Integrate all: hetnets in human disease
November 20, 2015
Asilomar Grounds
dhimmel on:
QBC Retreat
—Daniel Himmelstein

Sandler Neurosciences Center

Sergio
Founding Insight
-
context
bioinformatics → data explosion
-
goal
mine the data to advance human health
-
problem
high-throughput data tends to predict weakly
-
remedy
combine diverse datasets into a strong predictor
-
method
heterogeneous network (hetnet) edge prediction
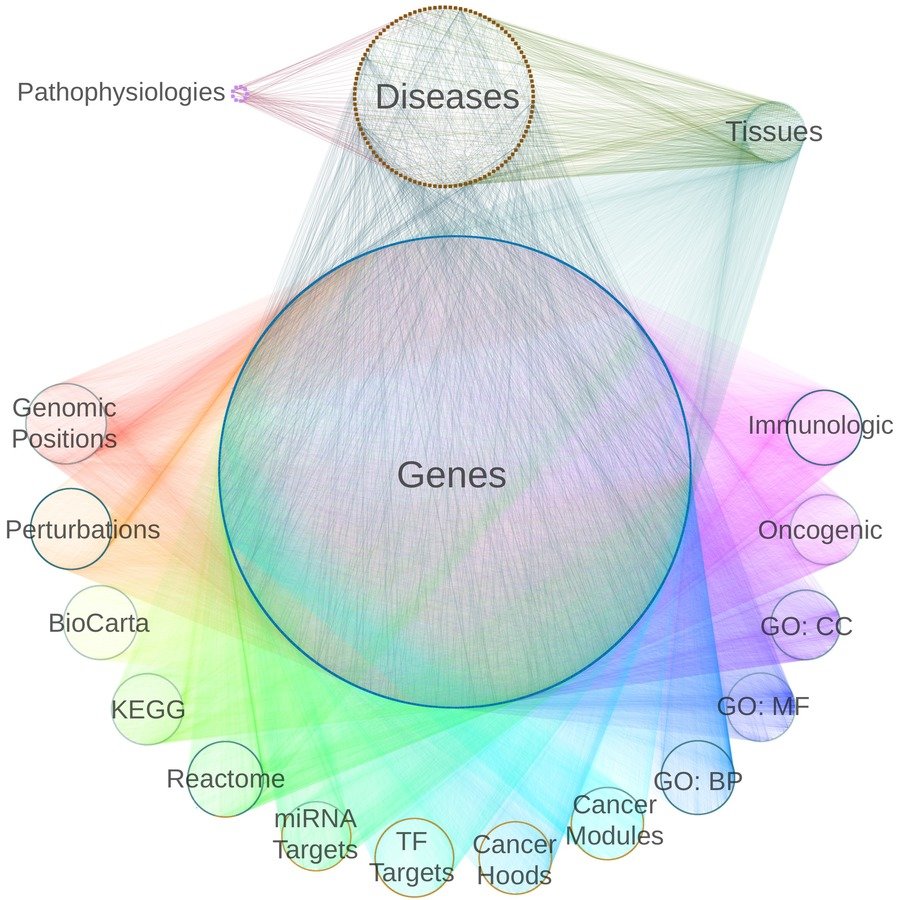
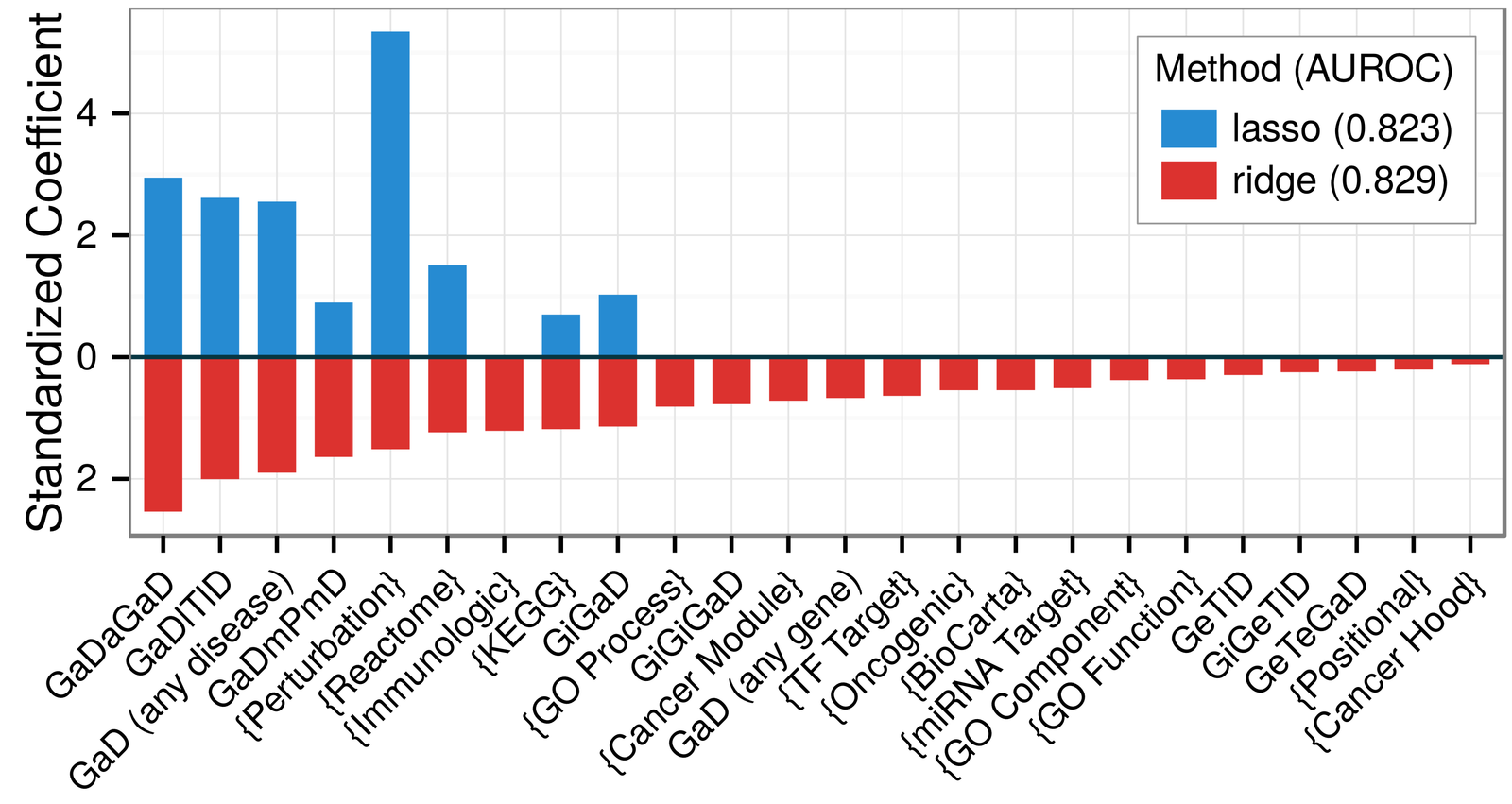
Predicting disease-associated genes
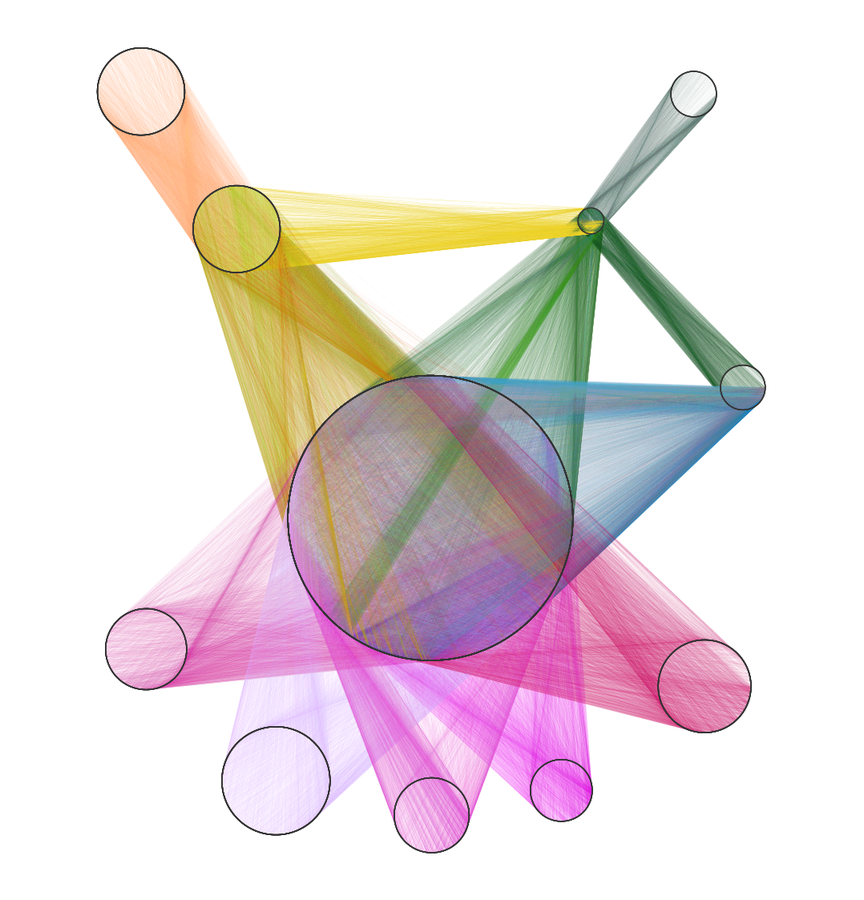
network of pathogenesis:
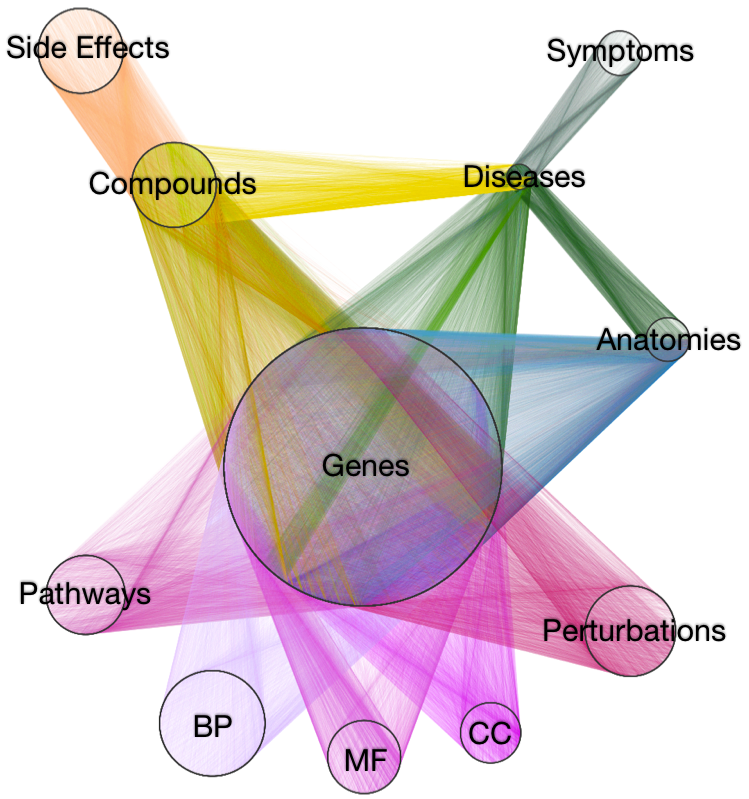
