Biodiversity Informatics (2017)
<!DOCTYPE html> <html> <head> </head> <body>
<p><img style="display: block; margin-left: auto; margin-right: auto;" src="59/download/inline" alt="" width="897" height="282" /></p>
<table style="background-color: #1f5301; table-layout: fixed; height: 64px; text-align: center; width: 100%; margin-left: auto; margin-right: auto;" border="1" cellspacing="2" cellpadding="2">
<tbody>
<tr style="text-align: center;">
<td style="text-align: center;"><strong><span style="color: #ffffff;"><a href="58"><span style="color: #ffffff;">Home</span></a></span></strong></td>
<td style="text-align: center;"><strong><span style="color: #ffffff;"><a href="68"><span style="color: #ffffff;">Plenary Speakers</span></a></span></strong></td>
<td style="text-align: center;"><strong><span style="color: #ffffff;"><a href="62"><span style="color: #ffffff;">Programme</span></a></span></strong></td>
<td style="text-align: center;"><strong><span style="color: #ffffff;"><a href="61"><span style="color: #ffffff;">Organisation</span></a></span></strong></td>
<td style="text-align: center;"><strong><span style="color: #ffffff;"><a href="65"><span style="color: #ffffff;">Registration</span></a></span></strong></td>
<td style="text-align: center;"><a href="82"><strong><span style="color: #ffffff;"><span style="color: #ffffff;">Sessions</span></span></strong></a></td>
</tr>
</tbody>
</table>
<p dir="ltr" style="line-height: 1.2; margin-top: 0pt; margin-bottom: 0pt; text-align: justify;"> </p>
<p><span style="font-size: 14pt;"><strong>Nature health benefits session</strong></span></p>
<p>Nature benefits human health in many ways. Examples are the importance of biodiversity to traditional and modern medicinal practice, and the utility of various species for medical research. Genetic and species diversity is functional to food production, and can play an important role in addressing issues of nutrition security including certain disease risks (e.g. obesity, diabetes) through dietary improvements. Biodiversity also plays a role in safeguarding air quality and access to fresh water, disaster risk reduction, and supports emergency responses and climate change adaptation. Furthermore, diverse natural environments may enhance experiences that reduce stress, support the development of cognitive resources, stimulate social contacts, attract people for physical activity, and support personal development throughout an individual’s lifespan. Moreover, recent studies show that declining contact with some forms of (microbiotic) life may contribute to the rapidly increasing prevalence of allergies and other chronic inflammatory diseases among urban populations worldwide (see other parallel session). Biodiversity thus can have an important contribution to both public health related ecosystem services and the reduction of health risks.</p>
<p> </p>
<p>In this session we will discuss a diversity of experiences, expectations, opportunities and challenges regarding nature health benefits work in science, policy and practice. Also will we discuss potential linkages between different topical foci within and beyond the realm of different nature health benefits.</p>
<p> </p>
<p>Introductory speakers:</p>
<p><a href="https://www.wageningenur.nl/en/Persons/dr.-S-Sjerp-de-Vries.htm">Sjerp De Vries</a> (Alterra): <a href="http://www.annualreviews.org/doi/abs/10.1146/annurev-publhealth-032013-182443">current methodological challenges on nature health benefits research</a> (confirmed)</p>
<p>Peter van den Hazel (<a href="http://www.phenotype.eu/">Phenotype project</a>): what we can learn from the project results (confirmed)</p>
<p><a href="http://www.ieep.eu/about-us/our-people/patrick-ten-brink-565" target="_blank">Patrick Ten Brink</a> (IEEP): overview of state of the art in science, policy and practice in Europe based on the <a href="http://ec.europa.eu/environment/nature/pdf/Study%20on%20Health%20and%20Social%20Benefits%20of%20Nature%20and%20Biodiversity%20Protection.pdf">Health and Social Benefits of Nature and Biodiversity Protection</a> project (confirmed)</p>
<p><a href="http://www.cdo.ugent.be/drupal-7.15/?q=profile/591">Patrick Van Damme</a> (UGhent): traditional medicine - medicinal plants (confirmed)</p>
<p> </p>
<p><strong><span style="font-size: 14pt;">Presentations</span></strong></p>
<p> </p>
<table>
<tbody>
<tr>
<td>
<p><strong>Name</strong></p>
</td>
<td>
<p><strong>Affiliation</strong></p>
</td>
<td>
<p><strong>Presentation</strong></p>
</td>
</tr>
<tr>
<td>
<p>Patrick ten Brink</p>
</td>
<td>
<p>Institute for European Environmental Policy</p>
</td>
<td>
<p>Health and Social Benefits of biodiversity and Nature Protection</p>
</td>
</tr>
<tr>
<td>
<p>Sjerp de Vries</p>
</td>
<td>
<p>Alterra, Wageningen UR (Netherlands)</p>
</td>
<td>
<p>Possible pathways linking nearby nature to human health and their relative importance</p>
</td>
</tr>
<tr>
<td>
<p>Peter Van den Hazel</p>
</td>
<td>
<p>Public health Services Gelderland-Midden (Netherlands)</p>
</td>
<td>
<p>Green and health in cities</p>
</td>
</tr>
<tr>
<td>
<p>Patrick Van Damme</p>
</td>
<td>
<p>Ghent University (Belgium)</p>
</td>
<td>
<p>Developing global medicinal plant markets: panacea or disaster ?</p>
</td>
</tr>
<tr>
<td>
<p>Chantal Shalukoma</p>
</td>
<td>
<p>Institut Congolais pour la Conservation de la Nature (Congo)</p>
</td>
<td>
<p>Typology of healers in traditional medicine around the Kahuzi-Biega national Park,DR Congo</p>
</td>
</tr>
<tr>
<td>
<p>Pierre Duez</p>
</td>
<td>
<p>University of Mons (UMONS)</p>
</td>
<td>
<p>The project PhytoKat in Lubumbashi, D.R. Congo: conditions for the integration of traditional medicine in modern healthcare</p>
</td>
</tr>
<tr>
<td>
<p>Julie Garnier</p>
</td>
<td>
<p>Odyssey Conservation Trust (France)</p>
</td>
<td>
<p>One Health and Conservation Areas: Benefits of Gender Sensitive Approach</p>
</td>
</tr>
<tr>
<td>
<p>Ben Somers</p>
</td>
<td>
<p>KU Leuven (Belgium)</p>
</td>
<td>
<p>Assessing spatio-temporal relationships between respiratory health and biodiversity using individual wearable technology - the Respirit project</p>
</td>
</tr>
<tr>
<td>
<p>Mariska Bauwelinck</p>
</td>
<td>
<p>VUB (Belgium)</p>
</td>
<td>
<p>Green space – health GRESP-H project Belgium</p>
</td>
</tr>
<tr>
<td>
<p>Xianwen Chen</p>
</td>
<td>
<p>NINA (Norway)</p>
</td>
<td>
<p>Urban Nature’s Health Effects and Monetary Valu-ation: A Systematic Review</p>
</td>
</tr>
<tr>
<td>
<p>Timo Assmuth</p>
</td>
<td>
<p>SYKE (Finland)</p>
</td>
<td>
<p>Multi-dimensional assessment of benefits and risks of nature to health – human and non-human</p>
</td>
</tr>
</tbody>
</table>
<p> </p>
<p><strong><span style="font-size: 14pt;">Posters</span></strong></p>
<p> </p>
<table>
<tbody>
<tr>
<td>
<p><strong>Authors</strong></p>
</td>
<td>
<p><strong>Poster</strong></p>
</td>
</tr>
<tr>
<td>
<p>Vitalija Povilaityte-Petri, Pierre Duez</p>
</td>
<td>
<p>Sustainable use of medicinal plants and their products</p>
</td>
</tr>
<tr>
<td>
<p>Daniela Penafiel, Celine Termote, Ramon Espinel, Patrick Van Damme</p>
</td>
<td>
<p>Traditional Foods in Guasaganda- Ecuador, Counting To The Nutrition Indicator for Biodiversity</p>
</td>
</tr>
<tr>
<td>
<p>Raf Aerts, An Van Nieuwenhuyse, Marijke Hendricks, Lucie Hoebeke, Nicolas Dendoncker, Catherine Linard, Sebastien Dujardin, Willem Verstraeten, Andy Delcloo, Rafiq Hamdi, Nelly Saenen, Tim Nawrot, Jean-Marie Aerts, Jos Van Orshoven, Ben Somers</p>
</td>
<td>
<p>Cumulative alpha diversity dose CADD as an integrated measure of human exposure to biodiversity</p>
</td>
</tr>
<tr>
<td>
<p>Marianne SCHLESSER</p>
</td>
<td>
<p>Biodiversity 2020, Update of Belgium's National Strategy</p>
</td>
</tr>
<tr>
<td>
<p>Bianca Ambrose-Oji, Liz O'Brien, Jack Forster, Tom Conolly</p>
</td>
<td>
<p>Wild horticulture can promote wellbeing and facilitate conservation learning and education</p>
</td>
</tr>
</tbody>
</table>
</body> </html>


