The Archive and Package (arcp) URI scheme
Stian Soiland-Reyes
eScience lab, The University of Manchester
Adapted from RDA talk on Research Objects
2018-03-22
This work is licensed under a
Creative Commons Attribution 4.0 International License.







A Research Object bundles and relates digital resources of a scientific experiment or investigation:
Data used and results produced in experimental study
Methods employed to produce and analyse that data
Provenance and settings for the experiments
People involved in the investigation
Annotations about these resources, to improve understanding and interpretation
Research Object
id: doi:10.15490/seek.1.investigation.56
createdOn: 2015-07-10T16:46:00Z
createdBy: http://orcid.org/0000-0001-9842-9718
aggregates:
- id: data/sequence/specimen5.bam
conformsTo: http://gemrb.org/iesdp/file_formats/ie_formats/bam_v1.htm
- id: http://example.com/blog/about-specimen5
authoredBy: http://orcid.org/0000-0001-7066-3350
- id: http://www.myexperiment.org/workflows/3355
history: provenance/workflow-evolution.ttl
annotations:
- about: data/sequence/specimen5.bam
content: annotations/specimen5-properties.jsonld
createdBy: http://orcid.org/0000-0001-7066-3350
- about: data/sequence/specimen5.bam
content: http://example.com/blog/about-specimen5
motivatedBy: oa:questioningResearch Object manifest
(simplified)
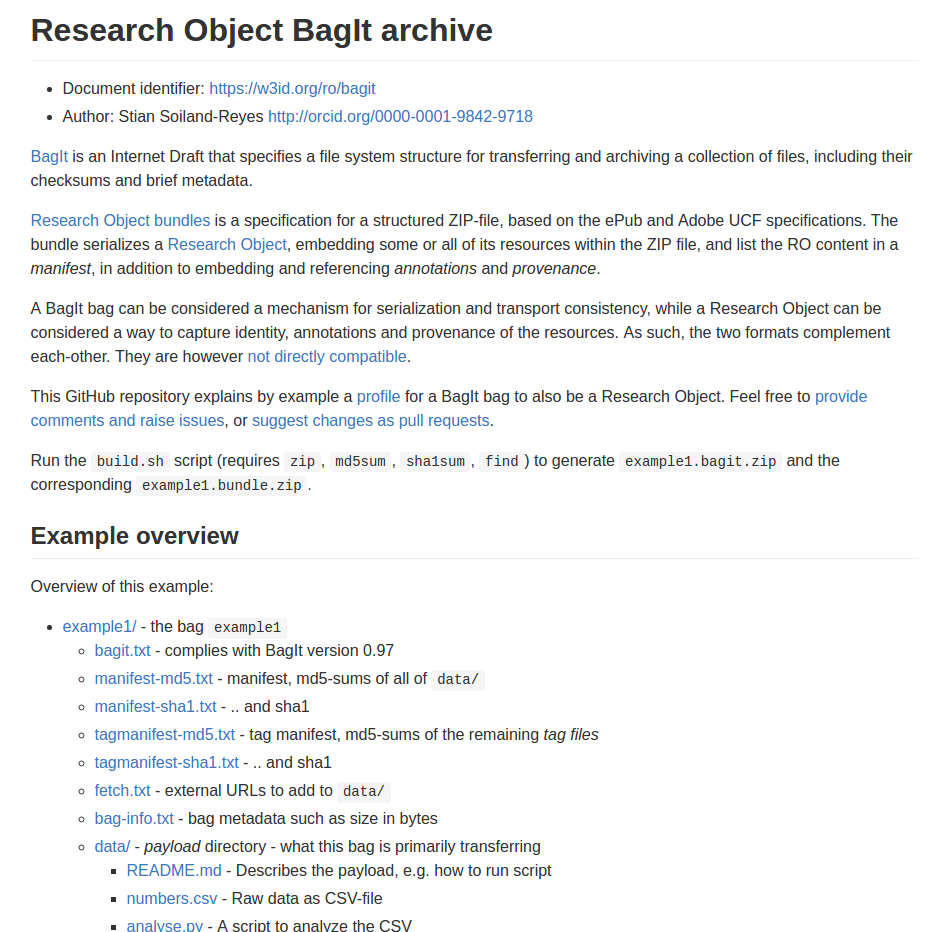
Reuse standards:
OAI-ORE, BagIt, W3C JSON-LD, PROV, Web Annotation Model

metadata/manifest.json
data/sequence/specimen5.bam
provenance/workflow-evolution.ttl
http://example.com/blog/about-specimen5
http://www.myexperiment.org/workflows/335
http://orcid.org/0000-0001-7066-3350
http://gemrb.org/iesdb/
file_formats_ie_formats_bam_v1.html



