Modeling Topological Polymers
/siam19
This talk!

Collaborators


Jason Cantarella
U. of Georgia
Tetsuo Deguchi
Ochanomizu U.
Erica Uehara
Ochanomizu U.
Funding: Simons Foundation
Linear polymers
A linear polymer is a chain of molecular units with free ends.
Polyethylene


Nicole Gordine [CC BY 3.0] from Wikimedia Commons
Shape of linear polymers
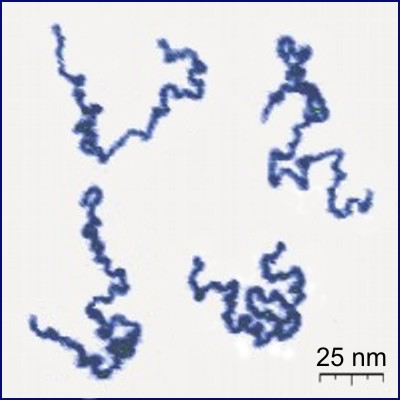
In solution, linear polymers become crumpled:
Protonated P2VP
Roiter–Minko, J. Am. Chem. Soc. 127 (2005), 15688-15689
[CC BY-SA 3.0], from Wikimedia Commons
Ring polymers
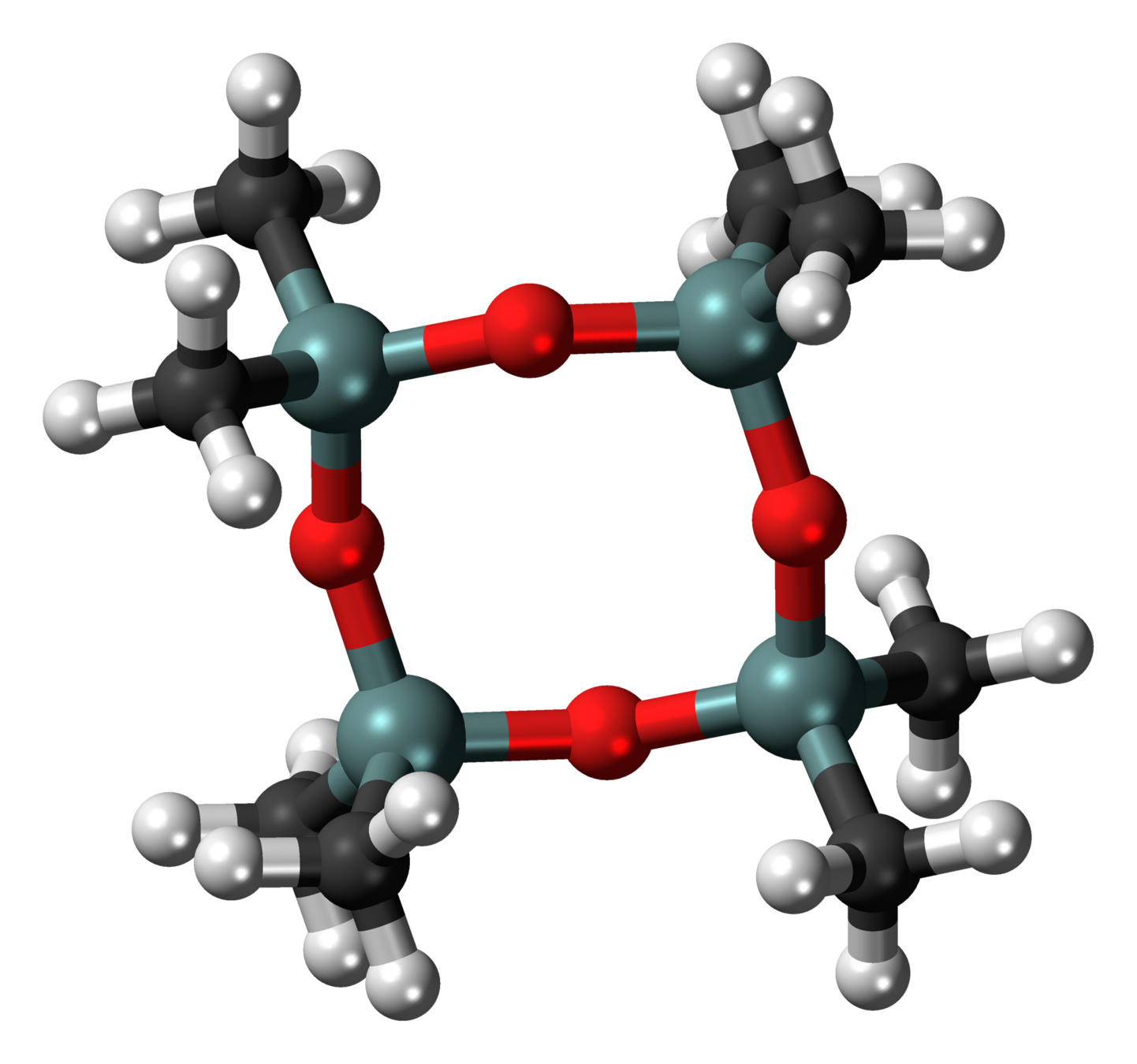
Octamethylcyclotetrasiloxane
(Common in cosmetics, bad for fish)
Ring biopolymers
Most known cyclic polymers are biological
Material properties
Ring polymers have weird properties; e.g.,
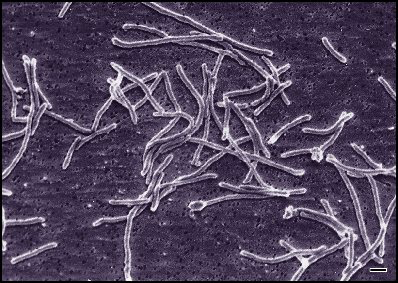
Thermus aquaticus
Uses cyclic archaeol, a heat-resistant lipid
Grand Prismatic Spring
Home of t. aquaticus; 170ºF
Topological polymers
A topological polymer joins monomers in some graph type:
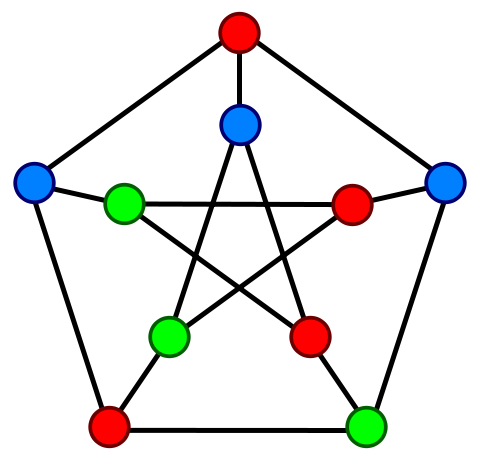
Petersen graph
In biology
Topological biopolymers have graph types that are extremely complicated (and thought to be random):
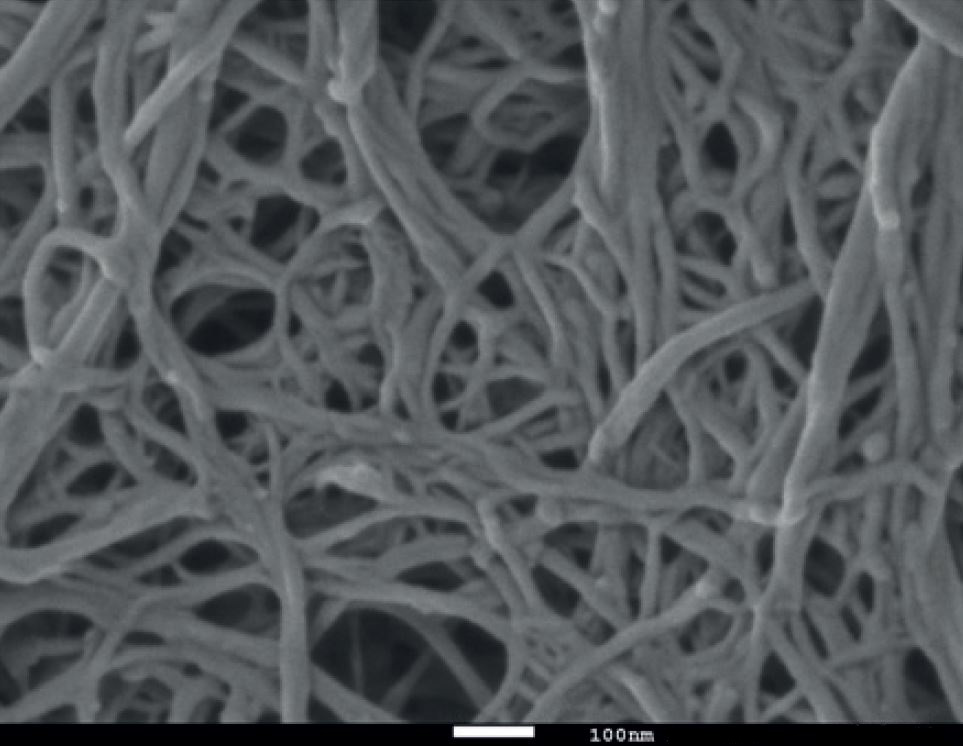
Wood-based nanofibrillated cellulose
Qspheroid4 [CC BY-SA 4.0], from Wikimedia Commons
Synthetic topological polymers
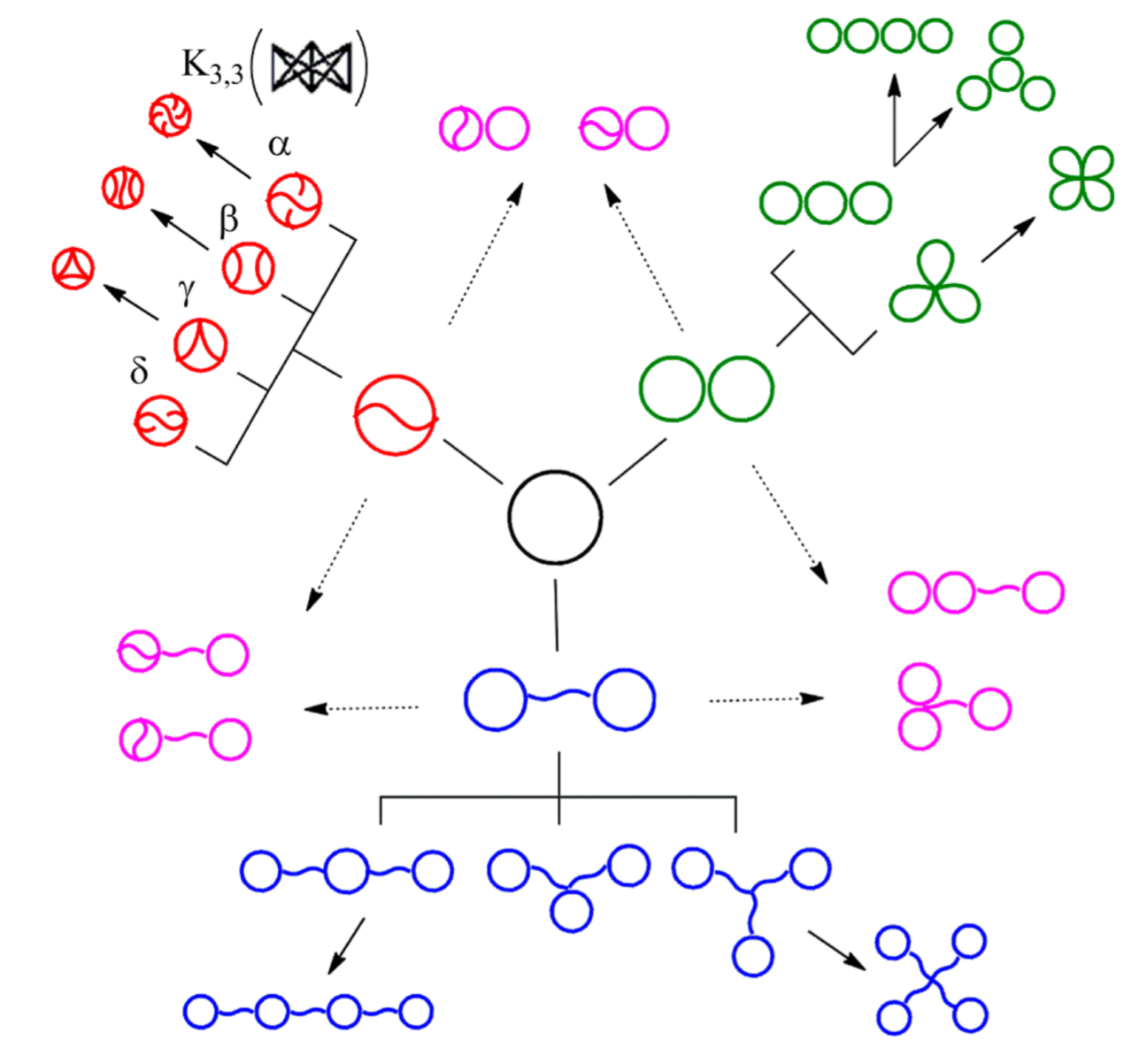
The Tezuka lab in Tokyo can now synthesize many topological polymers in usable quantities
Y. Tezuka, Acc. Chem. Res. 50 (2017), 2661–2672
Main Question
What is the probability distribution on the shapes of topological polymers in solution?
Ansatz
Linear polymers
Ring polymers
