
Computational Biology
Lecture 01:
Introduction to computational biology

(BIOSC 1540)
Aug 27, 2024

After today, you should be able to
1. Describe the course structure, expectations, and available resources for success.
2. Define computational biology and explain its interdisciplinary nature.
3. Identify key applications and recent advancements.
4. Understand the balance between applications and development.
5. Identify potential career paths and educational opportunities.
Meet your teaching team

Alex Maldonado, PhD
he/him/his
Acceptable ways to address me:
Alex (preferred)
Dr. Maldonado
Dr. Alex
Dr. M
Email: alex.maldonado@pitt.edu

Ph.D. in Chemical Engineering, 2023
University of Pittsburgh
B.S.E in Chemical Engineering, 2018
Western Michigan University
Justine Denby (she/her/hers)
Major: Computational biology
Reya Kundu (she/her/hers)
Major: Computational biology

Alex's fun facts


Every male in my (maternal) family played football
—I rebelled
Alex's fun facts
Part-time jobs
- Construction
- UPS package handler
- Kent County Traffic safety
- Jimmy John's delivery driver
- Wings West ice events



Get to know my ...
Single source of truth
All course materials will be posted on this website: pitt-biosc1540-2024f.oasci.org/
Why?
I am extra
There are few comprehensive resources for this rapidly changing field
Things that contain student information will be only on Canvas to be FERPA compliant
Assignments will be submitted on Gradescope
My course philosophy
Critical thinking is paramount and happens outside your comfort zone
How does this influence my teaching?
This is where my focus is (similar to computer science and engineering)
Few points
Many points
There are two types of programming languages
.gif)

Compiled
Interpreted
E.g., Mojo, Rust, Zig, Go, C, C++
E.g., Python and R
There are some exceptions: Java, Julia
*
Python is absolutely necessary for a career in computational biology
Programming is how you obtain, manage, and analyze data


Data
Results and insights
No coding will be necessary to successfully complete this course
Previous semesters used R or Python
We will emphasize learning the foundational principles instead of coding

There will be optional coding opportunities
Semester overview
2. Transcriptomics
1. Genomics
4. Computer-aided drug design
3. Molecular simulations
Bioinformatics
Modules
Computational Structural Biology
Special interests/ Python?
Subfields we are not able to cover in detail
Where do we get our insight from?


Scientific python
or
Assessments

We will have ...
- Eight homework assignments
- One hands-on project
- Two exams
An optional cumulative final will be provided to replace lowest exam grade
Attendance is not mandatory, but encouraged
Project: Computer-Aided Drug Design for a Novel Pathogen
DNA sequencing of Staphylococcus aureus
Assembled and annotated genome
Protein structure prediction
Protein-ligand docking



We will work through a complete, web-based workflow mirroring the steps researchers might take when confronted with a new pathogenic threat
Other policies
Please read the rest of the syllabus on your own
I can also answer any questions now

After today, you should be able to
1. Describe the course structure, expectations, and available resources for success.
2. Define computational biology and explain its interdisciplinary nature.
3. Identify key applications and recent advancements.
4. Understand the balance between applications and development.
5. Identify potential career paths and educational opportunities.
What is computational biology?

What is computational biology?
Any application of computational methods to obtain insight into biological phenomena.
My definition . . .
My main categories . . .
Bioinformatics
Computational structural biology
Bioinformatics deals with untangling big data for biological insight

