Agenda
Introduction
Adventages
Pipeline
Features
Results
Problems
Conculsions



Tumor CLASSIFICATION


MRI
WSI
Tumor CLASSIFICATION

WSI Tumor CLASSIFICATION
Adventages
Cellular level
A lot of information
Expert feedback
Great potential
Universal mechanisms
Adventages
Cellular level
A lot of information
Expert feedback
Great potential
Universal mechanisms
Cellular level resolution provides necessary information about the case. It is possible to precisely classify cells from overpopulated regions and predict type and stage of tumor

Adventages
Cellular level
A lot of information
Expert feedback
Great potential
Universal mechanisms
Every cell with it's surroundings contains a lot of relevant information. It can be used to simply classify tumor or healthy tissue. It can be also used to find unusual, atypical behavior to better understand the nature of the case

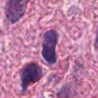


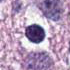



Adventages
Cellular level
A lot of information
Expert feedback
Great potential
Universal mechanisms
It is crucial to get professional feedback. Histopathologists are very solid source of fruitful feedback

Adventages
Cellular level
A lot of information
Expert feedback
Great potential
Universal mechanisms
The development of automatic WSI analysis can significantly improve efficiency and pace at which pathologists work. Moreover it seems possible to provide diagnosis without involvement of doctors

Adventages
Cellular level
A lot of information
Expert feedback
Great potential
Universal mechanisms
Vast majority of functions used for segmentation and feature extraction can be easily generalized to different cases and different tissue


Feature details
| Area |
| Circumference |
| Connectivity |
| Hu moments |
| Ellipse atributes |
| Density of nuclei |
| Fuzzy coefficient |
| Core of nucleous |
| Interior structure |
| - RGB |
| - Gray scale |
| - Hematoxylin and eosin |
Halo effect
Shape
Color

| Divide in Frames |
| ▼ |
| Frame segmentation |
| ▼ |
| Extract frames of interest |
| ▼ |
| Classify nuclei in a frame |
| ▼ |
| Classify each frame |
| ▼ |
| Classify the sample |

General procedure
| Divide in Frames |
| ▼ |
| Frame segmentation |
| ▼ |
| Extract frames of interest |
| ▼ |
| Classify nuclei in a frame |
| ▼ |
| Classify each frame |
| ▼ |
| Classify the sample |
General procedure

| Divide in Frames |
| ▼ |
| Frame segmentation |
| ▼ |
| Extract frames of interest |
| ▼ |
| Classify nuclei in a frame |
| ▼ |
| Classify each frame |
| ▼ |
| Classify the sample |

General procedure
| Divide in Frames |
| ▼ |
| Frame segmentation |
| ▼ |
| Extract frames of interest |
| ▼ |
| Classify nuclei in a frame |
| ▼ |
| Classify each frame |
| ▼ |
| Classify the sample |



General procedure



n = 774
n = 332
n = 815
n = 68
n = 774
n = 815
| Divide in Frames |
| ▼ |
| Frame segmentation |
| ▼ |
| Extract frames of interest |
| ▼ |
| Classify nuclei in a frame |
| ▼ |
| Classify each frame |
| ▼ |
| Classify the sample |


Degree of membership
Astrocyte
Oligodendrocyte
General procedure
| Divide in Frames |
| ▼ |
| Frame segmentation |
| ▼ |
| Extract frames of interest |
| ▼ |
| Classify nuclei in a frame |
| ▼ |
| Classify each frame |
| ▼ |
| Classify the sample |


| Oligo - 63% |
| Astro - 37% |
| Oligo - 24% |
| Astro - 76% |
General procedure
| Divide in Frames |
| ▼ |
| Frame segmentation |
| ▼ |
| Extract frames of interest |
| ▼ |
| Classify nuclei in a frame |
| ▼ |
| Classify each frame |
| ▼ |
| Classify the sample |


| Number of Oligodendroglioma frames > T |
|---|
| ▼ |
| Oligodendroglioma |
General procedure
Classify nuclei in a frame
Supported vector classification
Logistic regression
Random forests
Gaussian Naive Bayes
Supported vector machine
Explored classification algorithms
Explored classification algorithms
Supported vector classification
Logistic regression
Random forests
Gaussian Naive Bayes
Supported vector machine
Methodology
Model metaparameters were fitted with the use of genetic algorithm.
Cost function was calculated via score calculated on cases.
Classify nuclei in a frame
Classify each frame and classify the sample
| Degree of membership | |
| ▼ | |
| Frame summary | x 7 |
| ▼ | |
| Classification of the frame | x 7 |
| ▼ | |
| Frame set summary | x 7 |
| ▼ | |
| Thresholding | |
Results and voting method
| Crossvalidation |
| ▼ |
| Selecting best models |
| ▼ |
| Voting |
Results and voting method
32 cases total:
- 12 to teach nuclei classifier
- 10 to teach frame classifier
- 10 to test
| Crossvalidation |
| ▼ |
| Selecting best models |
| ▼ |
| Voting |
Results and voting method
Achieved about 73% average accuracy
| Crossvalidation |
| ▼ |
| Selecting best models |
| ▼ |
| Voting |
Results and voting method
| Crossvalidation |
| ▼ |
| Selecting best models |
| ▼ |
| Voting |
Final decision is based on the simple majority voting over the classifier output population
Problems
- Time
- Multitasking
- Segmentation accuracy
- Different data types
- Data mining problems

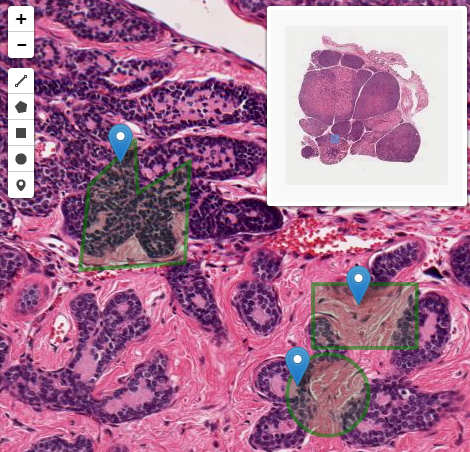



Conclusions
Can we help people?
Conclusions
Can we help people?
I hope you agree, that we can
Conclusions
We plan to:
- speed up computing (GPU) ,
- use more efficient segmentation methods,
- improve feature extraction methods,
- implement other classification algorithms,
but also:
- add MRI analysis as a significant part of the entire process,
- add medical history as a significant part of the process,
- expand our algorithm to deal with other brain tumor types




Our Team
Michał Błach
R&D Stermedia
Wrocław University of Technology
Witold Dyrka
R&D Stermedia
Wrocław University of Technology
Jakub Czakon
R&D Stermedia
Piotr Giedziun
R&D Stermedia


Piotr Krajewski
CIO Stermedia

Grzegorz Żurek
R&D Stermedia
Wrocław University of Technology



Łukasz Fuławka
Pathomorphology resident Lower Silesian Oncology Center
We are looking forward to collaborating with you

Contact us
info@stermedia.pl
jakub.czakon@stermedia.pl
grzegorz.zurek@stermedia.pl
Thank you for attention