Classifying EFO/MONDO diseases as low, medium, or high precision using nxontology-ml
slides released under CC BY 4.0
slides.com/dhimmel/efo-disease-precision
Prepared at nxontology-ml/issues/13
https://related.vc
nxontology software suite
-
nxontology
NetworkX-based Python library for representing ontologies. Uses fastobo, pronto, pygraphviz.
-
nxontology-data
Making ontologies accessible as simple JSON files (even MeSH). Uses rdflib, SPARQL, etc. -
nxontology-ml
Machine learning to classify ontology nodes.
Motivation
Posted to the obo-community slack by Philip Strömert generated with Midjourney prompt:
We cannot interpret our research data anymore because we did not annotate it with ontologies
Really easy access to popular biomedical ontologies based on JSON files that can be read into Python's networkx.
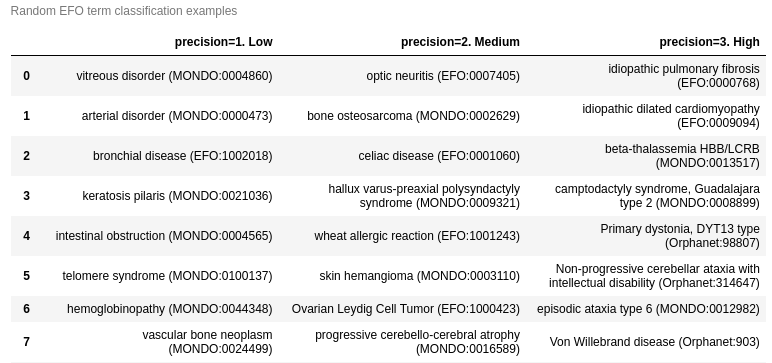
disease precision classification
- EFO OTAR Slim
- OTAR = Open Targets, see efo/issues/926
- Derivative of EFO focused on clinical interpretation
- Rooted by therapeutic class terms, other terms pruned
- Approach can be applied to MONDO
- Precision levels
- low
- medium
- high
precision levels
- Low
- group diseases with some, but often not many, shared characteristics
- indications in early stage clinical trials
- heterogenous patient population
- used by ontology for organization & completeness
- Medium
- indications in later stage clinical trials
- group patients with a condition with shared physiological or environmental origin
- diagnosable
- High
- small groups of relatively homogeneous patients
- greater diagnostic certainty
- represent the forefront of clinical practice (precision medicine)
early feedback
- Is Bardet-Biedl syndrome a single disease entity?
- Is Alzheimer disease a single disease?
- But Alzheimer's has a number of different genetic etiologies - are you sure it's a single disease?
- How many diseases are there?
- Is there an existing treatment for SLE Type 16? Downward propagation SLE treatments makes sense, but probably not a treatment for connective tissue disease.
- What diseases should we predict treatments for?
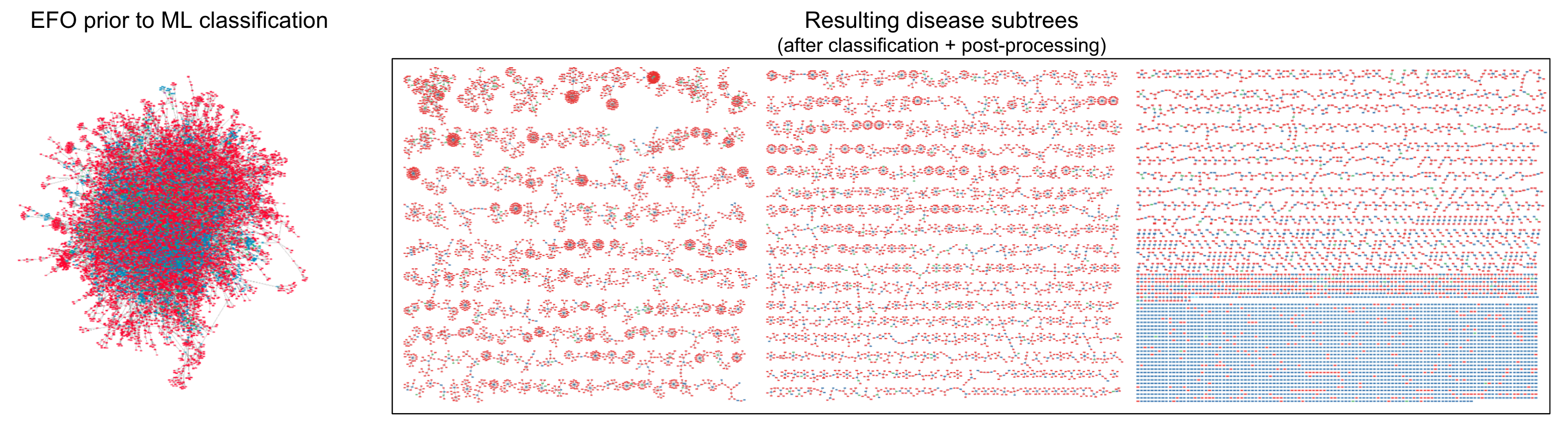
- Untangling the hairball.