- integrate diverse data to provide context
- 18 metanodes
40,343 nodes
- 19 metaedges
1,608,168 edges
type is essential when operating on hetnets
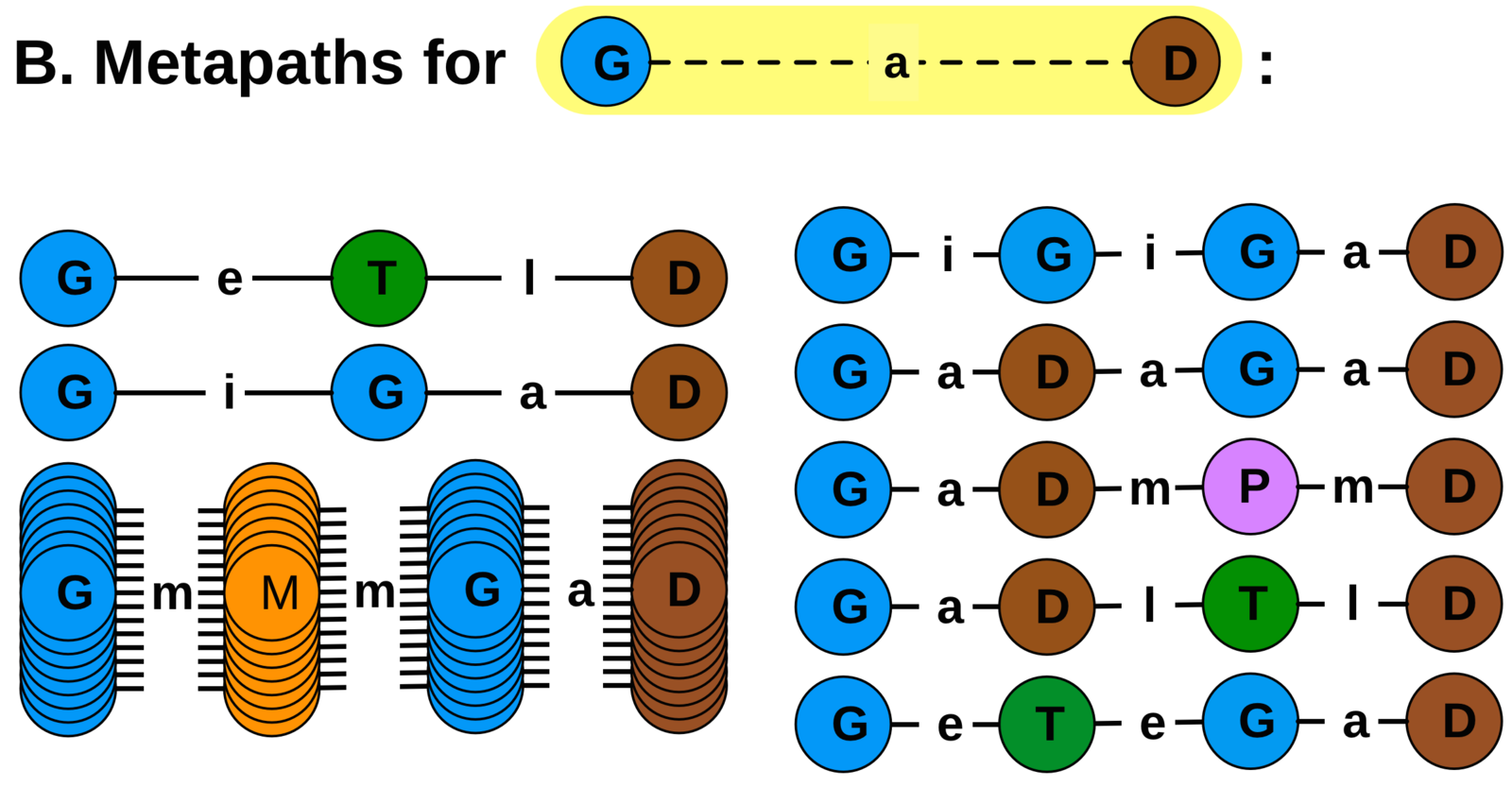
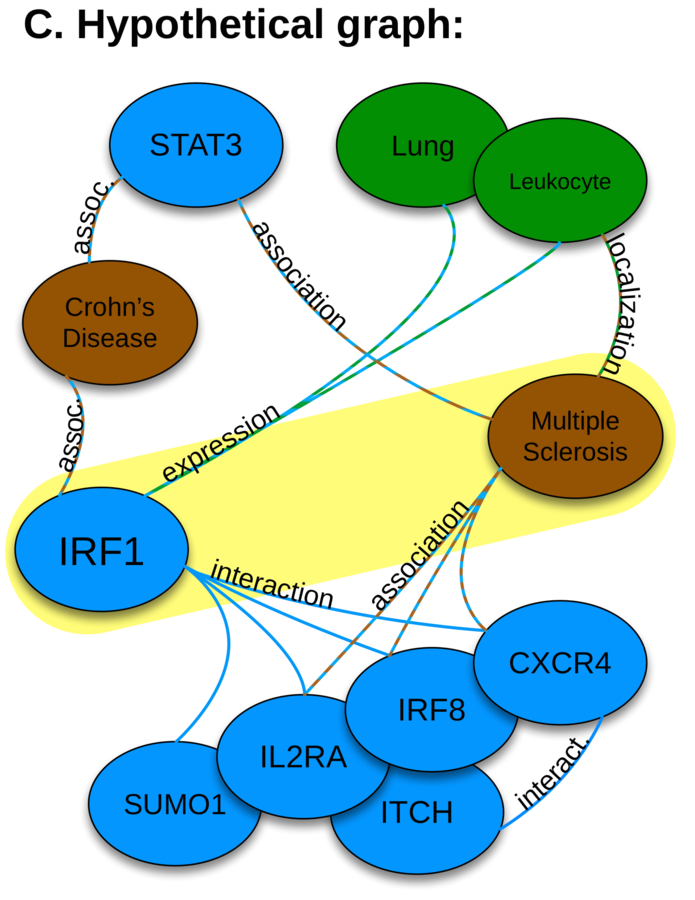
metapath-based approach
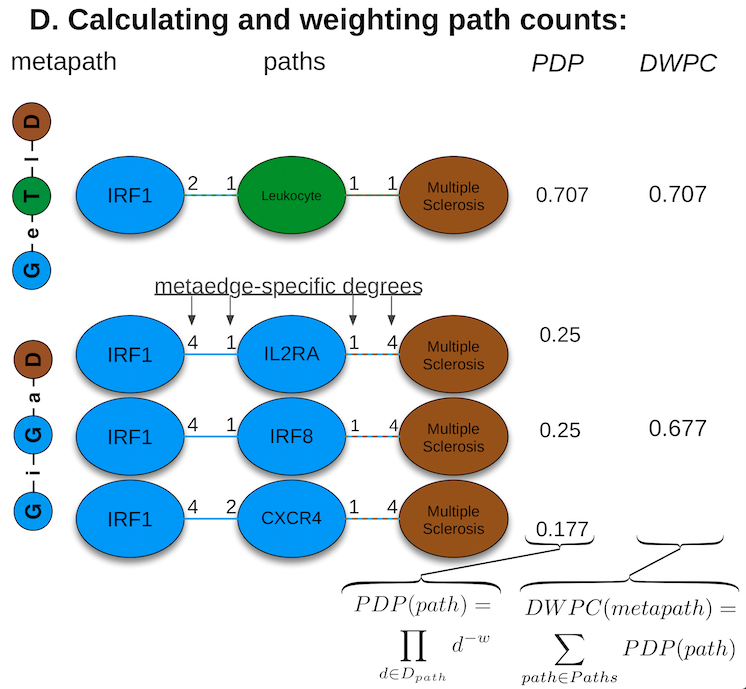
feature extraction: the DWPC
learning to classify:
regularized logistic regression
mechanisms of pathogenesis:
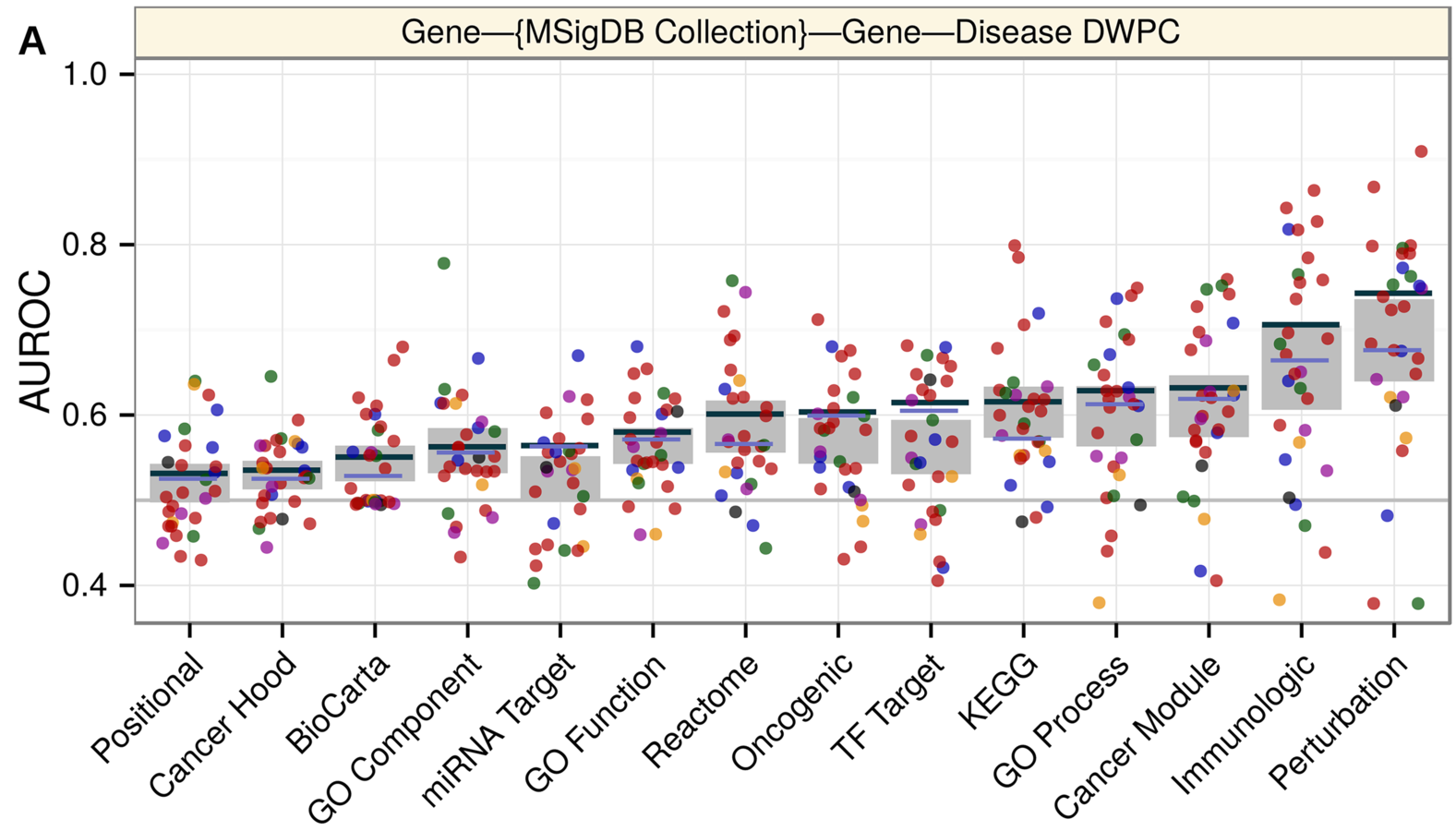
comparing gene set collections
extras
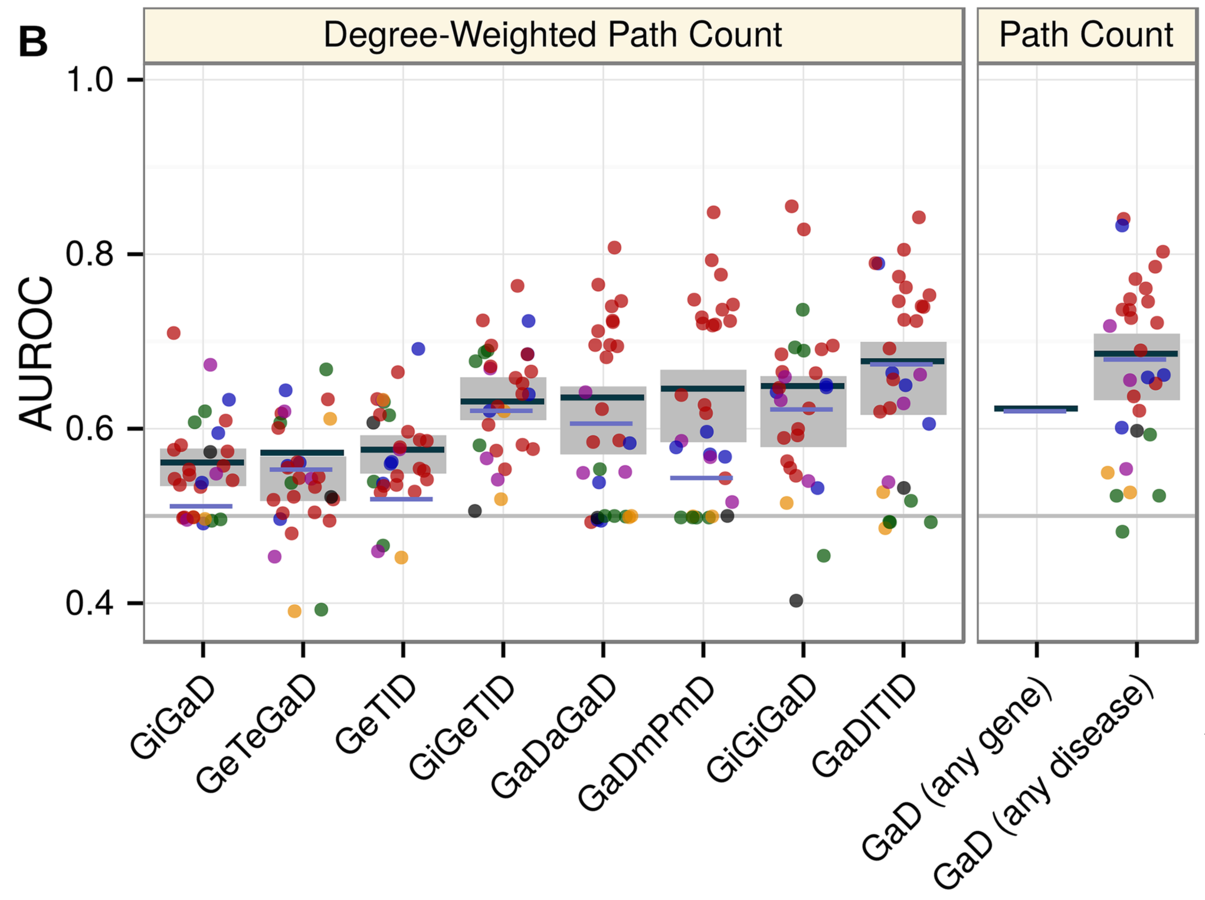
mechanisms of pathogenesis:
comparing metapaths
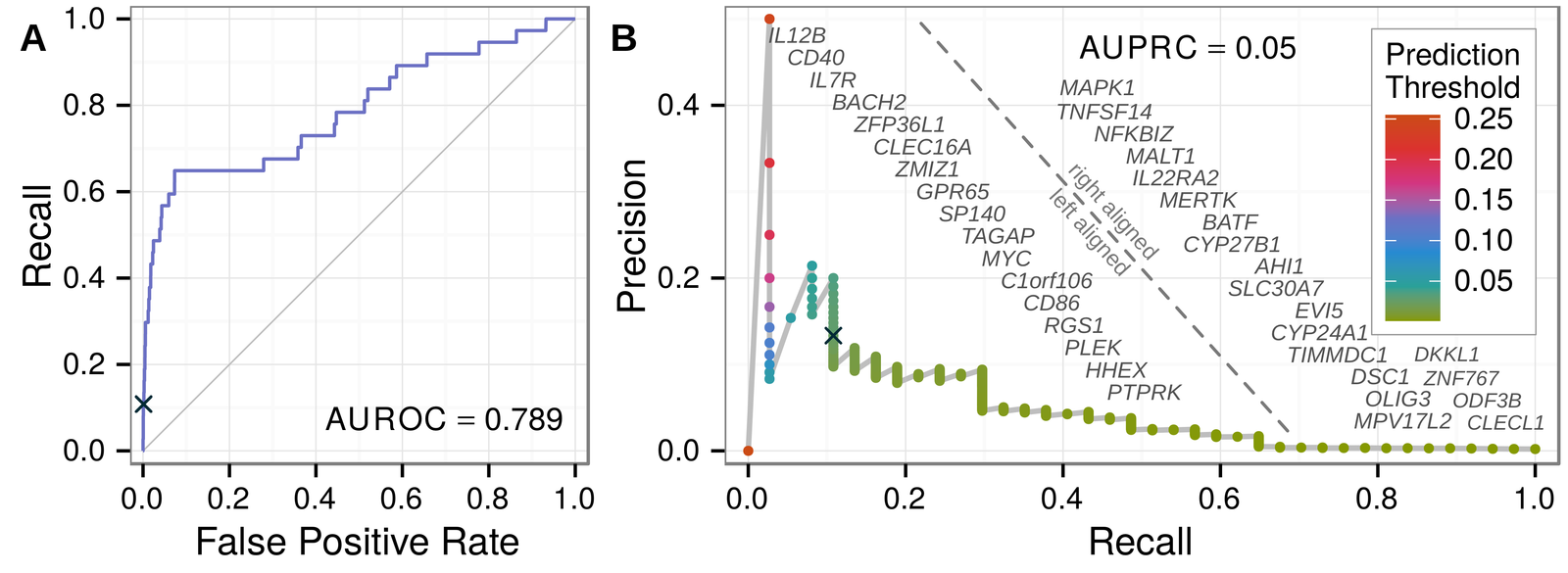

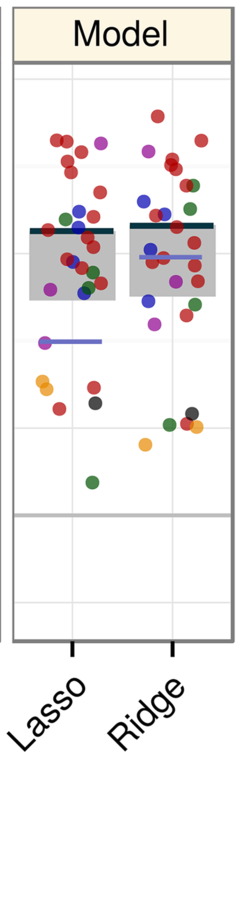