dimitri.brosens@inbo.be





We Deal With
















Institute for Nature and Forest Research (INBO)
•Policy support
•Nature management
•International reporting
•(Natura2000 Monitoring)
•MERS
•(EV)INBO
•EU projects
•Flemish Projects
•Belgian Projects
•Open data institute (2015)
We deal with
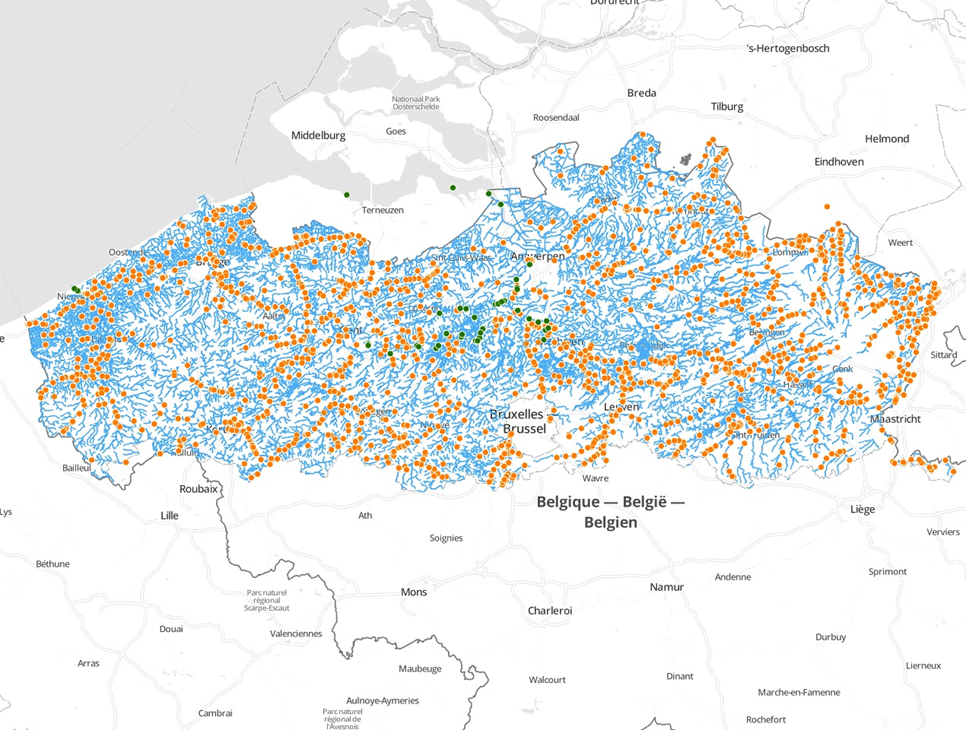
Human observations
Going back 20 years and more
Chinese mittencrab invasion in Flanders