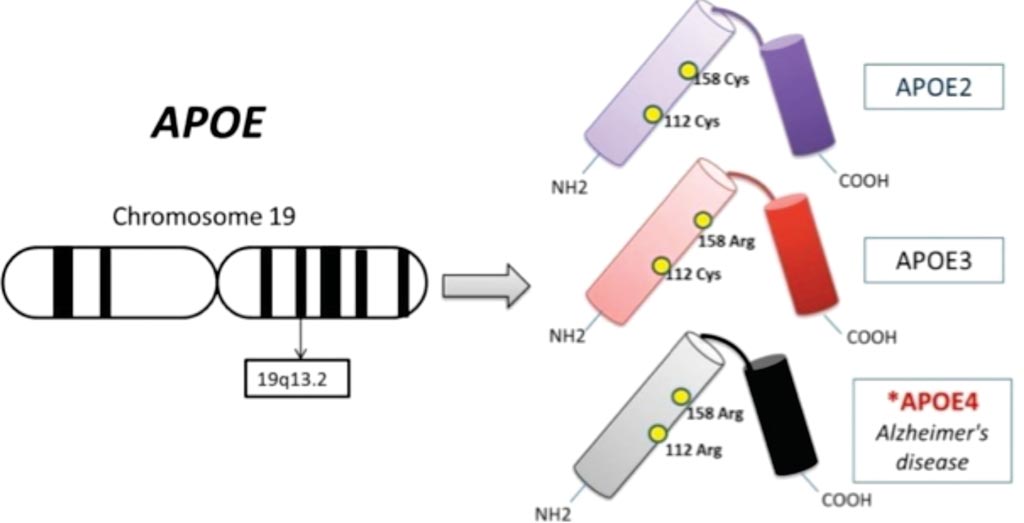
Genetic sequences of healthy and Alzheimer patients
Find genetic risk factors
Data
Information
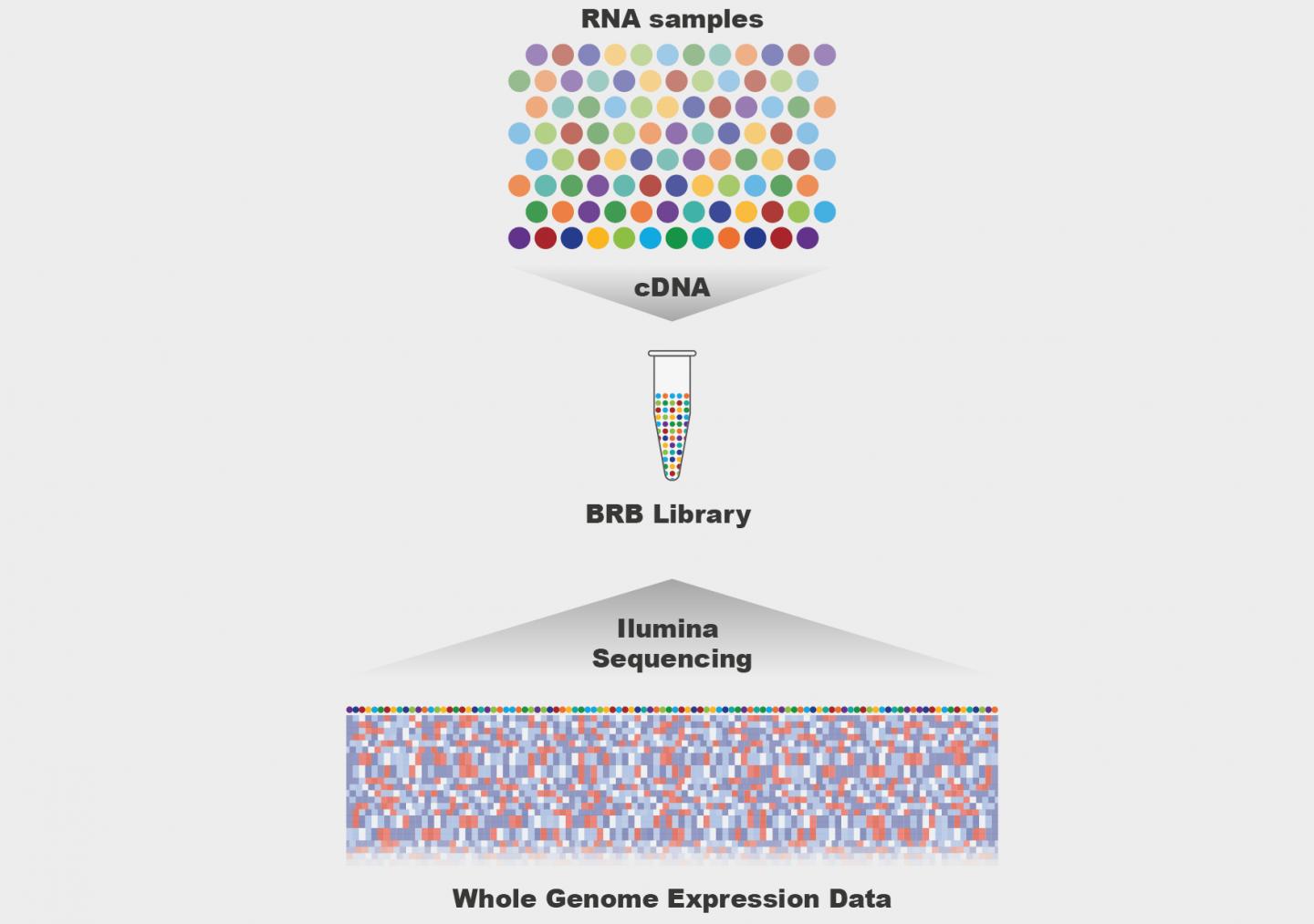
Bioinformatics deals with untangling big data for biological insight