applications

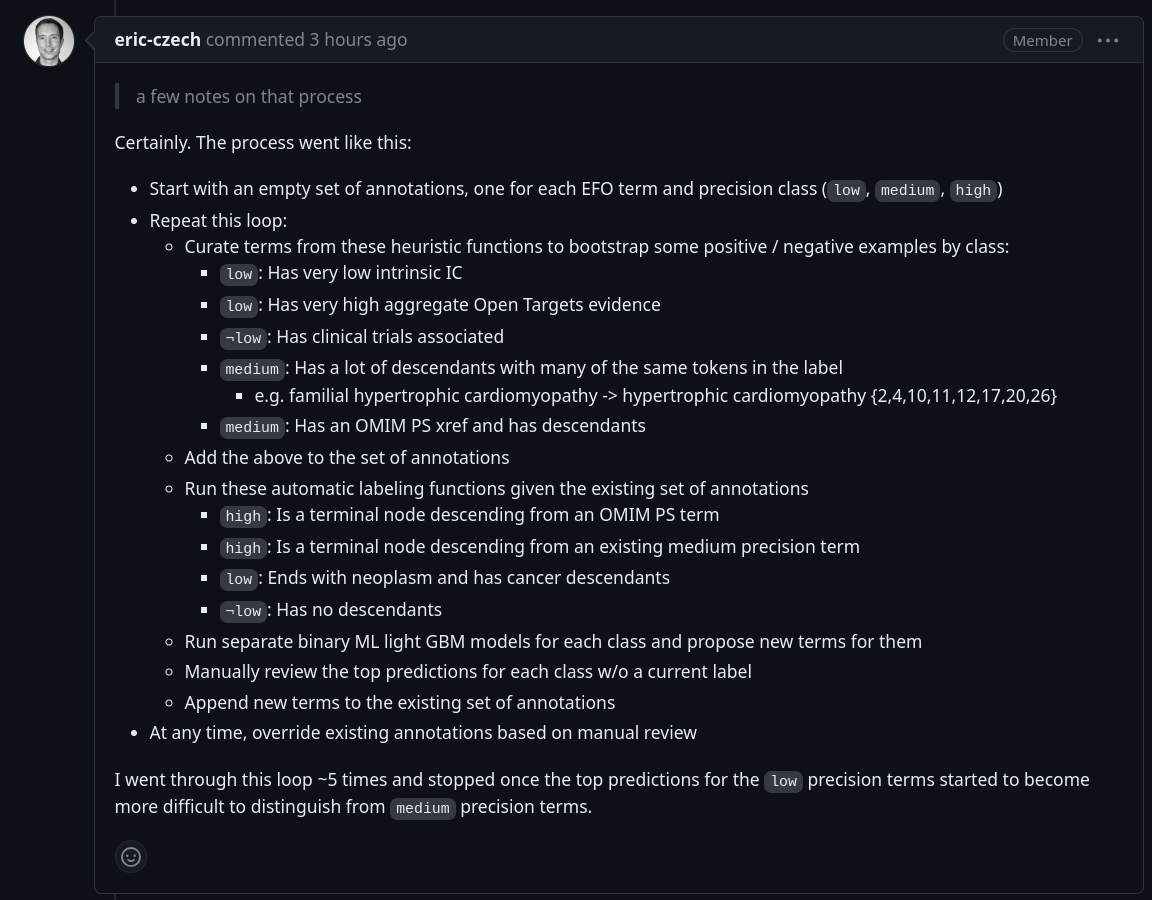
Training labels
- Semi-automated hand labeling of terms in v3.43.0
features
feature groups:
- topology
- cross-references
- prefixes
- subsets
- descriptions
- gpt tags
- gwas
topology features
derived solely from the topology of the directed acyclic graph
- depth
- n_ancestors
- n_descendants
- n_parents
- n_roots
- n_children
- n_leaves
- intrinsic_ic
- intrinsic_ic_scaled
- intrinsic_ic_sanchez
- intrinsic_ic_sanchez_scaled

cross-reference features
- doid
- gard
- icd10
- icd9
- meddra
- mesh
- mondo
- ncit
- omim
- omim.ps
- orphanet
- snomedct
- umls
cross-references (xref) counts by external database.
nxontology-data performs bioregistry normalization of xref prefixes
prefix features
- EFO: 12,312
- MONDO: 9,061
- Orphanet: 2,052
- HP: 1,384
- GO: 335
- OBA: 40
- OTAR: 9
- DOID: 6
- OBI: 4
- NCIT: 3
- MP: 2
- OGMS: 1
EFO includes terms imported from other ontologies.