activity(run:2e1287e0-6dfb-11e7-8acf-0242ac110002, , ,
[prov:type='wfprov:WorkflowRun', prov:label="Run of workflow/packed.cwl#main"])
// main workflow run started outside somehow (we're don't know how)
wasStartedBy(run:4305467e-6dfb-11e7-885d-0242ac110002, -, -,
-, 2017-10-27T15:00:00Z)
// ...
// step is a nested workflow, so also a WorkflowRun
activity(run:4305467e-6dfb-11e7-885d-0242ac110002, -, -,
[prov:type='wfprov:WorkflowRun', prov:label="Run of workflow/packed.cwl#main/nested1"])
// started by the mother activity
wasStartedBy(run:4305467e-6dfb-11e7-885d-0242ac110002, -, -,
run:2e1287e0-6dfb-11e7-8acf-0242ac110002, 2017-10-27T15:00:30Z)
// inner step of nested workflow, ProcessRun as this is a command line execution
activity(run:c42dc36e-6dfd-11e7-bc24-0242ac110002, -, -
[prov:type='wfprov:ProcessRun', prov:label="Run of workflow/packed.cwl#nested/innerStep1"])
wasStartedBy(run:c42dc36e-6dfd-11e7-bc24-0242ac110002, -, -,
run:4305467e-6dfb-11e7-885d-0242ac110002, 2017-10-27T15:01:00Z)
// ...Identifying intermediate data
Output 1B file is also Input 2C and Input 3D downstream
Simple filenames -> duplications
./data/step1/outputB.txt
./data/step2/inputC.txt
./data/step3/inputD.txt
Content-adressable
SHA-256 hash of bytes as filename:
./data/51/51fb8af0c4ae0422fbe88340d91880ecb9d7537cf57339c1cf1256b7ca58f32d
RFC6920 URI as global identifier:
nih:sha-256;51fb8af0c4ae0422fbe88340d91880ecb9d7537cf57339c1cf1256b7ca58f32d
Randomly generated
arcp://uuid,32a423d6-52ab-47e3-a9cd-54f418a48571/css/base.css>>> uuid.uuid4()
UUID('32a423d6-52ab-47e3-a9cd-54f418a48571')UUID v4
External-Identifier: urn:uuid:32a423d6-52ab-47e3-a9cd-54f418a48571Self-declared UUID in bagit-info.txt
{ "id" "urn:uuid:32a423d6-52ab-47e3-a9cd-54f418a48571",
"name": "HCV1a [taxonomy:31646 ledipasvir"
... }from bco.json ?
Hashed from archive download URL
arcp://uuid,b7749d0b-0e47-5fc4-999d-f154abe68065/pics/flower.jpeg>>> uuid.uuid5(uuid.NAMESPACE_URL, "http://example.com/data.zip")
UUID('b7749d0b-0e47-5fc4-999d-f154abe68065')UUID v5
Location-independent archive identifier (BDBag)
>>> uuid.uuid5(uuid.NAMESPACE_URL, "http://identifiers.org/ark/ark:/57799/b91w9r")
UUID('4f11f216-e2dc-57cd-a714-300409a430ce')

stain@biggie:~$ sha256sum archive.zip
7f83b1657ff1fc53b92dc18148a1d65dfc2d4b1fa3d677284addd200126d9069RFC6920 (Naming Thing with Hashes) URI
>>> urlsafe_b64encode("7f83b1657ff1fc53b92dc18148a1d65dfc2d4b1fa3d677284addd200126d9069"
.decode("hex"))
'f4OxZX_x_FO5LcGBSKHWXfwtSx-j1ncoSt3SABJtkGk='
arcp://ni,sha-256;f4OxZX_x_FO5LcGBSKHWXfwtSx-j1ncoSt3SABJtkGk/src/luhn.cHash checksum of archive
ni:///sha-256;f4OxZX_x_FO5LcGBSKHWXfwtSx-j1ncoSt3SABJtkGk/wget http://repo.example.com/.well-known/
ni/sha-256/f4OxZX_x_FO5LcGBSKHWXfwtSx-j1ncoSt3SABJtkGk/Retrievable/verifiable
arcp URI Python library
>>> from arcp import *
>>> arcp_random()
'arcp://uuid,dcd6b1e8-b3a2-43c9-930b-0119cf0dc538/'
>>> arcp_random("/foaf.ttl", fragment="me")
'arcp://uuid,dcd6b1e8-b3a2-43c9-930b-0119cf0dc538/foaf.ttl#me'
>>> arcp_hash(b"Hello World!", "/folder/")
'arcp://ni,sha-256;f4OxZX_x_FO5LcGBSKHWXfwtSx-j1ncoSt3SABJtkGk/folder/'
>>> arcp_location("http://example.com/data.zip", "/file.txt")
'arcp://uuid,b7749d0b-0e47-5fc4-999d-f154abe68065/file.txt'
pip install arcp
Parsing arcp URIs
>>> is_arcp_uri("arcp://uuid,b7749d0b-0e47-5fc4-999d-f154abe68065/file.txt")
True
>>> u = parse_arcp("arcp://uuid,b7749d0b-0e47-5fc4-999d-f154abe68065/file.txt")
ARCPSplitResult(scheme='arcp',prefix='uuid',
name='b7749d0b-0e47-5fc4-999d-f154abe68065',
uuid='b7749d0b-0e47-5fc4-999d-f154abe68065',
path='/file.txt',query='',fragment='')
>>> u.path
'/file.txt'
>>> u.prefix
'uuid'
>>> u.uuid
UUID('b7749d0b-0e47-5fc4-999d-f154abe68065')
>>> u.uuid.version
5
>>> parse_arcp("arcp://ni,sha-256;f4OxZX_x_FO5LcGBSKHWXfwtSx-j1ncoSt3SABJtkGk/folder/")
.hash
('sha-256', '7f83b1657ff1fc53b92dc18148a1d65dfc2d4b1fa3d677284addd200126d9069')