITH
[
size((n0)-[:BINDS_CbG]-()),
size(()-[:BINDS_CbG]-(n1)),
size((n1)-[:PARTICIPATES_GpPW]-()),
size(()-[:PARTICIPATES_GpPW]-(n2)),
size((n2)-[:PARTICIPATES_GpPW]-()),
size(()-[:PARTICIPATES_GpPW]-(n3)),
size((n3)-[:ASSOCIATES_DaG]-()),
size(()-[:ASSOCIATES_DaG]-(n4))
] AS degrees, path
RETURN
path,
reduce(pdp = 1.0, d in degrees| pdp * d ^ -0.4) AS path_weight
ORDER BY path_weight DESC
LIMIT 10Cypher query to find the top CbGbPWaD paths
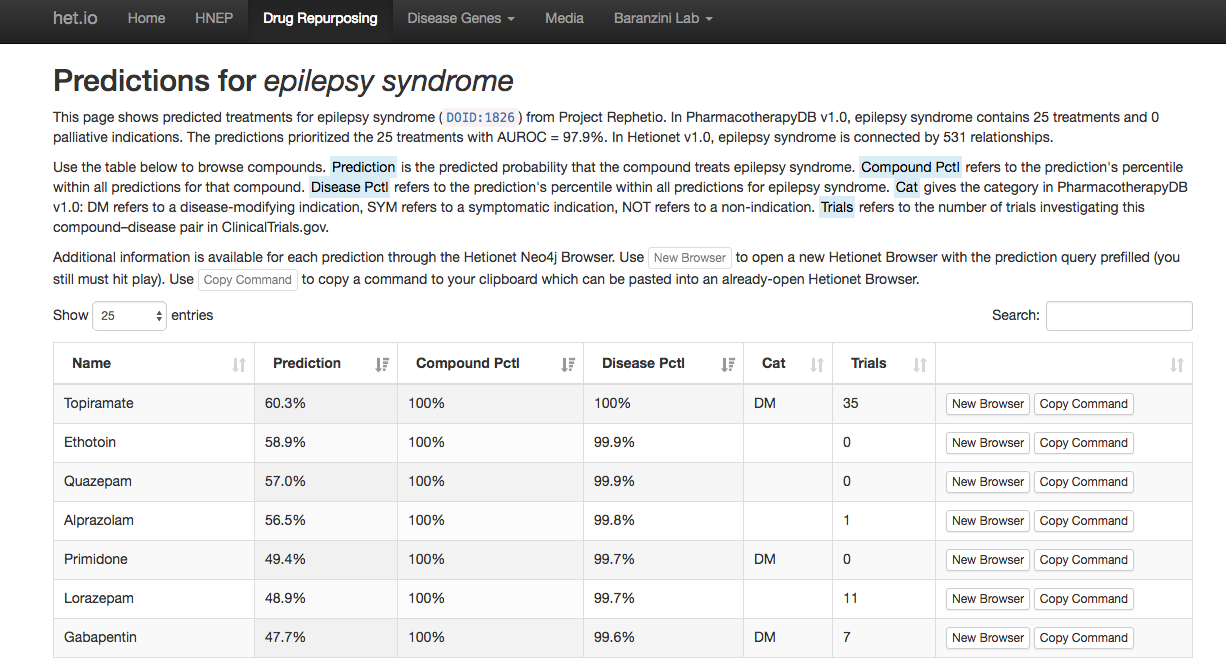
Epilepsy predictions
(browse all predictions at het.io/repurpose)
Discuss at thinklab.com/d/224
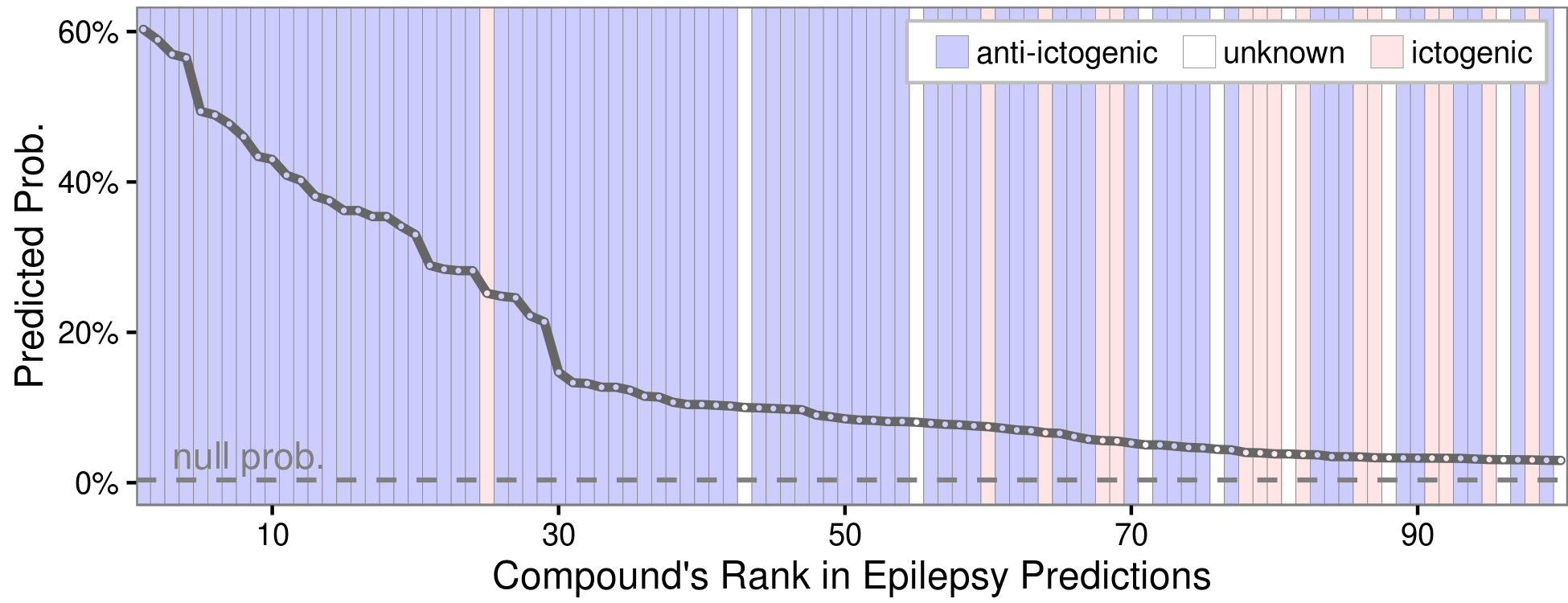
Evaluating the top 100 epilepsy predictions
Discuss at thinklab.com/d/224#5