Topological polymers
Random walks with independent steps
Random walks with steps conditioned on closure
Random walks with steps conditioned on ???
Functions and vector fields
Suppose \(\mathcal{G}\) is a directed graph with \(\mathcal{V}\) vertices and \(\mathcal{E}\) edges.
Definition. A function on \(\mathcal{G}\) is a map \(f:\{v_1,\dots , v_\mathcal{V}\} \to \mathbb{R}\). Functions are vectors in \(\mathbb{R}^\mathcal{V}\).
Definition. A vector field on \(\mathcal{G}\) is a map \(w:\{e_1,\dots , e_\mathcal{E}\} \to \mathbb{R}\). Vector fields are vectors in \(\mathbb{R}^\mathcal{E}\).
Gradient and divergence
By analogy with vector calculus:
Definition. The gradient of a function \(f\) is the vector field
Definition. The divergence of a vector field \(w\) is the function
Gradient and divergence as matrices
So if \(B = \operatorname{div}\), which is \(\mathcal{V} \times \mathcal{E}\), then \(\operatorname{grad} = B^T\).
\(-B B^T = L\), the graph Laplacian.
Helmholtz’s Theorem
Fact.
The space \(\mathbb{R}^\mathcal{E}\) of vector fields on \(\mathcal{G}\) has an orthogonal decomposition
Corollary.
A vector field \(w\) is a gradient (conservative field) if and only if the (signed) sum of \(w\) around every loop in \(\mathcal{G}\) vanishes.
Gaussian embeddings
Definition.
A function \(f:\{v_i\} \to \mathbb{R}^d\) determines an embedding of \(\mathfrak{G}\) into \(\mathbb{R}^d\). The displacement vectors between adjacent vertices are given by \(\operatorname{grad}f\).
A Gaussian random embedding of \(\mathcal{G}\) has displacements sampled from a standard multivariate Gaussian on \((\text{gradient fields})^d\subset \left(\mathbb{R}^\mathcal{E}\right)^d\).
