Research Objects
Stian Soiland-Reyes
eScience lab, University of Manchester
http://orcid.org/0000-0001-9842-9718
http://slides.com/soilandreyes/
#BioCompute2017 FDA workshop
NIH, Bethesda, 2017-03-17
This work is licensed under a
Creative Commons Attribution 4.0 International License.



What is in a Research Object?
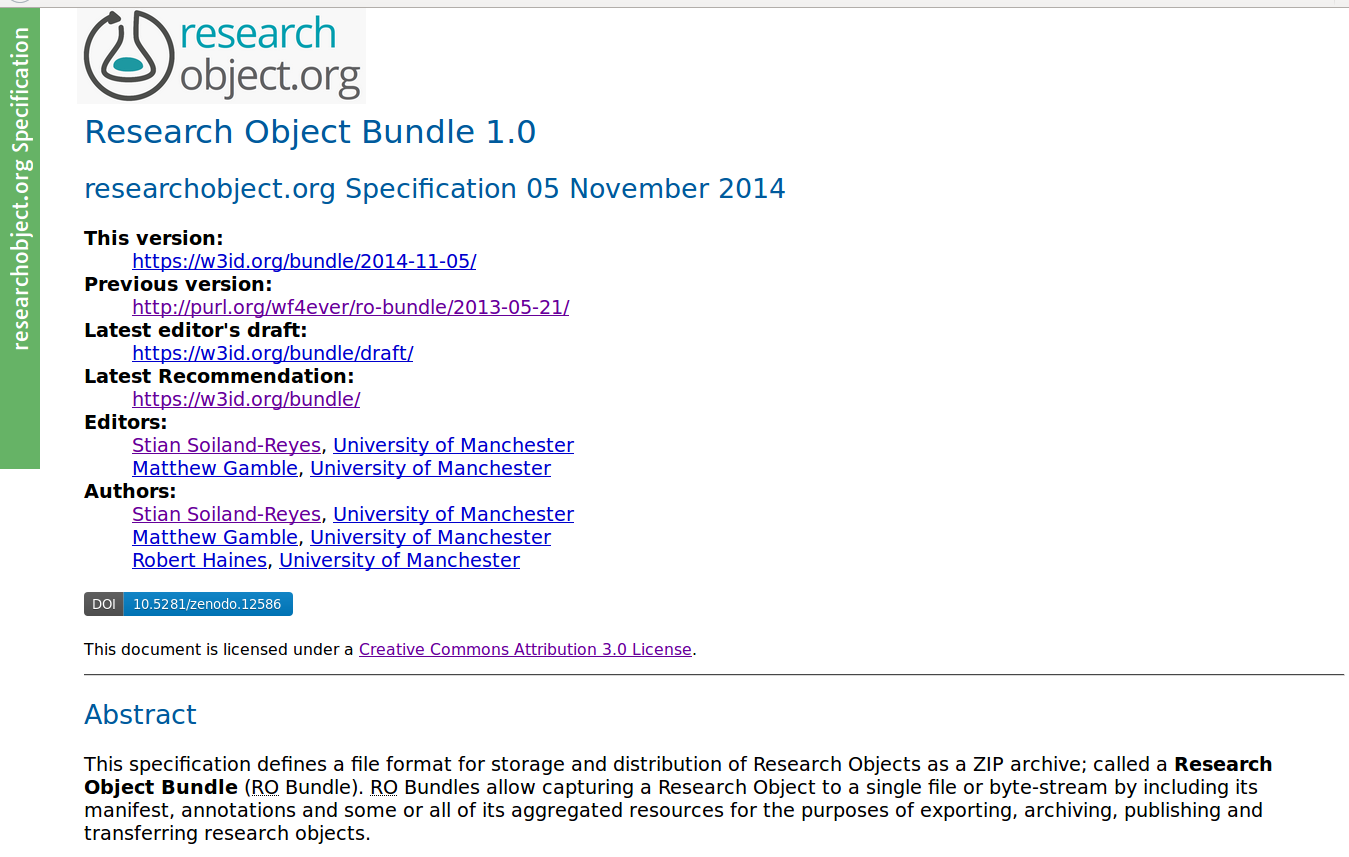
A Research Object bundles and relates digital resources of a scientific experiment or investigation:
Data used and results produced in experimental study
Methods employed to produce and analyse that data
Provenance and settings for the experiments
People involved in the investigation
Annotations about these resources, to improve understanding and interpretation