Counts from EFO Otar Slim v3.57.0
from collections import Counter
from nxontology import NXOntology
url = "https://github.com/related-sciences/nxontology-data/raw/2ce01d8495024d46cbc54fb0c26a92500ad717e0/efo_otar_slim.json"
nxo = NXOntology.read_node_link_json(url)
prefix_counts = Counter()
for node in nxo.graph.nodes:
prefix, _ = node.split(":")
prefix_counts[prefix] += 1
for prefix, count in prefix_counts.most_common():
print(f"{prefix}: {count:,}")
subset features
- mondo#disease_grouping
- mondo#gard_rare
- mondo#ordo_etiological_subtype
- mondo#ordo_group_of_disorders
EFO includes subsets, many imported from MONDO. We're not actually sure how to find documentation for these subsets, but they sound relevant.
GWAS feature
EFO notes whether a term is a GWAS trait.
description features
Large "Embedding" Vector (768 demensions)
EFO Node description (term name & definition)
Compressed "Embedding" Vector (64 dimensions)
Transformer Model ("BioLinkBERT")
Dimensionality reduction (e.g. PCA, LDA)
MONDO:0005301 🡒 multiple sclerosis: A progressive autoimmune disorder affecting the central nervous system resulting in demyelination. Patients develop physical and cognitive impairments that correspond with the affected nerve fibers. [ NCIT : P378 ]
ChatGPT features
A list of records will be provided from an ontology of disease terms. Each record will contain information describing a single term.
Assign a `precision` label to each of these terms that captures the extent to which they correspond to patient populations with distinguishing clinical, demographic, physiological or molecular characteristics. Use exactly one of the following values for this label:
- `high`: High precision terms have the greatest ontological specificity, sometimes (but not necessarily) correspond to small groups of relatively homogeneous patients, often have greater diagnostic certainty and typically represent the forefront of clinical practice.
- `medium`: Medium precision terms are the ontological ancestors of `high` precision terms (if any are known), often include indications in later stage clinical trials and generally reflect groups of patients assumed to be suffering from a condition with a shared, or at least similar, physiological or environmental origin.