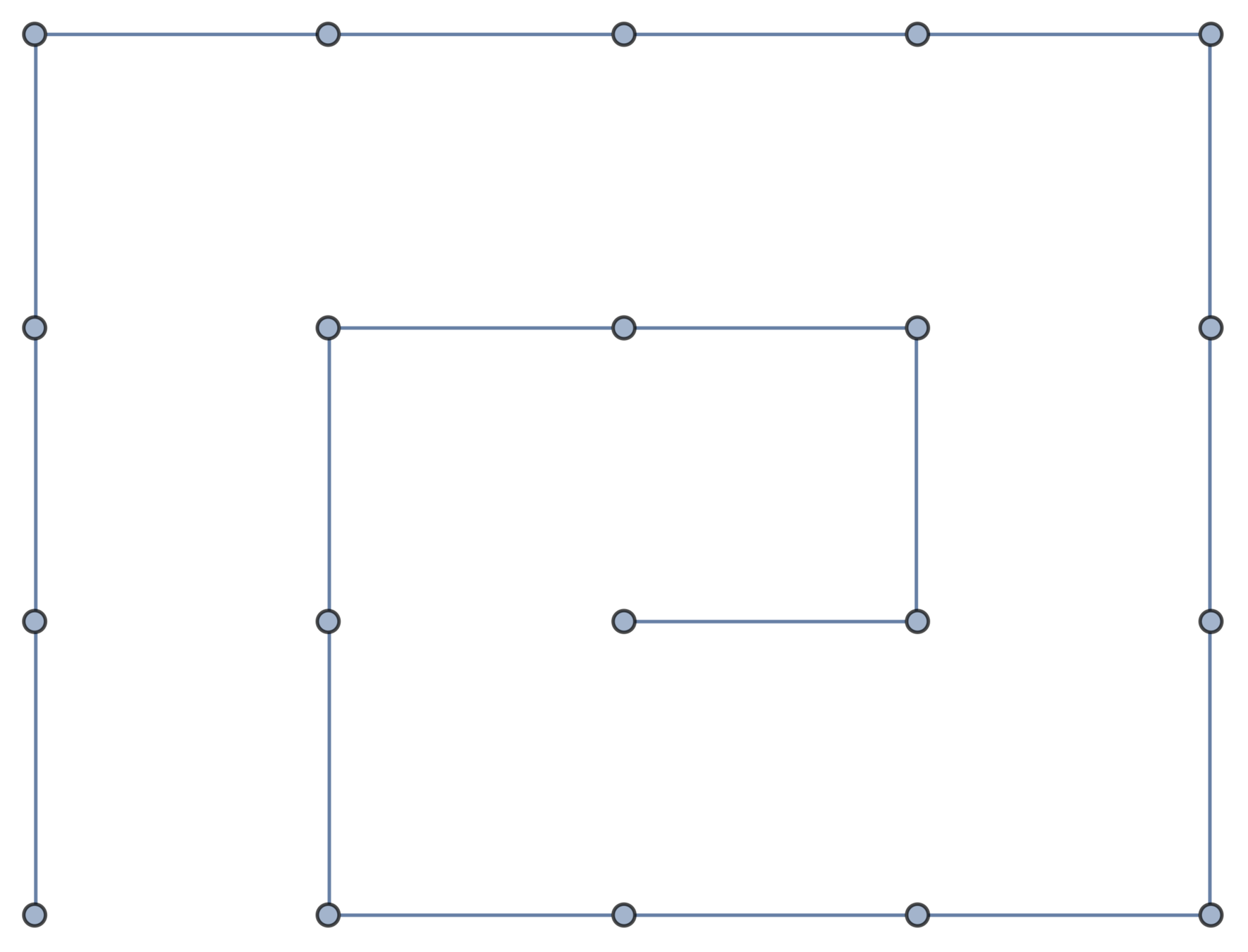
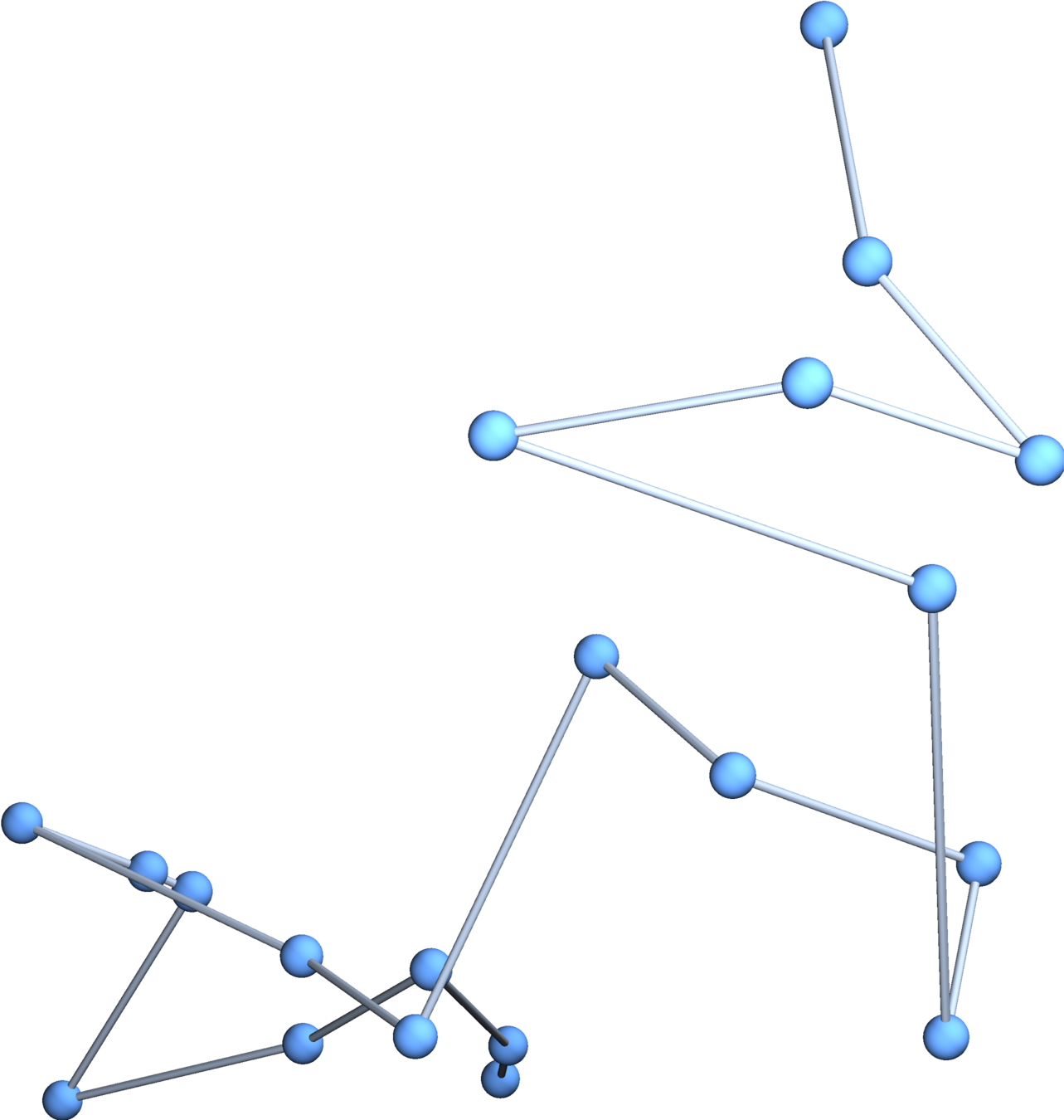
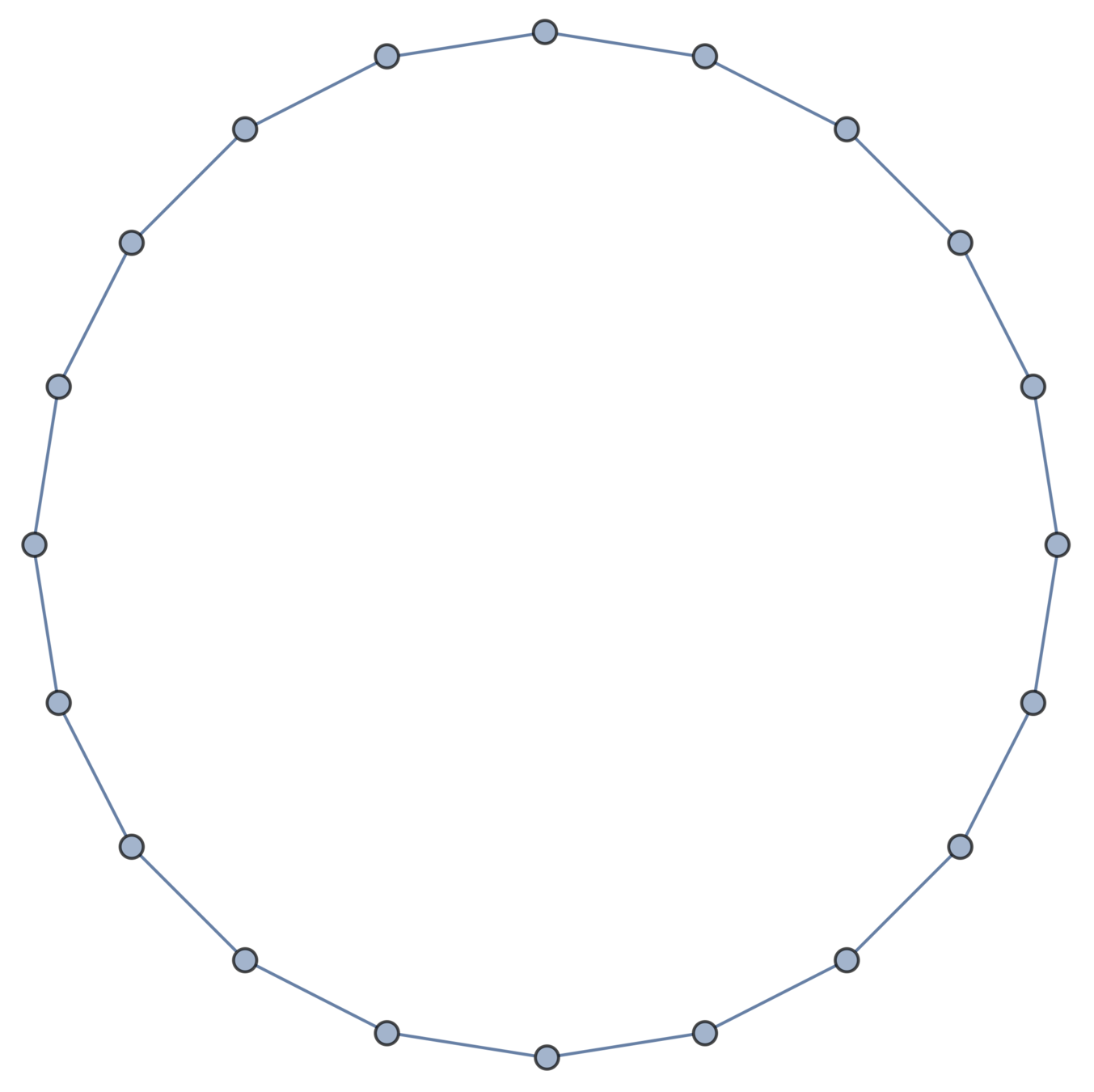
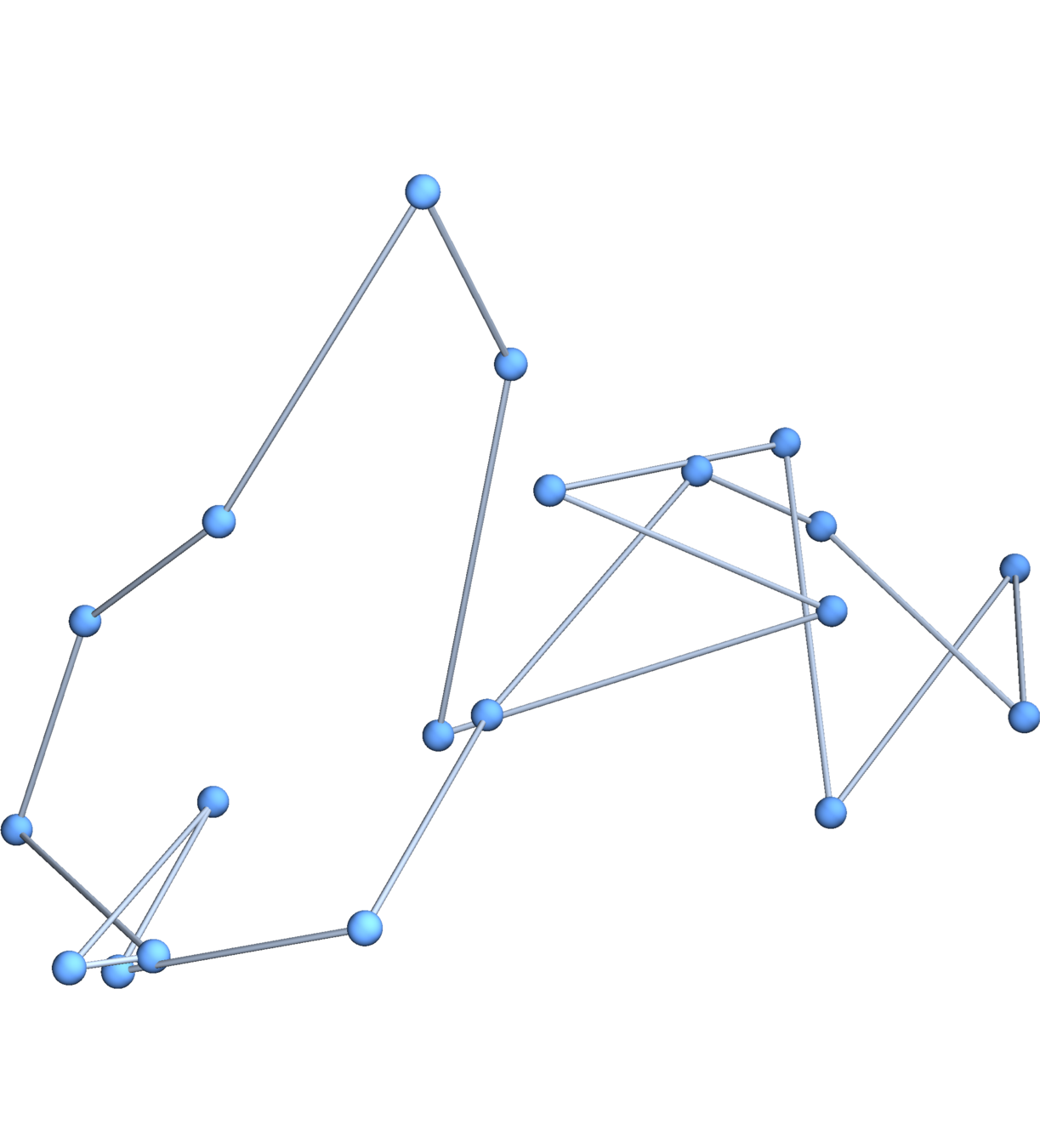
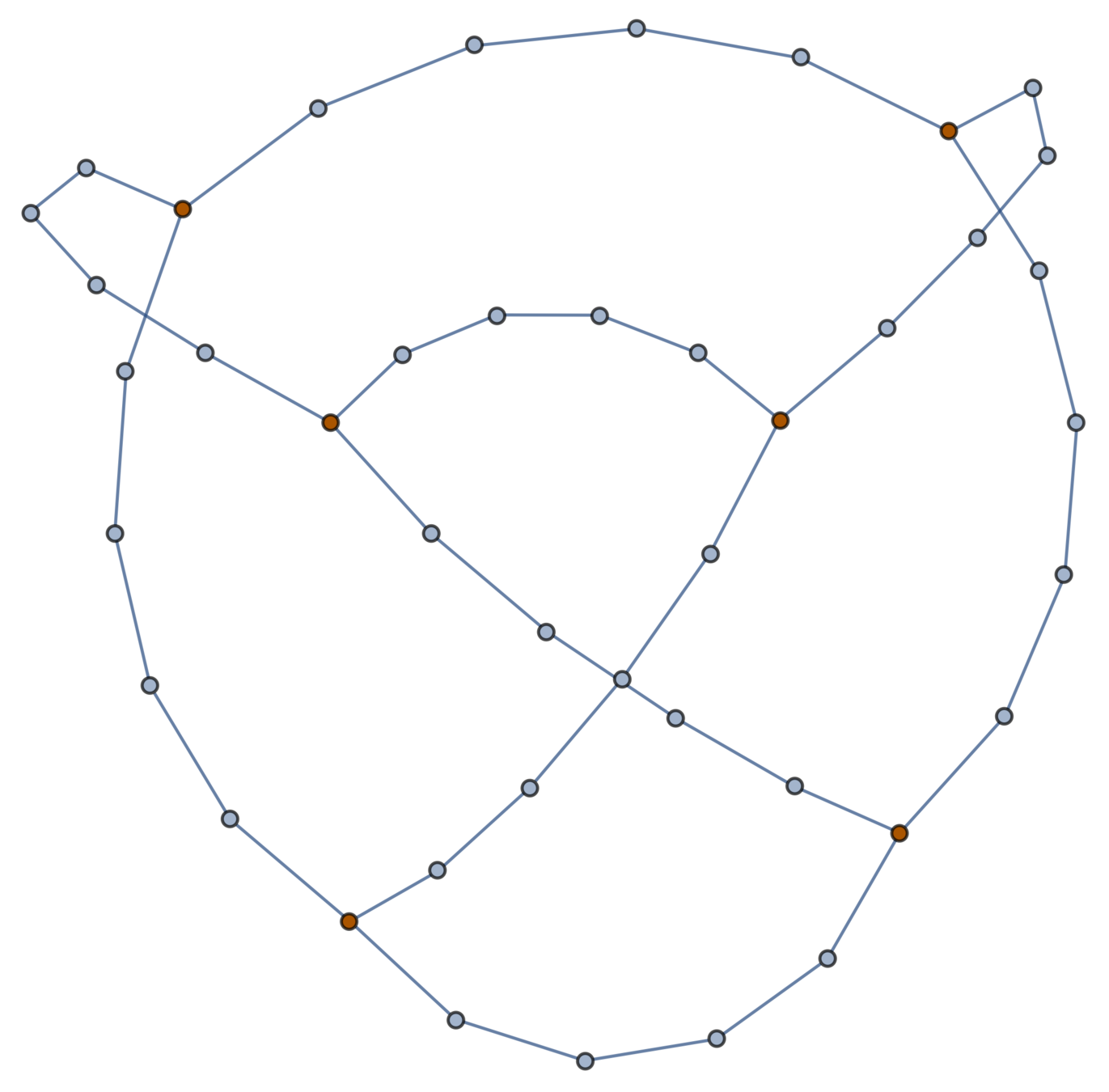
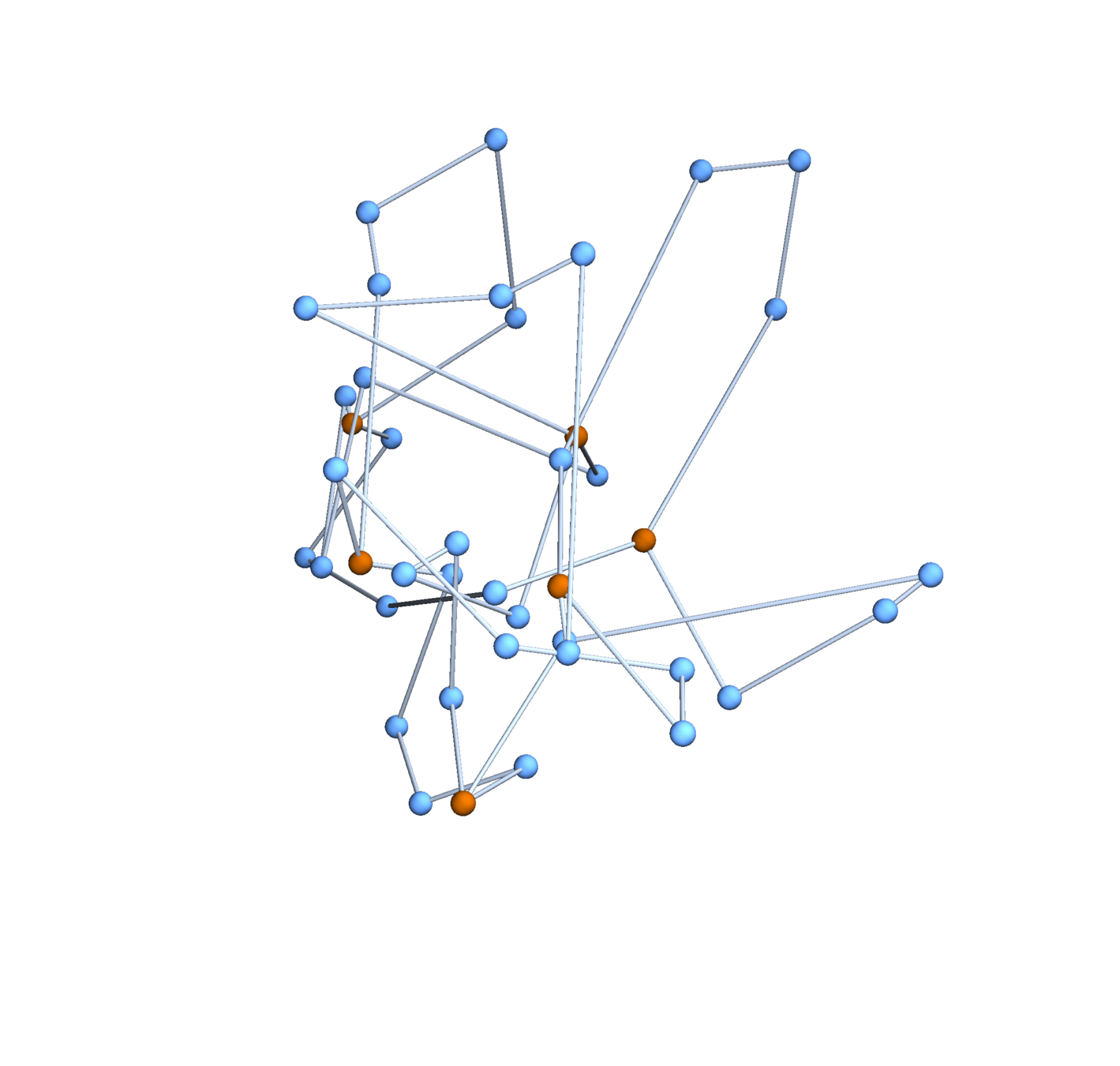
Expected radius of gyration
Theorem (w/ Cantarella, Deguchi, & Uehara; also Estrada–Hatano)
If \(\lambda_i\) are the nonzero eigenvalues of \(L\), the expected squared radius of gyration of a Gaussian random embedding of \(\mathcal{G}\) in \(\mathbb{R}^d\) is
This quantity is called the Kirchhoff index of \(\mathcal{G}\).
Suppose \(\mathcal{G}\) is a graph with \(\mathcal{V}\) vertices. Let \(L\) be the graph Laplacian of \(\mathcal{G}\).

Subdivisions
Current topological polymers are subdivisions of relatively simple graphs.