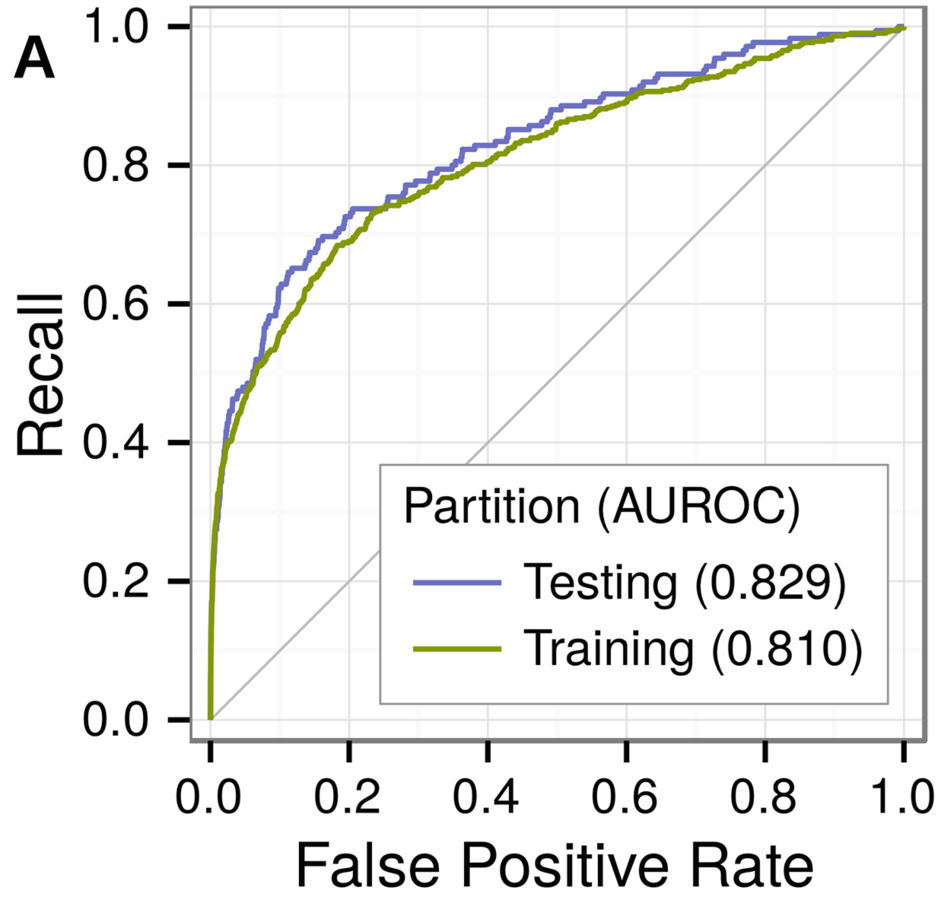
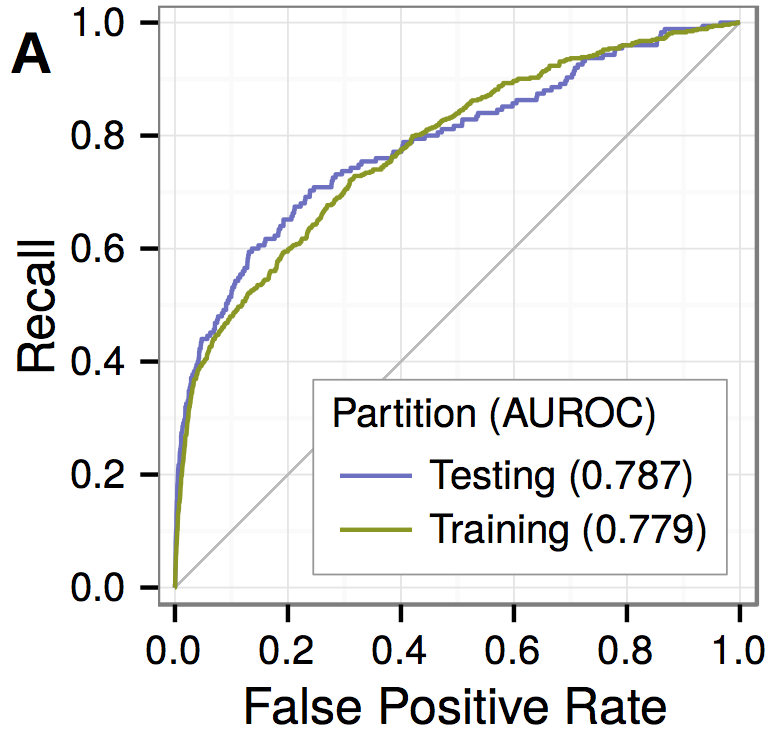
Predicting withheld MS associations
Novel MS associations
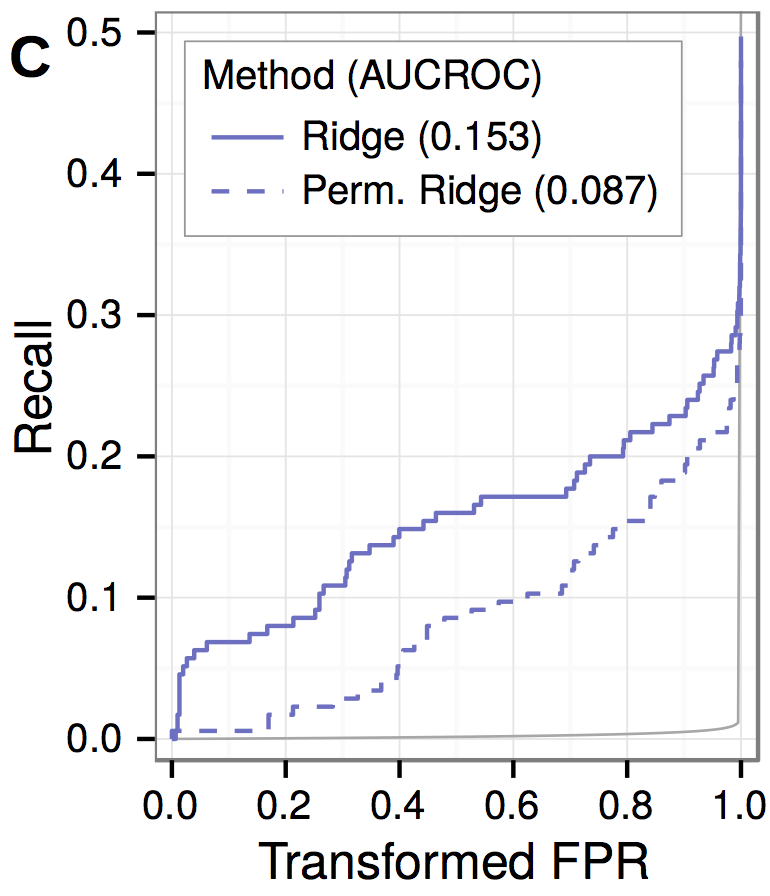
performance and permuation
Metagraph for predicting drug repurposing
Network for drug repurposing
- 50k nodes
10 types
- 3M edges
27 types
-
28 public resources
- open for reuse & reproducibility
1. copyright
limited by:
-
fair use
-
originality (excludes facts)
2. contract
agreement entered into to receive access to a resource
- can impose restrictions beyond copyright
restrictions on data
automatically granted to "original works of authorship" giving the exclusive right to:
-
copy
-
distribute
-
create derivatives
1. ∅ license
3. ∅ distribute
- MSigDB — publicly-funded project from the Broad
- publication data supplements
complications
4. standard
- 9 resources
- all rights reserved
- upon contact:
-
- 1 permission
- 0 licenses added
2. unclear
- 4 resources
- clarification after laborious and slow permission requests
- 11 resources
- incompatibilities
5. government
- 4 resources
- public domain
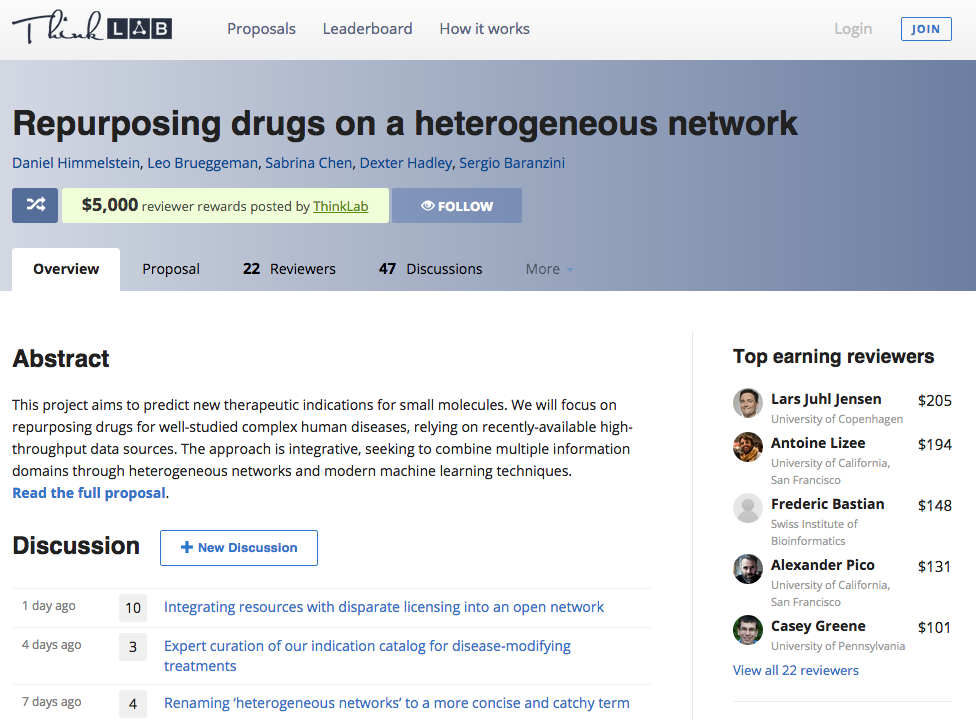
Resolution after months & 5000+ word discussion: mixed approach
massively collaborative open science
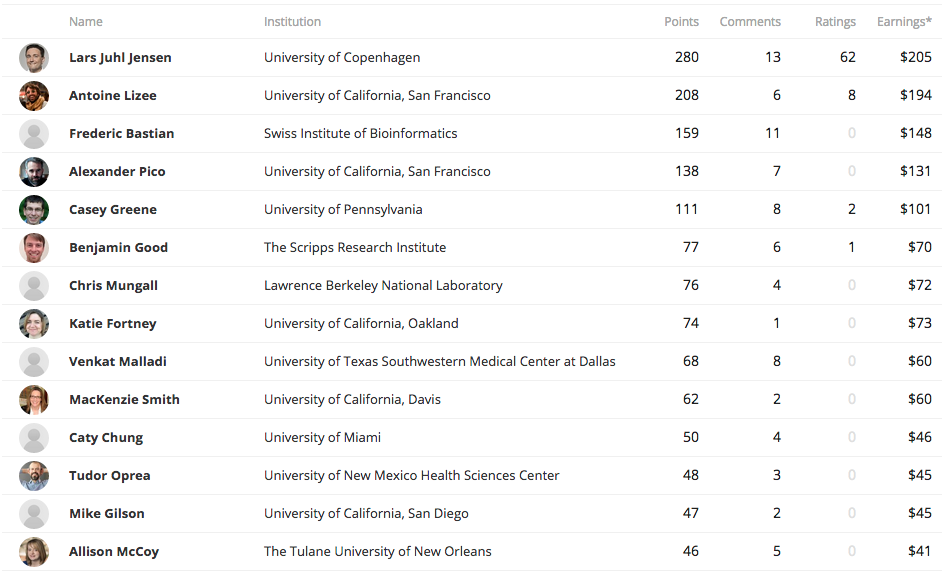
22 reviewers, 53 discussions, 293 comments