- `low`: Low precision terms are the ontological ancestors of both `medium` and `high` precision terms, group collections of diseases with *some* shared characteristics and typically connote a relatively heterogenous patient population. They are often terms used within the ontology for organizational purposes.
The records provided will already have the following fields:
- `id`: A string identifier for the term
- `label`: A descriptive name for the term
- `description`: A longer, possibly truncated description of what the term is; may be NA (i.e. absent)
Here is a list of such records (in YAML format) where the `precision` label is already assigned for 3 examples at each level of precision:
Examples:
- Prompt: https://gist.github.com/yonromai/daa0bd3c8e9f81250a68c3f6b614598d
- Completion 1: https://gist.github.com/yonromai/68e74c7a8032d9f59e427f6196652d91
- Completion 2: https://gist.github.com/yonromai/f2b852e20f42bfea2da13b3e755267fe
- Completion 3: https://gist.github.com/yonromai/66ff9de3141cb4dcd35cd11cd06c936e
- Each prompt request generates 3 responses, used as votes
-
poll
Which feature group has the greatest influence on the outcome?
Slido: 1342 945
- topology
- cross-references
- prefixes
- subsets
- descriptions
- gpt tags
- gwas

ML pipeline overview
catboost slide
- todo

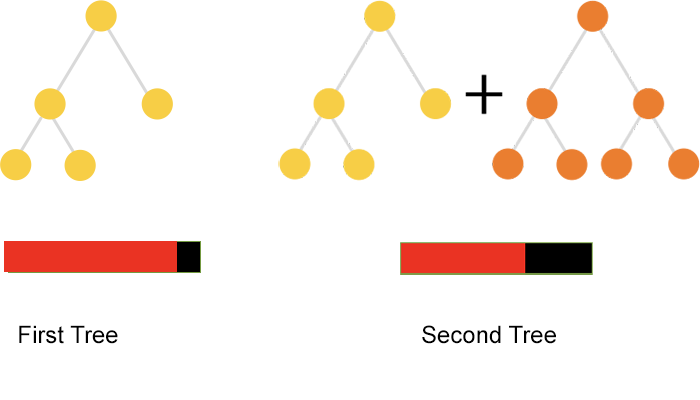
Source: https://catboost.ai/news/catboost-enables-fast-gradient-boosting-on-decision-trees-using-gpus
model performance
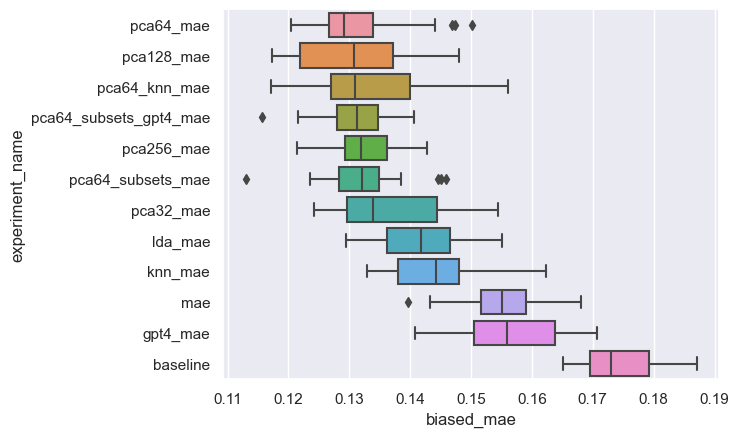
feature group importance
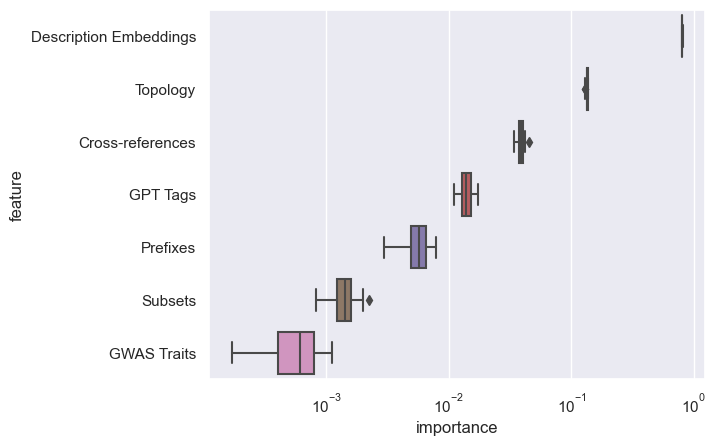
feature group importance
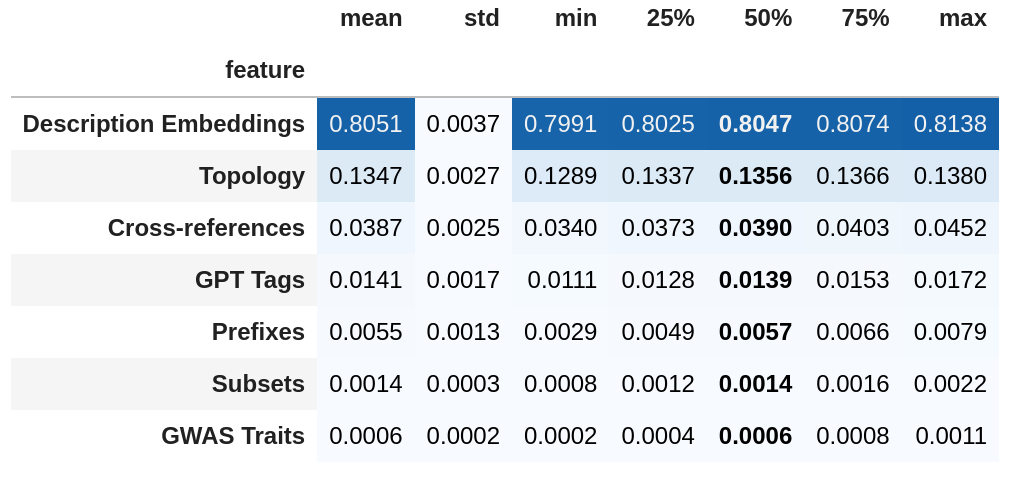
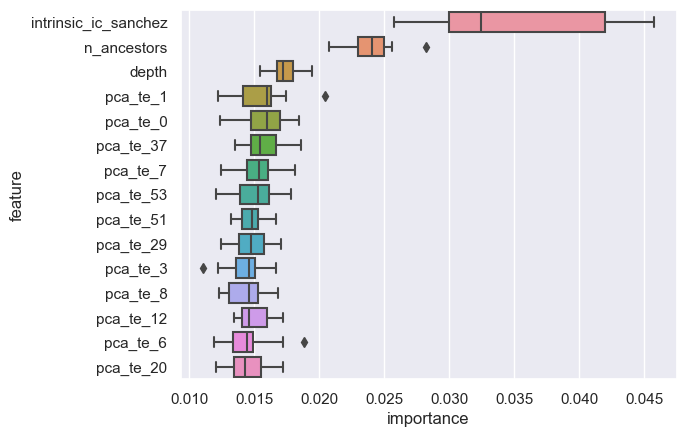
top individual features
intrinsic_ic_sanchez source
Predictions & Availability
Visualization: precision by border
- low = dotted
- medium = dashed
- high = solid

questions
next steps
- data users
- collaborators
- contributors
- predictions internalized to EFO / MONDO?