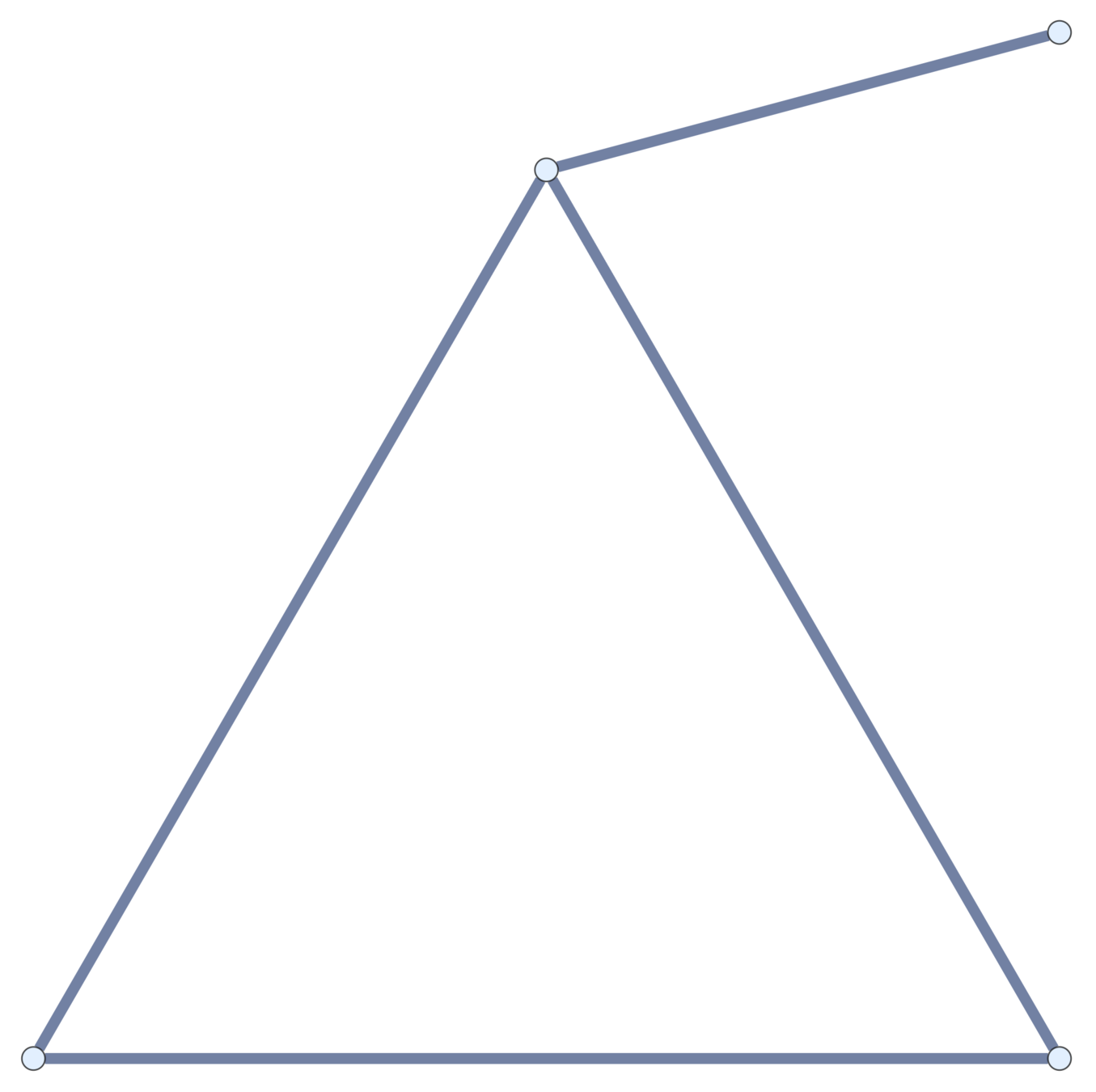
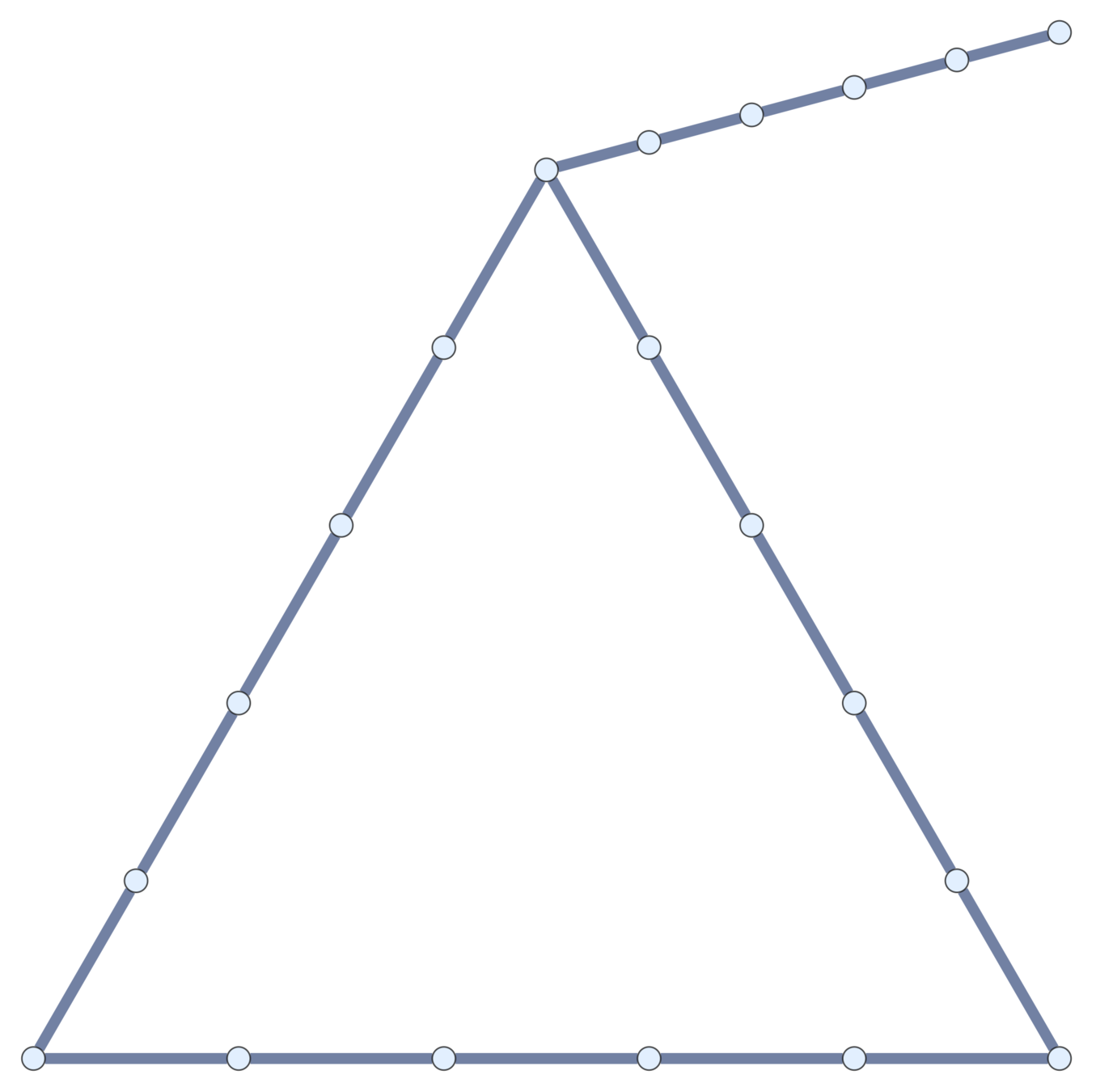
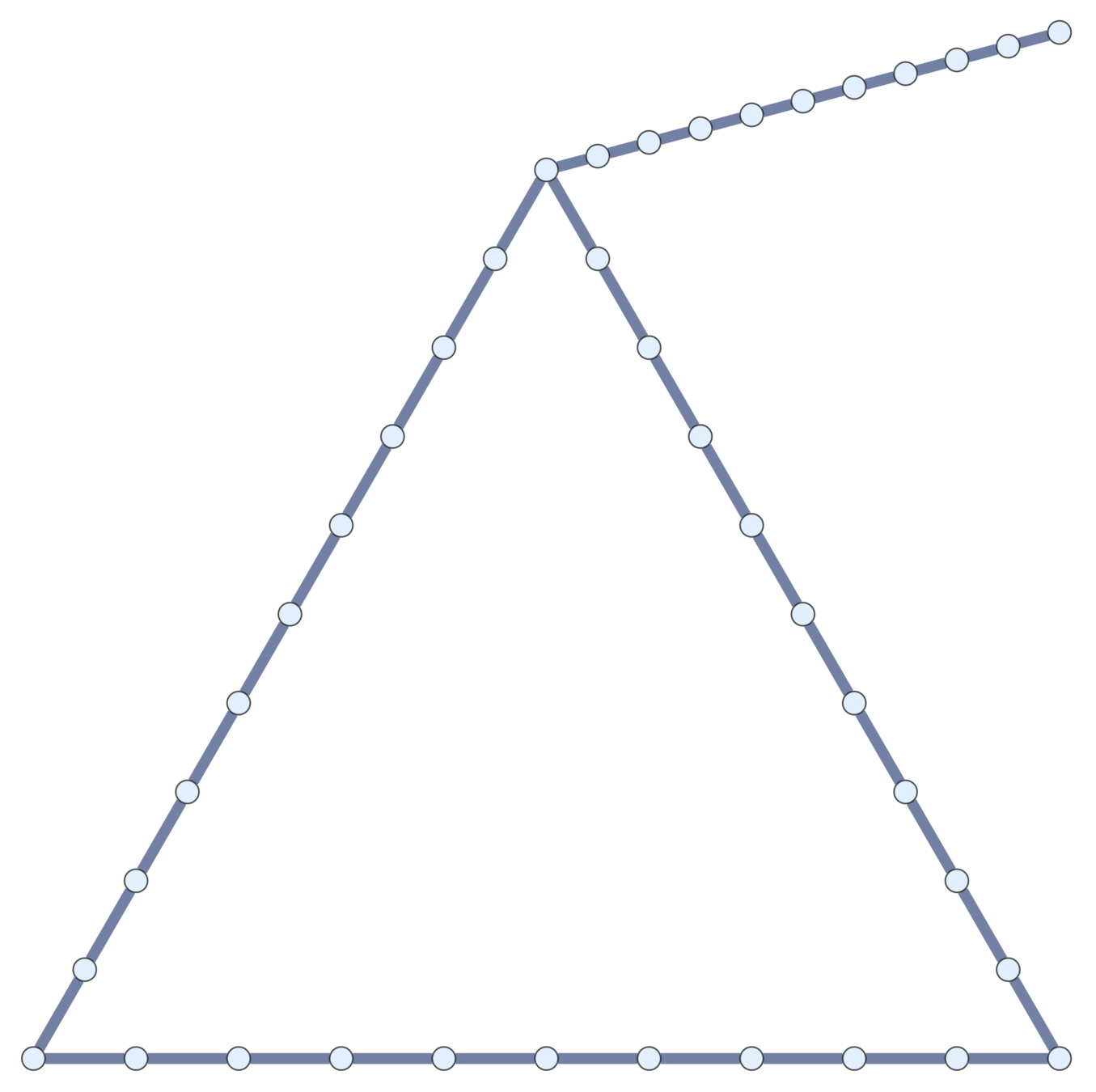
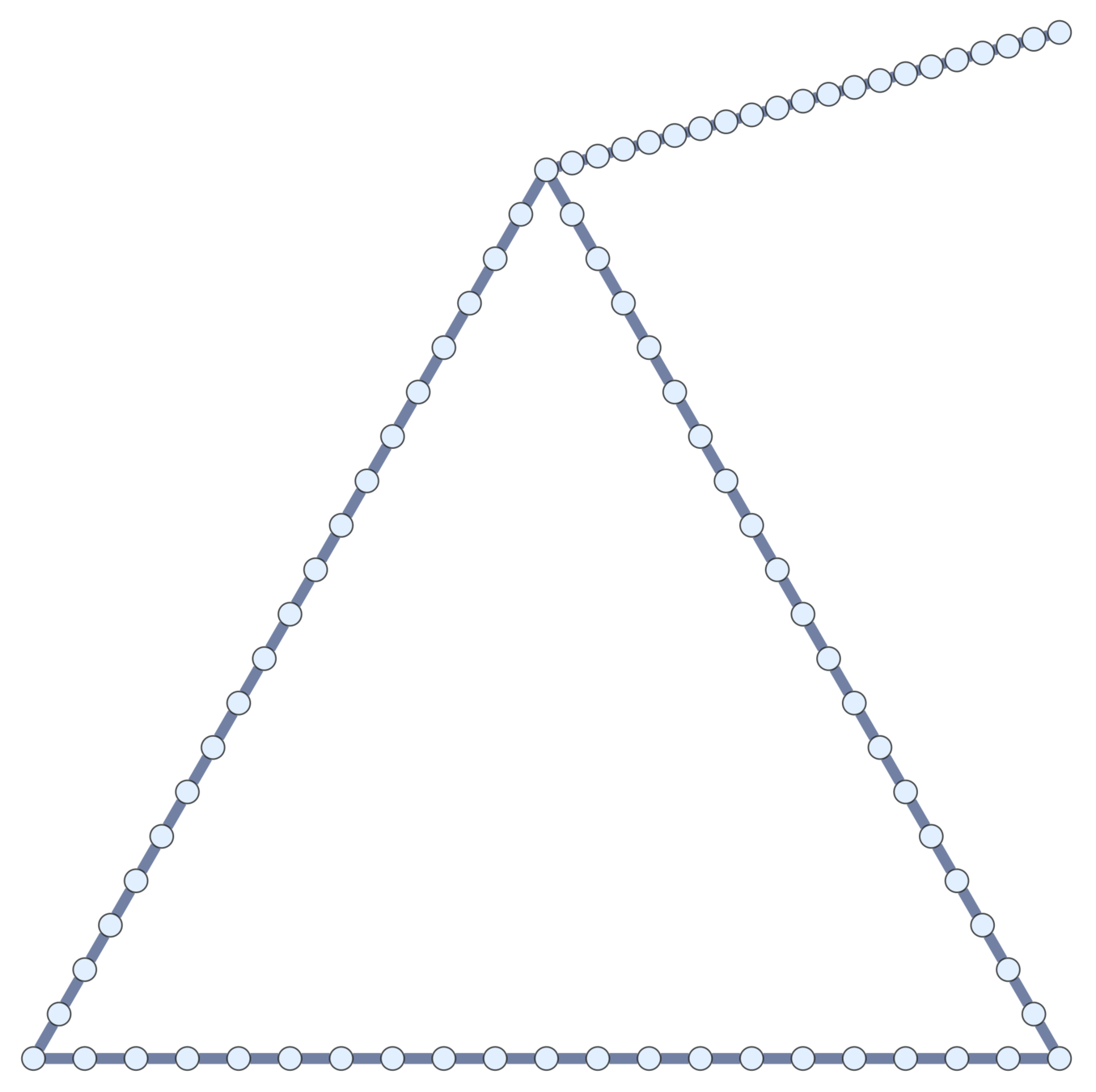
Notation. If \(\mathcal{G}\) is a (multi-)graph, \(\mathcal{G}_n\) is its \(n\)th subdivision.
Subdivision Theorem
Theorem (w/ Cantarella, Deguchi, & Uehara)
where the \(\lambda_k'\) are the nonzero eigenvalues of the normalized graph Laplacian \(\mathcal{L}\) of \(\mathcal{G}\).
\(T\) is the diagonal matrix of degrees of vertices of \(\mathcal{G}\).