stats from 2015-10-20
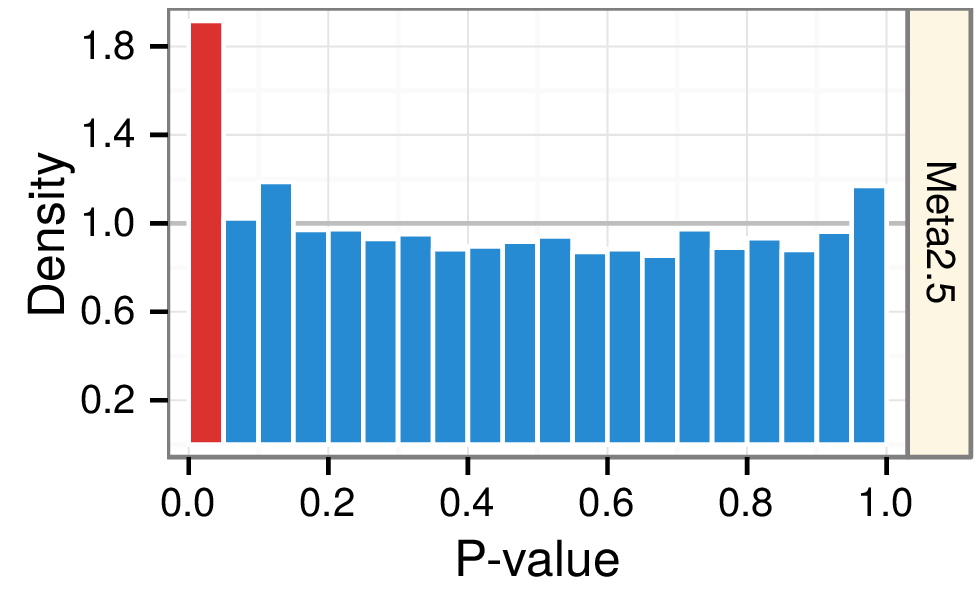
Mining knowledge from 69 years of biomedical publication
Discuss on
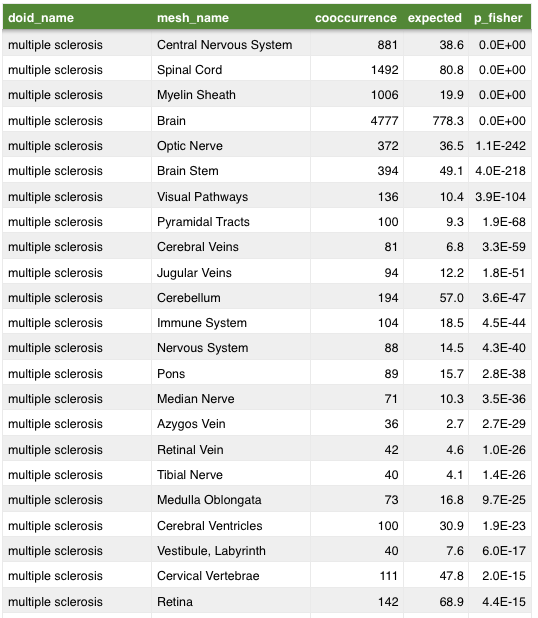
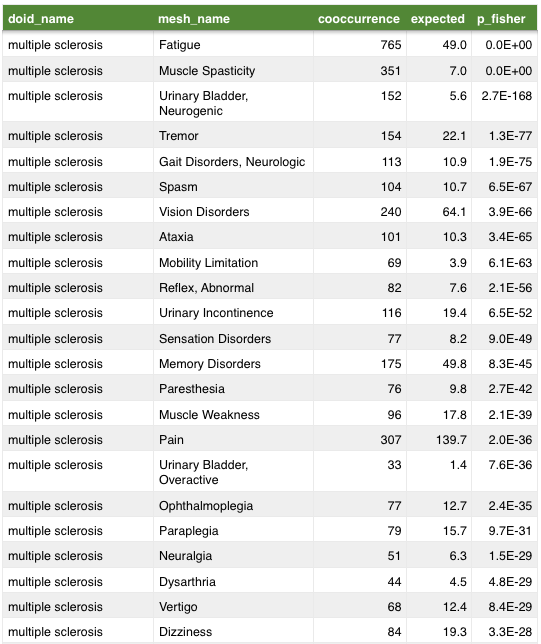
anatomy
symptom
Mining MEDLINE for disease context
LINCS L1000: ~20,000 small molecules
We combine all signatures for each DrugBank compound to get a consensus signature

Brueggeman
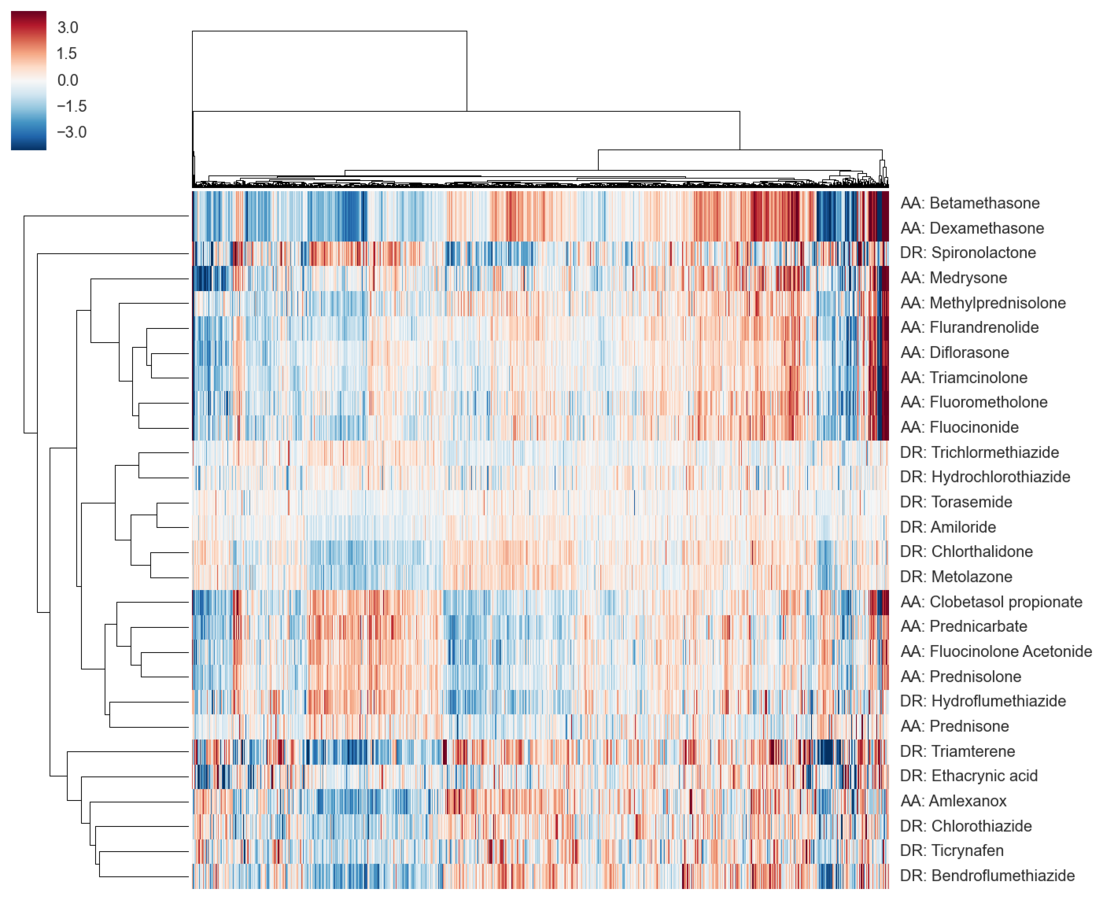
transcriptional signatures discriminate
diuretics (DR) & anti-Inflammatories (AA)
← genes →
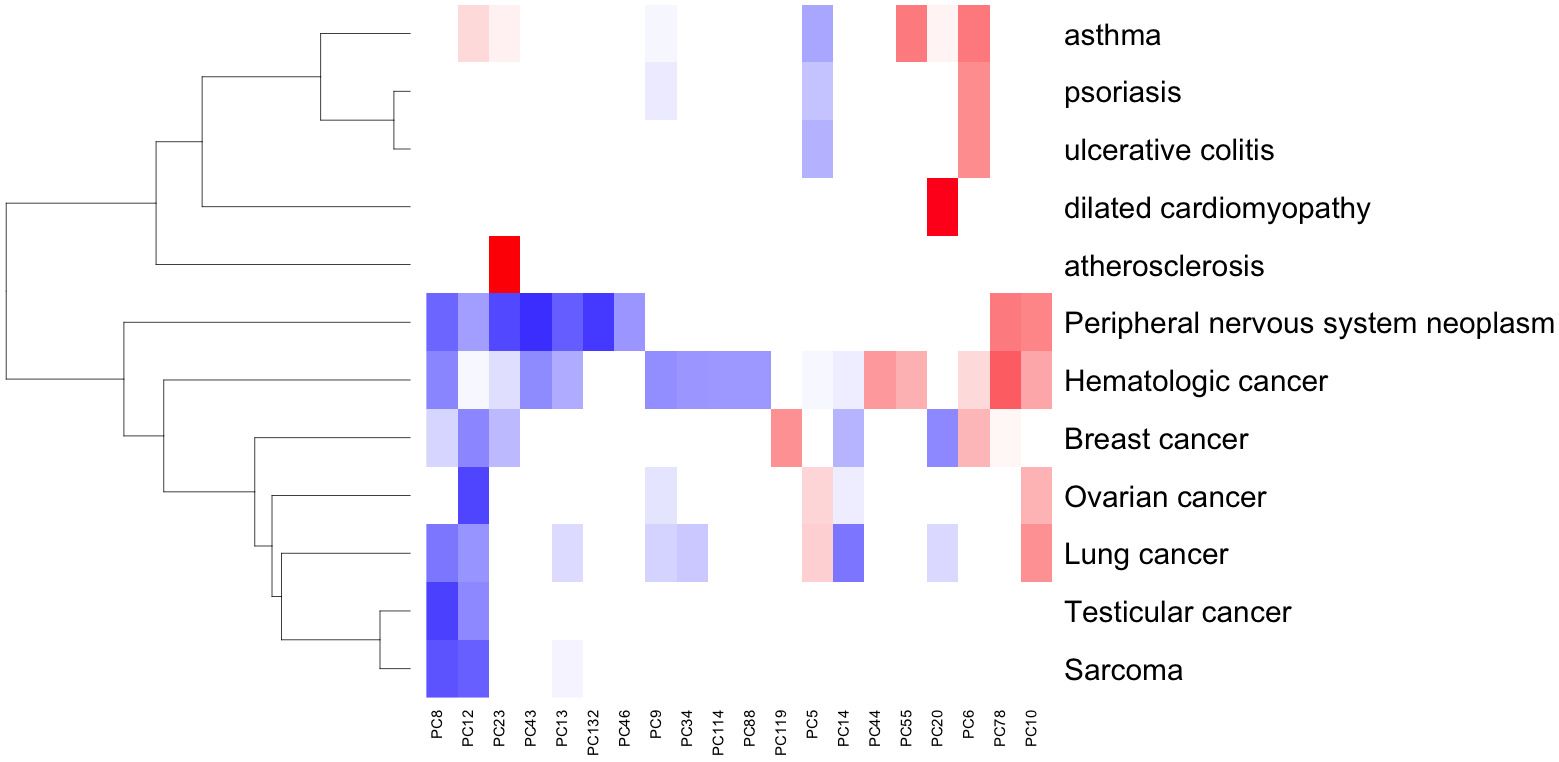
Disease-specific models uncover therapeutic signatures
-
MEDI-HPS:
- RxNorm