Top 100 epilepsy predictions & their chemical structure
Discuss at thinklab.com/d/224#5
Top 100 epilepsy predictions & their drug targets
Discuss at thinklab.com/d/230#14
Project Rephetio contributions on Thinklab
(see thinklab.com/p/rephetio/leaderboard)
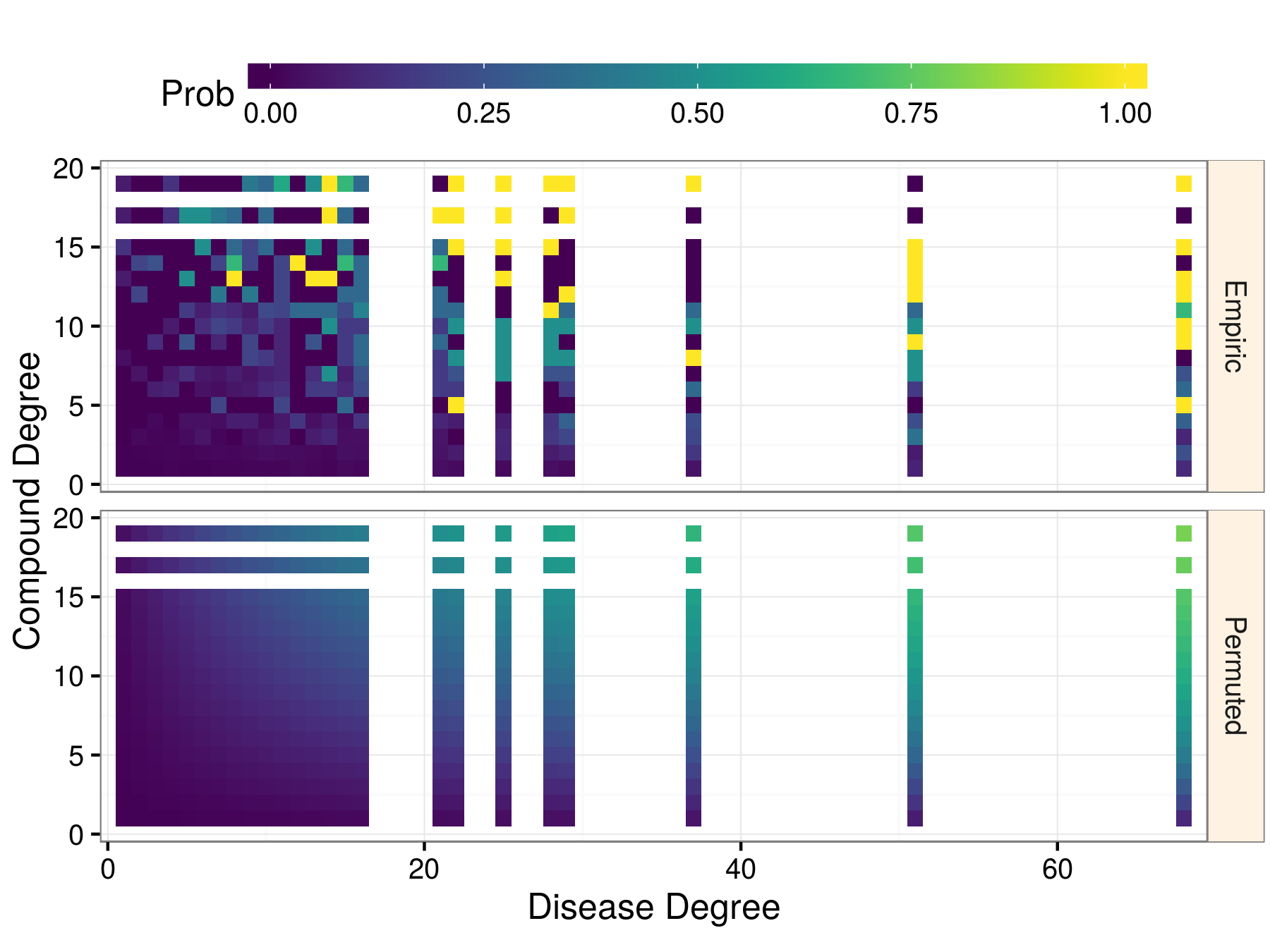
Prior probability of treatment
Methotrexate treats 19 diseases and hypertension is treated by 68 compounds. Methotrexate received a 79.6% prior probability of treating hypertension, whereas a compound and disease that both had only one treatment received a prior of 0.12%.
Questions
Slides at slides.com/dhimmel/big-data-seminar

https://github.com/cognoma/cognoma
Advertisement: Cognoma Meetup with DataPhilly & Code for Philly