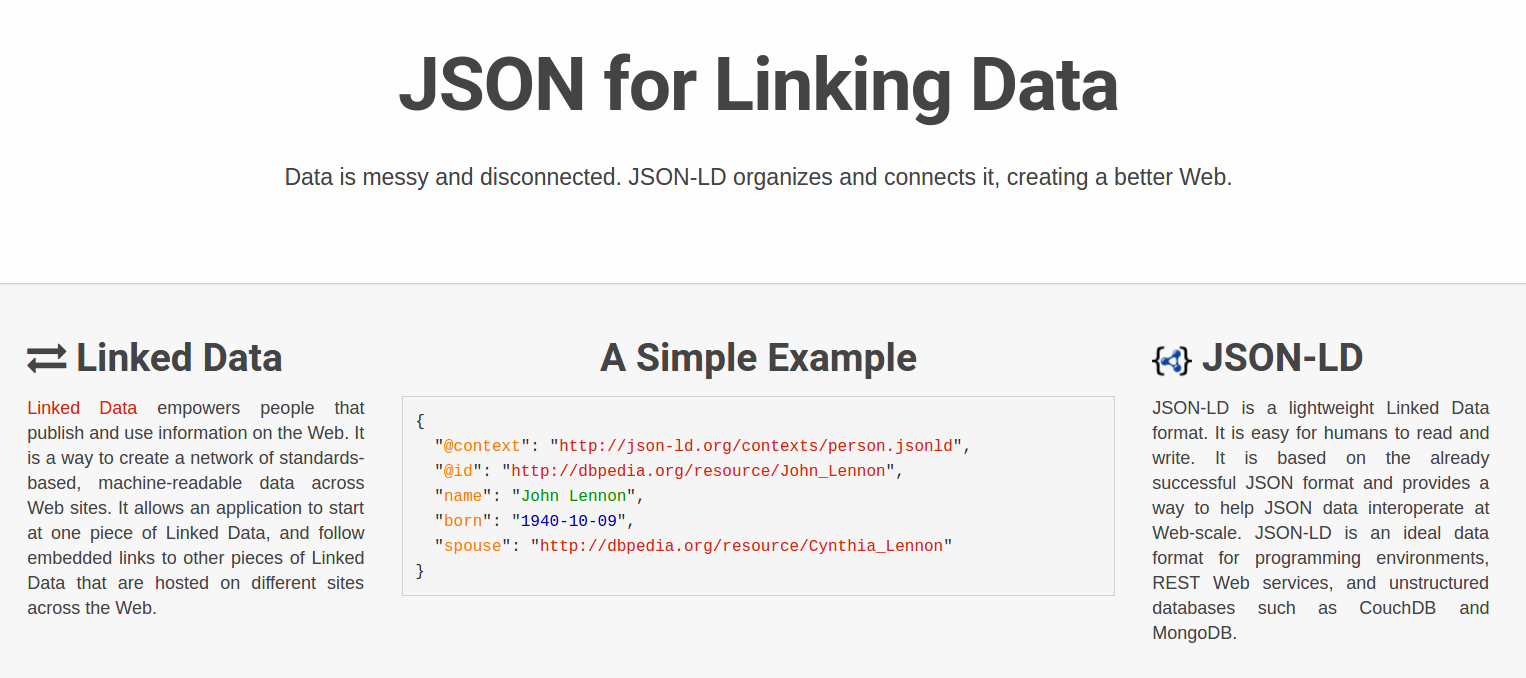
id: doi:10.15490/seek.1.investigation.56
createdOn: 2015-07-10T16:46:00Z
createdBy: http://orcid.org/0000-0001-9842-9718
aggregates:
- id: /sequence/specimen5.bam
conformsTo: http://gemrb.org/iesdp/file_formats/ie_formats/bam_v1.htm
- id: http://example.com/blog/about-specimen5
authoredBy: http://orcid.org/0000-0001-7066-3350
- id: http://www.myexperiment.org/workflows/3355
history: provenance/workflow-evolution.ttl
annotations:
- about: /sequence/specimen5.bam
content: annotations/specimen5-properties.jsonld
createdBy: http://orcid.org/0000-0001-7066-3350
- about: /sequence/specimen5.bam
content: http://example.com/blog/about-specimen5
motivatedBy: oa:questioningWho is using Research Objects?









| Versions | Provenance | Dependencies | Checklists |
|---|---|---|---|
|
|
|
|
|
|
|
|
||
|
|
|
|
PROV
PAV
wfprov
ProvONE
Community-based
Any Research Object
Project/lab-specific





