Machine observations
Mainly via lifewatch.inbo.be




A lot of DATA
What is "Biodiversity Informatics"
- The Computerized management of any aspects of Biodiversity
- To handle the huge amount of data and information generated by the study of Biodiversity
- Create global access to information on biological species and their role in nature

Not to confuse with “Bioinformatics” ◦interdisciplinary field that develops and improves on methods for storing, retrieving, organizing and analyzing biological data. (sequence analysis, genome annotation, protein expression analysis…
Why “Biodiversity Informatics”
- Improved management; discovery & access
- New ways to
- view and analyse existing data
- create models
- Work with huge loads of data (Big Data)
- Integration data from different sources
- CoL + GBIF + EoL…
- Compare data from different sources
- Occurrence data + taxonomical data + …
To provide answers on large biodiversity questions

What questions does it “potentially” solve
Global Change Biology
- Changes in species and population distribution and diversity over time
- How intrinsic factors and extrinsic factors interact to determine species responses
Biota: wide picture of diversification and interactions
Future communities
- Predict combinations of species not previously experienced
- Alien species interactions
•Integrating phenotype and genotype
•Synthetic conservation planning
- Take advantage of up-to-date modern taxonomic information to define units of biodiversity
Model the world
Thresholds:
Data Quality
-
reliable
- occurrence data
- coördinates
- dates
- identifications
- information
-
metadata
- complete
Data Quantity
-
Critical amount of
- records
- information
Data Interoperability
-
Correct use of
- standards
- mapping
- licenses
Fit for use!





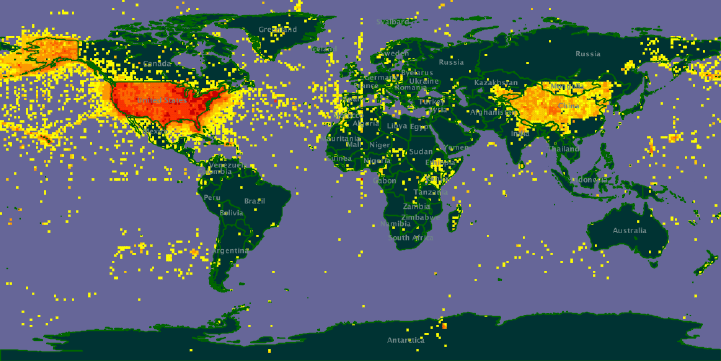

Global biodiversity informatics outlook
The Global Biodiversity Informatics Outlook (GBIO) offers a framework for reaching a much deeper understanding of the world’s biodiversity, and through that understanding the means to conserve it better and to use it more sustainably.
Global list of all species
Catalogue of Life Plus (CoL+), latest ATTEMPT to tie all information related to a scientific name
Creating an open, shared, and sustainable consensus taxonomy
Current Biodiversity Informatics issues
Problems with genus and species scientific names as unique and persistent identifiers
Linnaean system has many advantages, but also many problems (homonyms, synonyms...)
One proposed solution to this problem is the usage of Life Science Identifiers (LSIDs) for machine-machine communication purposes
Achieving a consensus classification of organisms

Some major Global Biodiversity Informatics projects

4 Broad Activity categories
- Data extraction and capture
- Data compilation and serving
- Data display and visualization
- Data analysis (workflows)

Another view on Connecting biodiversity projects
the way to linked open data
Some Important Biodiversity Informatics related INITIATIVES



Georeferenced data
Distributions
Taxonomic backbone
Species information
Occurrences
Classifications
Type information
Location
Webservices
Pictures
Some Important Biodiversity Informatics related INITIATIVES

Georeferenced data
Distributions
Taxonomic backbone
Species information
Occurrences
Classifications
Type information
Location
Webservices
Pictures
Some Important Biodiversity Informatics related INITIATIVES

Some Important Biodiversity Informatics related INITIATIVES

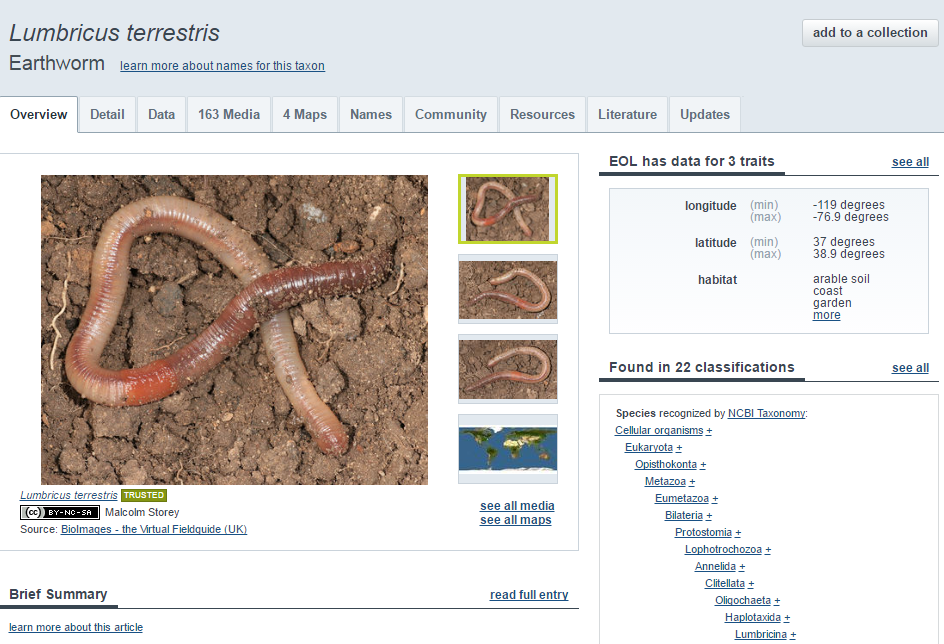
Overview
Detail
Data
Media
Maps
Community
Resources
Literature
Updates
Webservices
Some Important Biodiversity Informatics related INITIATIVES