- MedlinePlus
- SIDER 2
- Wikipedia
- ehrlink: linked data from health records
- LabeledIn: expert and MTurk curated drug labels
- PREDICT:
- UMLS links
- drugs.com
- drug labels
catalog of indications
aggregated 4 databases:
yielding 1,388 indications
| drug | disease | ajg | csh | eq |
|---|---|---|---|---|
| Acetylsalicylic acid | systemic lupus erythematosus | DM | DM | 1 |
| Alprazolam | systemic scleroderma | SYM | SYM | 1 |
| Baclofen | multiple sclerosis | SYM | SYM | 1 |
| Bupropion | panic disorder | SYM | DM | 0 |
| Captopril | rheumatoid arthritis | NOT | NOT | 1 |
| Cisplatin | hematologic cancer | DM | DM | 1 |
| Cladribine | hematologic cancer | DM | DM | 1 |
| Clopidogrel | coronary artery disease | DM | DM | 1 |
| Cocaine | dental caries | NOT | SYM | 0 |
| class | ajh | csh |
|---|---|---|
| DM | 26 | 32 |
| SYM | 20 | 17 |
| NOT | 4 | 1 |
curation pilot results
- ~58% disease modifying
- ~37% symptomatic
- ~5% non-indications
- discuss on
66% ✓
| metapath | nonzero | auroc |
|---|---|---|
| CcSEcCiD | 0.590 | 0.897 |
| CiDiCiD | 0.255 | 0.840 |
| CiDaGaD | 0.411 | 0.820 |
| CtGtCiD | 0.227 | 0.807 |
| CiDpSpD | 0.410 | 0.801 |
| CiDlAlD | 0.400 | 0.797 |
| CtGiGaD | 0.335 | 0.710 |
| CuG<kuGaD | 0.425 | 0.692 |
| CtGvD | 0.009 | 0.514 |
Results for all metapaths ≤ length 3
feature performance
(including symptomatic indications)
common side effects
shared indications
shared gene associations
shared targets
shared symptoms
knowledge-bias free
genetics

-
graph database
-
designed for property graphs
natively supports hetnets
-
cypher query language
MATCH path = (source:Disease)--(:Symptom)--(target:Disease)
WHERE
source.name = 'multiple sclerosis' AND
target.name = 'psoriasis'
RETURN pathCommon symptoms between MS and psoriasis

MATCH path =
(source:Compound)-[:TARGET|BINDING]-
(:Gene)-[:VARIATION]-(target:Disease)
WHERE target.name = 'multiple sclerosis'
RETURN pathCompounds that target MS-associated genes


DWPC in Cypher