Research Object profiles






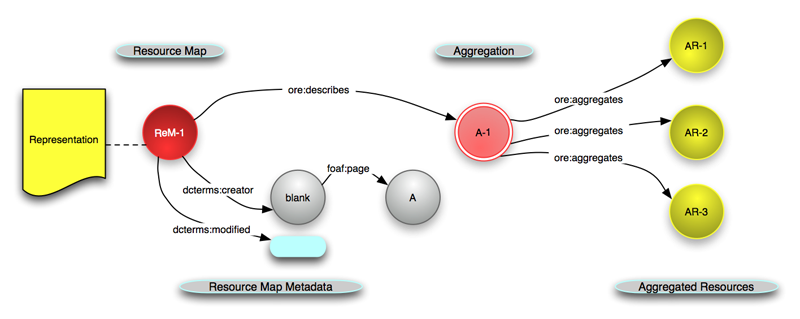
Open Archives Initiative Object Reuse and Exchange (OAI-ORE)
Copyright © 2017 W3C® (MIT, ERCIM, Keio, Beihang). W3C liability, trademark and document use rules apply.
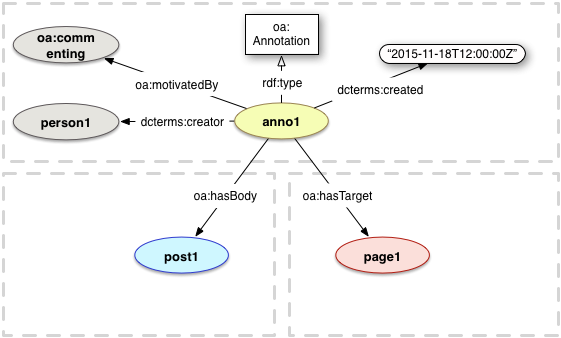

Copyright © 2013 W3C® (MIT, ERCIM, Keio, Beihang), All Rights Reserved. W3C liability, trademark and document use rules apply.
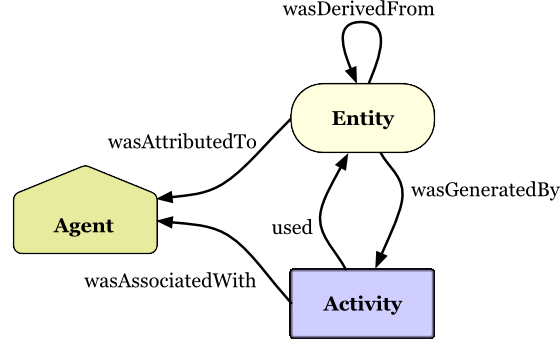
PROV Model Primer
W3C Working Group Note 30 April 2013