Some Important Biodiversity Informatics related INITIATIVES

The gateway to our online database of the world's known species of animals, plants, fungi and micro-organisms.
Species Checklist
Taxonomic Hierarchy
Names Information
Relationships
Distribution
Webservices
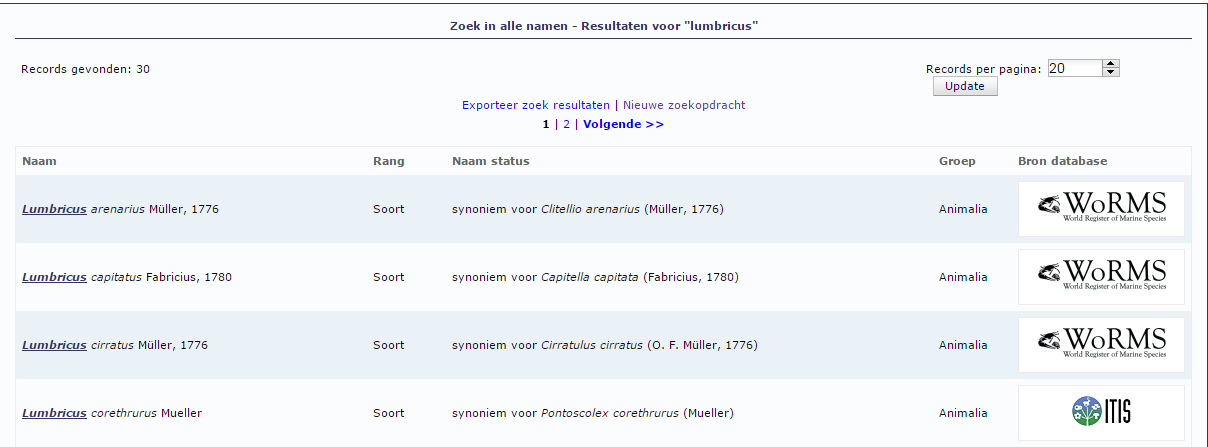
Some Important Biodiversity Informatics related INITIATIVES

Some Important Biodiversity Informatics related INITIATIVES

biodiversity literature
Books
Journals
Authors
Subjects
Scientific names
Webservices

Some Important Biodiversity Informatics related INITIATIVES

Some Important Biodiversity Informatics related INITIATIVES





Some Important Biodiversity Informatics related INITIATIVES
Overview
Common Names
Taxonomy
Distribution
Conditions (T°, [NaCl], Depth)
Occurrences
Data
DataSets
Webservices

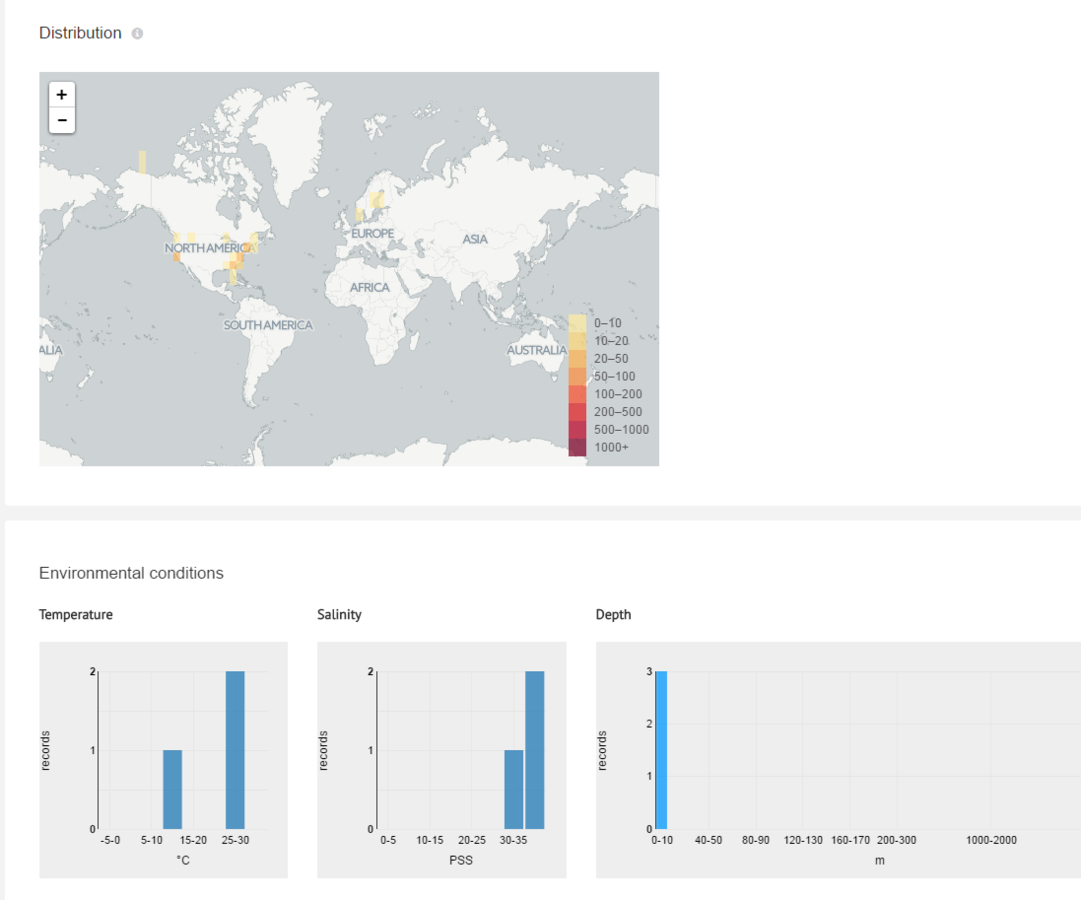


Some Important Biodiversity Informatics related INITIATIVES


Some Important Biodiversity Informatics related INITIATIVES
Metadatabase
Data Portal
Atlas
Traits database
Tools
Resources
Policies
Networks
Blog