Topological polymers
Size exclusion chromatograph
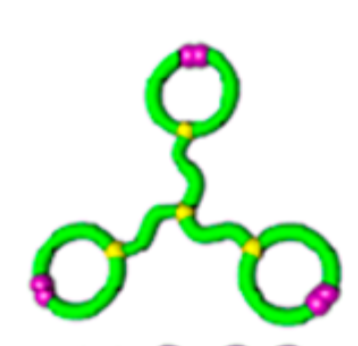
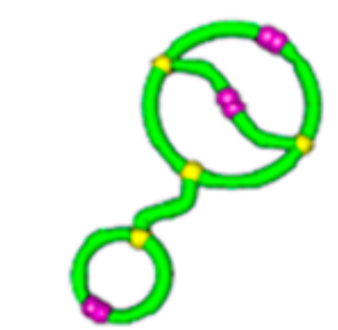
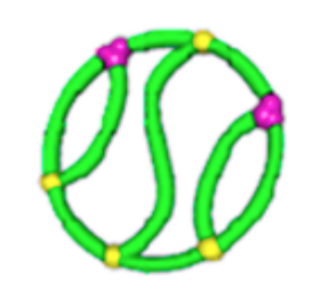
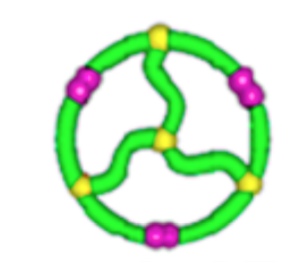
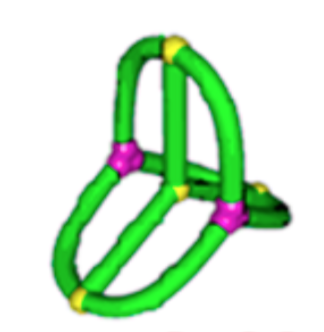
\(\lim_{n \to \infty}\frac{1}{\mathcal{V}(\mathcal{G}_n)} \mathbb{E}[R_g^2(\mathcal{G}_n)]\)
\(\frac{17}{162}\approx 0.105\)
\(\frac{107}{810}\approx 0.132\)
\(\frac{109}{810}\approx 0.135\)
\(\frac{31}{162}\approx 0.191\)
\(\frac{43}{162}\approx 0.265\)
\(\frac{49}{162}\approx 0.302\)
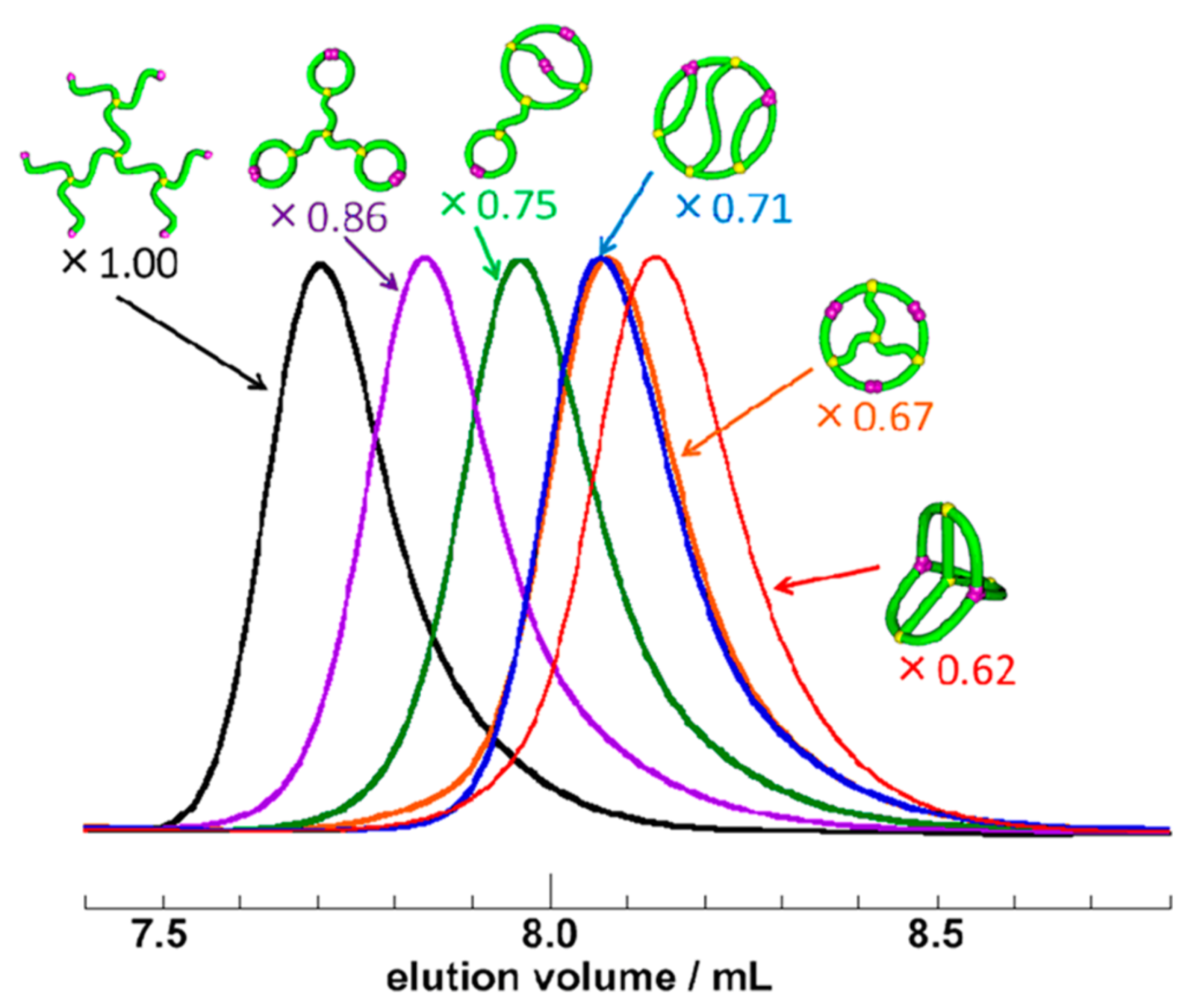
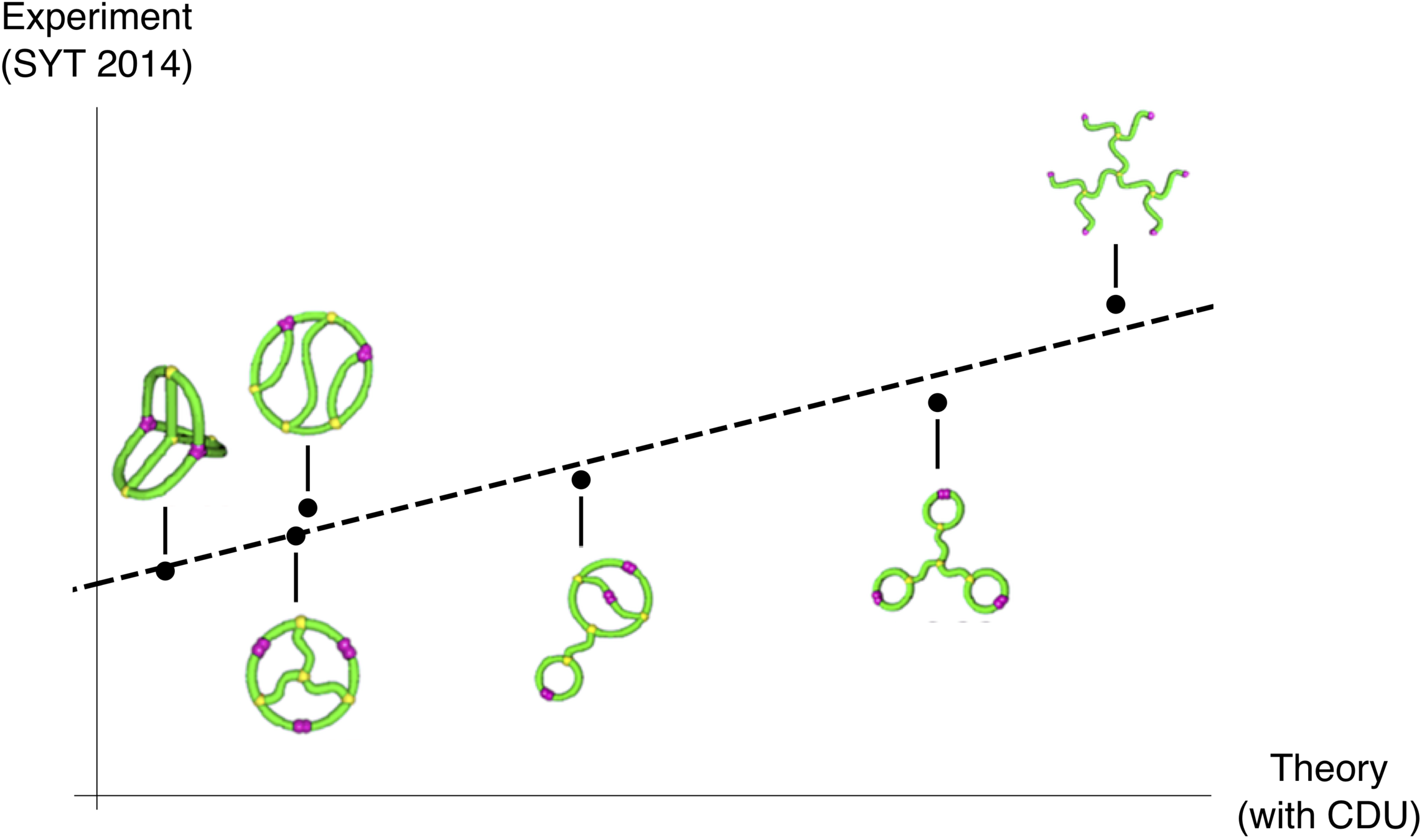
“an extremely compact 3D conformation, achieving exceptionally thermostable bioactivities”
Open questions
- Topological type of graph embedding?
- What if the graph is a random graph?