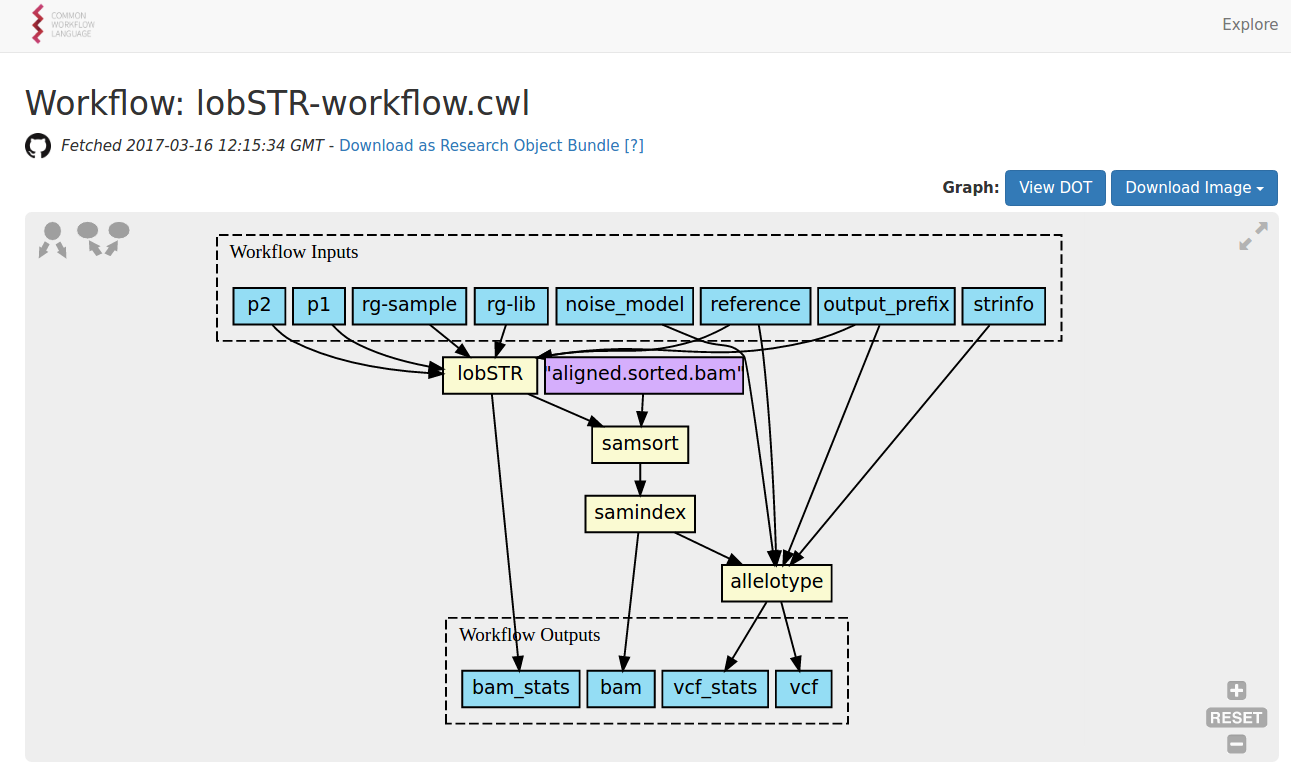
http://www.commonwl.org/
cwlVersion: v1.0
class: Workflow
inputs:
inp: File
ex: string
outputs:
classout:
type: File
outputSource: compile/classfile
steps:
untar:
run: tar-param.cwl
in:
tarfile: inp
extractfile: ex
out: [example_out]
compile:
run: arguments.cwl
in:
src: untar/example_out
out: [classfile]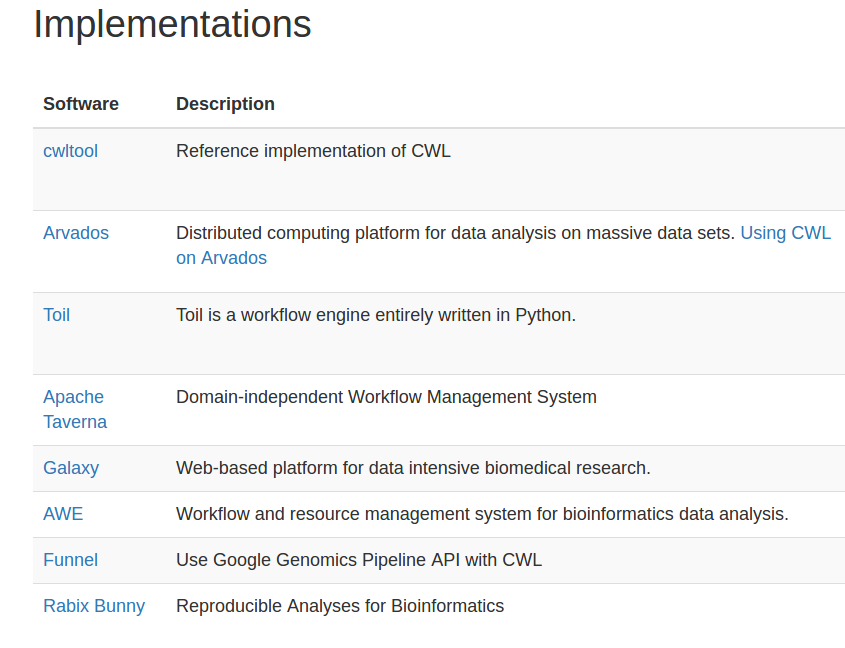
BCO + CWL + RO = 💕💕
BCO: Scientific overview
Domain-specific metadata
Parameters, taxonomies, keywords
TODO: not everything in single JSON,
point to other files for workflow/provenance
CWL: Analytical pipeline
Computational workflow and tool composition
#1: site-specific/template CWL ⇾ descriptive
#2: native CWL ⇾ executable, portable
RO: Packaging
Gather data, resources and tools ⇾ archive, transfer
Generic metadata
Deep attribution, linking, provenance
Questions?


This work has been done as part of the BioExcel CoE (www.bioexcel.eu), a project funded by the European Union contract H2020-EINFRA-2015-1-675728.