Some Important Biodiversity Informatics related INITIATIVES

Some Important Biodiversity Informatics related INITIATIVES
Collections
Training & Workshops
Taxonomy
Datasets
Webservices



Some Important Biodiversity Informatics related INITIATIVES


Some Important Biodiversity Informatics related INITIATIVES
Funded by Belspo
Focus on Antarctica
Data Portal & Services
- Taxonomy
- Metadata
- Environmental
RAMS


Some Important Biodiversity Informatics related INITIATIVES
Funded by Belspo
Focus on Belgium
Data Portal & Services
- Taxonomy
- Metadata
GBIF based


Some Important Biodiversity Informatics related INITIATIVES
Focus on Australia
Data Portal & Services
- Taxonomy
- Metadata
- Collections
- Maps
API



Some Important Biodiversity Informatics related INITIATIVES
ETC...










Some Important Biodiversity Informatics related PROJECTS




Lifewatch







Citizen Science & Observations


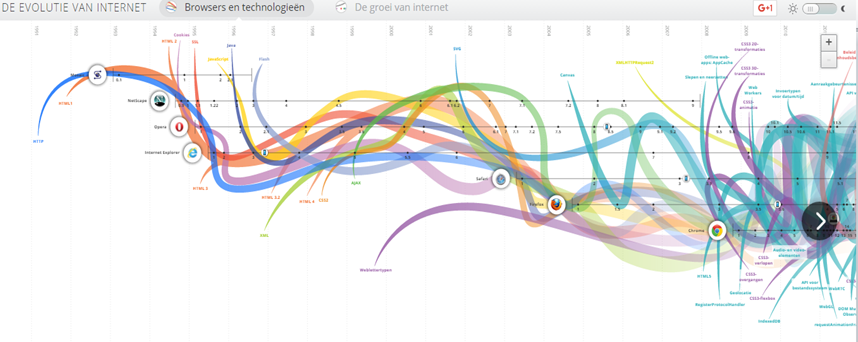
Biodiversity Informatics and the evolution of the Internet

Evolution of The Web
- WEB (1.0): Static web pages, informative
- WEB (2.0): User generated content, usable, interoperability, communities (No technical update)
- WEB (3.0): The semantic web, interactive and participative, connecting data, web through standards, shared & linked data...
IN Biodiversity










2000-2005

2006-2012

2012-2014

2014-2016

The semantic web (3.0)
- The semantic web, a framework where data can be shared and reused
- Need for open data*!
- Machine-to-machine interacting (webservices)
- Certain data should be freely available for anyone to use -> Open Data
- No restrictions in copyright, patents or other mechanisms (CC0)
*By data we mean specimen, observation or checklist datasets published as a Darwin Core Archive and any derivatives. This does not include code, pictures, poems and movies…

Web services: how computers talk…
- A Web service is a method of communication between two electronic devices over World Wide Web
- A Web service is a software function provided at a network address over the web or the cloud; it is a service that is "always on" as in the concept of utility computing (http://en.wikipedia.org/wiki/Web_service)
-
It has an interface described in a machine-processable format
- Different protocols
- REST, SOAP http://webservice.catalogueoflife.org/
- API (Application Programming Interface); a set of Hypertext Transfer Protocol (HTTP) request messages

The Internet and his tools to
TO Create Open SCIENCE

Open Data

“Open means anyone can freely access, use, modify, and share for any purpose(subject, at most, to requirements that preserve provenance and openness).”
Acceptable:
- The license may require distributions of the work to include attribution of contributors, rights holders, sponsors, and creators as long as any such prescriptions are not onerous.
- The license may require that modified versions of a licensed work carry a different name or version number from the original work or otherwise indicate what changes have been made.
- The license may require distributions of the work to remain under the same license or a similar license.
- ...



Why Should we Publish Under OPEN CC0
There is very little copyright in occurrence & checklist data
- You cannot copyright facts
- Different rules in different countries
Other licenses are too restrictive or do not apply to factual data -> no use in using them
With restricted data you can't do anything.!!


The Blue list: elements of taxonomic information that are not subject to copyright.
A hierarchical organization (= classification)
A life-form ordering of taxa.
Scientific names of genera or other uninomial taxa, species epithets of species names, binomial combinations as species names, or names of infraspecific taxa.
Information about the etymology of the name; statements as to the correct, alternate or erroneous spellings.
Rank, composition and/or apomorphy of taxon.
Lists of synonyms and/or chresonyms or concepts, including analyses and/or reasoning as to the status or validity of each.
Citations of publications that include taxonomic and nomenclatural acts, including typifications.
Reference to the type species of a genus or to other type taxa.
References to type material, including current or previous location of type material, collection name or abbreviation thereof, specimen codes, and status of type.
The Blue list: elements of taxonomic information that are not subject to copyright.
Data about materials examined.
References to image(s) or other media with information about the taxon.
Information on overall distribution and ecology, perhaps with a map.
Known uses, common names, and conservation status (including Red List status recommendation).
Description and / or circumscription of the taxon (features or traits together with the applicable values), diagnostic characters of taxon, possibly with the means (such as a key) by which the taxon can be distinguished from relatives.
General information including but not limited to: taxonomic history, morphology and anatomy, reproductive biology, ecology and habitat, biogeography, conservation status, systematic position and phylogenetic relationships of and within the taxon, and references to relevant literature.
•All rights reserved -> Data unusable
(So why bother publishing)
•Open Data Commons Public Domain Dedication and License (PDDL)
•Creative Commons Attribution-NoDerivs (CC BY-ND)
•Creative Commons Attribution-NonCommercial (CC BY-NC)
•Creative Commons Attribution-ShareAlike (CC BY-SA) or Open Data Commons Open Database License (ODbL)
•Creative Commons Attribution (CC BY) or Open Data Commons Attribution License (ODC-By)





Other Licenses
OPEN SOURCE & Code
Generally, open source refers to a computer program in which the source code is available to the general public for use and/or modification from its original design. Open-source code is meant to be a collaborative effort, where programmers improve upon the source code and share the changes within the community.
https://en.wikipedia.org/wiki/Open_source
Open research is research conducted in the spirit of free and open source software. Much like open source schemes that are built around a source code that is made public, the central theme of open research is to make clear accounts of the methodology freely available via the internet, along with any data or results extracted or derived from them.

OPEN METHODOLOGY


OPEN PEER REVIEW
Open peer review (also called "public peer review", "transparent peer review") denotes several, closely related forms of scholarly peer review: Open-identity or attributed peer review (as opposed to anonymous peer review) Open-disclosure or public peer review, where the peer review contents are publicly available.

OPEN ACCESS

Open access (OA) refers to online research outputs that are free of all restrictions on access (e.g. access tolls) and free of many restrictions on use (e.g. certain copyright and license restrictions).[1] Open access can be applied to all forms of published research output, including peer-reviewed and non peer-reviewed academic journal articles, conference papers, theses,[2] book chapters,[1] and monographs.[3]

By Petr Knoth and Nancy Pontika - Own work, CC BY 3.0, https://en.wikipedia.org/w/index.php?curid=50529549
BUT.....

•Biologists (not only) are usually very reluctant towards open data.
•Biologists are reluctant towards the use of data & code generated by others.
à Not a problem? What is not known, cannot be loved!
Biologists & Data Publication

The Solution is in the combination
-
In the Research Institute for Nature and Forest we publish occurrence data:
- Open (CC0 waiver)
- Norms for data use
-
GBIF is evolving towards 3 licenses only:
- CC0, under which data are made available for any use without restriction or particular requirements on the part of users
- CC-BY, under which data are made available for any use provided that attribution is appropriately given for the sources of data used
- CC-BY-NC, under which data are made available for any use provided that attribution is appropriately given and provided the use is not for commercial purposes

OPEN Data: Some examples
OPEN Data: Some examples
OPEN Data: Some examples

dealing with data
Some Definitions
-
Raw data
-
Data collected from a source
-
Human observation, a collection, electronic generated data
-
-
Primary species occurrence data
-
When; what; where
-
-
Accuracy & precision
-
High accuracy: occurrence within the bounding box
-
Low accuracy: not sure if the occurrence was within the bounding box
-
High precision: very precise coordinate
-
(lat: 51,23564 ;long: 3,25644)
-
-
Low precision: less precise coordinate
-
(lat: 51,2; long 3,2)
-
-
-
Data quality
-
“Fitness for use” or “potential use”
-
-
Data model
-
Visual projection of all the entities, relations and restrictions in a database
-
-
Data standard
-
Convention used in the context of “Data”
-




dealing with data
The Data Life Cycle


dealing with data
Data Management

-
Follow the Data Policy
-
Long term goals; guiding principles for management & publication & metadata
-
-
Identify Clear roles
- Data collector; metadata generator; data analyzer; database administrator; administrative support staff; archiving; ICT staff
-
Plan and document your database
- Complete Metadata
- Define procedures for updates
- Hardware; software; formats; storage & data
-
Standardize and Store data Criteria for data access
- Publish data
-> Create a Data Management Plan

dealing with data
DataBase design & specs

RESEARCH QUESTION?
- Worksheet
- Existing Database
- Extend a Database
- Develop a Database
No datamodel
Existing datamodel
Change a datamodel
New datamodel
•Think before you begin
•Keep it simple
•Use standards if possible
•Ask advice or become a biogeek

dealing with data
Data Management Plan


dealing with data
Data Management Plan


dealing with data
Data Management Plan


dealing with data
Tips & Tricks

- Manage your data for yourself:
- Keep yourself organized – be able to find your files (data inputs, analytic scripts, outputs at various stages of the analytic process, version control, etc)
- Track your science processes for reproducibility – be able to match up your outputs with exact inputs and transformations that produced them
- Control your versions of the Quality control your data more efficiently
- Make backups to avoid data loss
- Format your data for re-use (by yourself or others)
- Be prepared: Document your data for your own recollection, accountability, and re-use (by yourself or others)
- Prepare it to share it – gain credibility and recognition for your scientific efforts!


dealing with data
Data Quality



Impact on data quality and “Fitness for use”
- Data capture and recording at the time of collecting
- Data manipulation prior to digitization (label preparation, copying of data to a ledger, etc.),
- Identification of the collection (specimen, observation) and its recording,
- Digitization (import)of the data,
- Documentation of the data (capturing and recording the metadata)
- Data validation
- Data storage and archiving,
- Data presentation and dissemination (paper and electronic publications, web-enabled databases, etc.),
- Use of the data (analysis and manipulation).





Dealing with data



Data Capture and recording
- The “collector” has fundamental responsibility
- Record all needed parameters, clear and accurate
- Record readable and unambiguous
- Complete all required fields
- Note clear field notes
Data Import
- The “curator” has fundamental responsibility
- data is correctly imported into the database
- the correct quality control procedures are implemented and exercised
- all the documentation is created
- metadata is recorded
- validation checks are routinely carried out on the data
- carried out validation checks are well documented
- the data is stored in a suitable manner
- earlier versions are systematically stored and archived to allow comparisons and return to the “uncleaned original” data
- data integrity is maintained
- data can be exported in a correct manner.
DATA Quality, Capture and Import
dealing with data
Data Quality



DATA VALIDATION AND CLEANING
- Improve the overall quality of the dataset
- Improve the fitness for use
- Determine if data are
- Inaccurate, incomplete or unreasonable
- Flag inaccurate data
- Create automatic validation
- Fix errors
dealing with data
Data Quality



DATA Visualisation
TOOLS:
- GIS or Webgis tools
- CartoDB, QGIS....

dealing with data
MetaData




- Data about data
- Structural metadata
- The story of the database
- Descriptive metadata
- The story of the data
- When, what, who, where, how…
- The story of the data
- Structural metadata
- Metadata standards
- Dublin core (books)
- Inspire (EU) (geographical dataset; maps)
- EML (ecological metadata language)
- Metadata in GBIF
- GBIF metadata profile
dealing with data
Data Paper




http://zookeys.pensoft.net/articles.php?id=4575
International
- Bring technologic, economic and societal benefits
- Harmonize technical specifications of products and services
- www.iso.org
Biodiversity
- Deals with biodiversity standards and standardization
- develop, adopt and promote standards and guidelines for the recording and exchange of data about organisms
- act as a forum for discussion
- www.tdwg.org
Standards and Standardization


Standards and Standardization
The Darwin Core Standard



- Is a bag of terms
- Inspired on DublinCore
- Facilitates the exchange of information about organisms
- CORE: Collections | Observations | sampleBased | Checklists
- Stable standard for sharing information on biological diversity
- Very simple flat .txt files
Standards and Standardization
The Darwin Core Standard + extensions

- Simple structure, extensions possible
- vernacularNames
- measurementsOrFacts
- speciesDistribution
- references
- one to many relations!

http://rs.gbif.org/
Standards and Standardization
The Darwin Core Standard + extensions

http://rs.gbif.org/
Standards and Standardization
The Darwin Core Standard + extensions

http://rs.gbif.org/
Standards and Standardization
The Darwin Core Standard + extensions

http://rs.gbif.org/

Standards and Standardization
The Darwin Core Standard
Sample Based Data

http://rs.gbif.org/

Recap
http://rs.gbif.org/
- In web (3.0) websites can communicate
- Many (tools, websites, data portals..) related to biodiversity do make use of this
- To make this work, we need data to be available
- Data needs to be managed properly (Might need a DMP)
- Data needs to be standardized
- Data needs metadata
- We need qualitative data
- Data needs to be published in an open format
- We need to make use of standards to make data talk
- A dataset is a 'heap' of information in one package
Standards and Standardization
The ABCD Standard
http://rs.gbif.org/
- Access to Biological Collections Databases
- Exchange of data between collection databases worldwide
- Same goal as the Darwin Core
- Less rigid
- 1200 concept fields
- Very big bag of concepts
- ABCDEFG
- Access to Biological Collections Databases Extended For Geosciences
- ABCDDNA
- Access to Biological Collections Databases for genetic material
- Xml document with ABCD schema
- For GBIF, the Darwin Core is mostly promoted

Standards and Standardization
Darwin Core Archives
Data-Exchange-Format

http://rs.gbif.org/

Standards and Standardization
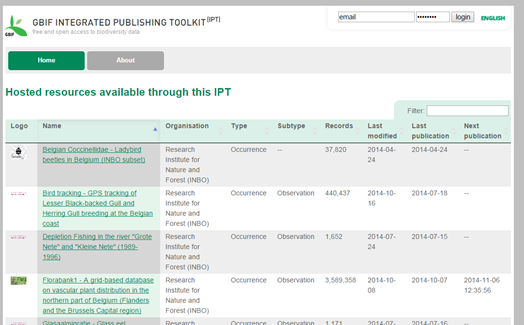
Publication TOOL: Integrated Publishing Toolkit

http://rs.gbif.org/
Standards and Standardization
Publication TOOL: BioCase

http://rs.gbif.org/

What can 'Biodiversity' Informatics do for you
http://rs.gbif.org/
All the "Biodiversity Informatics" tools, standards, portals, webservices, checklists, communities, programs, code, visualisation, GIS tools, databases, data
are there to help you to
improve, present, discover, explore, analyse and disseminate your research










Check and improve your data
On line taxonomic Checklists
- Catalogue of Life
- Global Names Initiative
- Pesi
- Fauna Europaea
- Worms
- Fishbase
- Fada
- GBIF Names Service
- Encyclopedia of Life
https://github.com/inbo/inbo-pyutils/tree/master/gbif/gbif_name_match
Match a set of species names with the GBIF taxonomic backbone
Introduction
Working with different partners/institutes/researchers results in a diversity of taxonomic names to define species. This hardens comparison amongst datasets, as in many occasions, aggrgeation is aimed for or filtering on specific species. By translating all species names to a common taxonomic backbone (ensuring unique ID's for each species name), this can be done.
Aim
This small utility provides the functionality to add the species information from the GBIF backbone to any data table (CSV-style or a Pandas dataframe) by requesting this information via the GBIF API. For each match, the corresponding accepted name is looked for. Nevertheless there will always be errors and control is still essential, the acceptedkeys provide the ability to compare species names from different data sources.
Functionality
The functionality can be loaded within Python itself by importin the function extract_species_information or by running the script from the command line.
Command line function
To check the functionality of the command line function, request for help as follows:
python gbif_species_name_match.py --helpCheck and improve your data
GBIF taxonomic Backbone
Pyton Tools Example

Match a set of species names with the GBIF taxonomic backbone
1. This small utility provides the functionality to add the species information from the GBIF backbone to any data table (CSV-style or a Pandas dataframe) by requesting this information via the GBIF API. 2. The functionality can be loaded within Python itself by importin the function extract_species_information or by running the script from the command line.
Check and improve your data
GBIF taxonomic Backbone
Jupyter Notebook Example

Check and improve your data
GBIF taxonomic Backbone
Exploratory Example

Check and improve your data
GBIF taxonomic Backbone
Open Refine

Check and visualize your data
Carto.com
https://dimitri.carto.com/dashboard/
On line literature resources
The Biodiversity Heritage Library works collaboratively to make biodiversity literature openly
available to the world as part of a global biodiversity community.
Light up your mind
access to more than 1,500 archival journals on JSTOR
scholarly journals, primary sources, and now books


GBIF
The Global Biodiversity Information FAcility
The Global Biodiversity Information Facility (GBIF) is an international open data infrastructure, funded by governments.
The Belgian biodiversity Platform is the Belgian node for GBIF
http://projects.biodiversity.be/ifbl
It allows anyone, anywhere to access data about all types of life on Earth, shared across national boundaries via the Internet.
By encouraging and helping institutions to publish data according to common standards, GBIF enables research not possible before, and informs better decisions to conserve and sustainably use the biological resources of the planet.
GBIF operates through a network of nodes, coordinating the biodiversity information facilities of Participant countries and organizations, collaborating with each other and the Secretariat to share skills, experiences and technical capacity.

GBIF
The Global Biodiversity Information FAcility

Some facts about GBIF
It provides a single point of access (through this portal and its web services) to more than 600 million records, shared freely by hundreds of institutions worldwide, making it the biggest biodiversity database on the Internet.
◦
The data accessible through GBIF relate to evidence about more than one million species, collected over three centuries of natural history exploration and including current observations from citizen scientists, researchers and automated monitoring programmes
◦
More than 4000 peer-reviewed research publications have cited GBIF as a source of data, in studies spanning the impacts of climate change, the spread of pests and diseases, priority areas for conservation and food security. About 20 such papers are published each month.
Many GBIF Participant countries have set up national portals using tools, codes and data freely available through GBIF to better inform their citizens and policy makers about their own biodiversity.
GBIF
The Global Biodiversity Information FAcility

Biodiversity Informatics in practice
The Global Biodiversity Information FAcility
Using the GBIF portal


Go to www.gbif.org
Go to “Get Data” and choose:
Explore occurrences
Number of Animalia records (scientificName)
Number of Animalia Specimens in GBIF (basisOfRecord)
How many Pieris japonica occurrences in GBIF (a butterfly)
◦
Explore species
Look for Polypodium virginianum
How many infraspecies you can find on GBIF
Give one synonym
Vernacular name in “English, Swedish and German”
Where can you find the Holotype? (Occurrences TypeStatus)
Explore the Portal


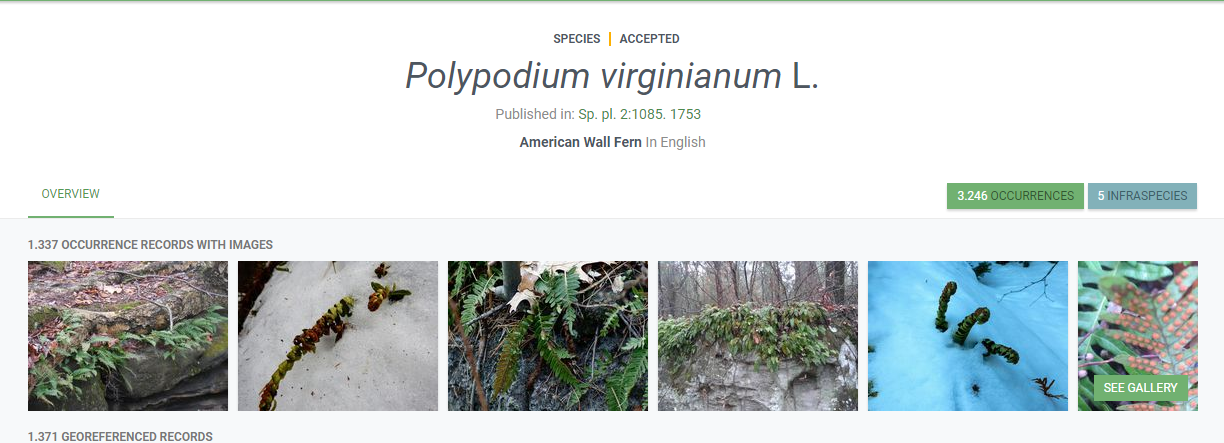



Explore datasets
- Find Florabank1
- What is the CC license?
- Follow the DOI for the datapaper…
Explore by country
- Go to Belgium
- How many datasets are published by Belgium
- How many datasets contain data about Belgium
- How many specimens are digitally available about Belgium (in Belgium)
Explore About GBIF Belgium on the Homepage, can you find the same page for Holland (tip: NL)
Check the BE country Report

Look for this dataset “Loopkevers aan de grensmaas-Carabid beetles near the river Meuse” View “occurrences” ◦How many occurrences did you find? Add a filter for ScientificName ◦Carabidae ◦Try some other filters ◦How many occurrences did you find? Download the dataset from GBIF or Download the data from INBO IPT (external Data) Unzip the dataset Or download the data from GBIF (Simple CSV file) Google “Loopkevers aan de grensmaas-Carabid beetles near the river Meuse” Check the “RTF” file for metadata; Can you find any other access points for this dataset? http://Data.inbo.be/ipt Find the same data beginning from Explore occurrences (use the filters)