Data
Information
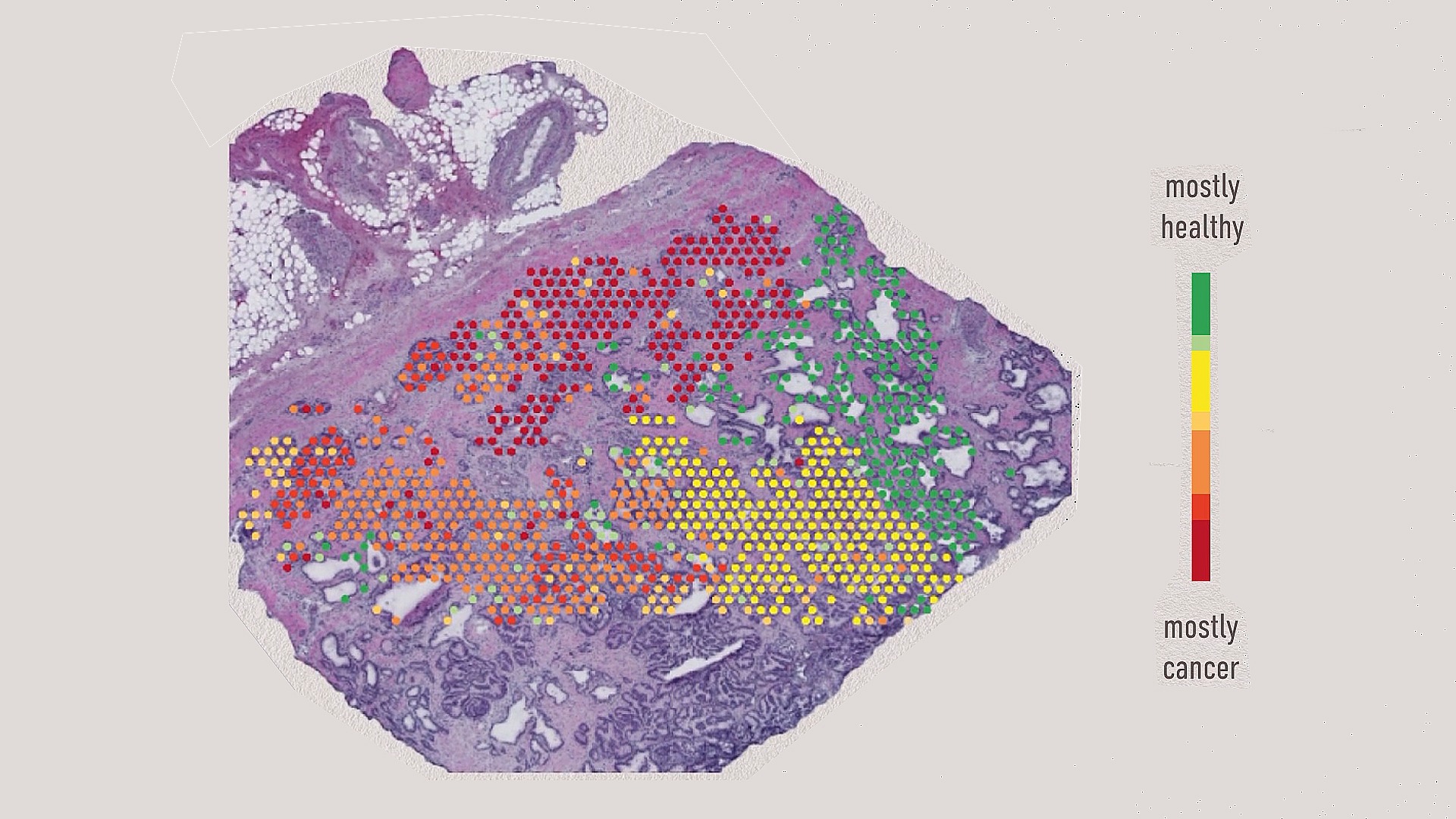
mRNA of cancer cells in a tumor
Early detection of benign to cancerous cell transition

Phenomena
Representation
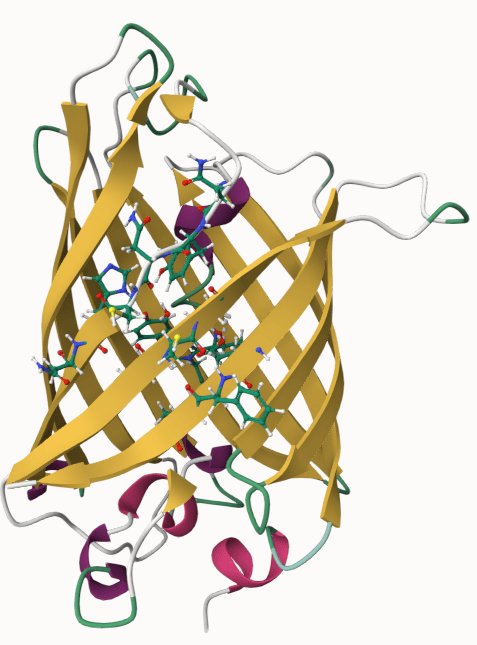
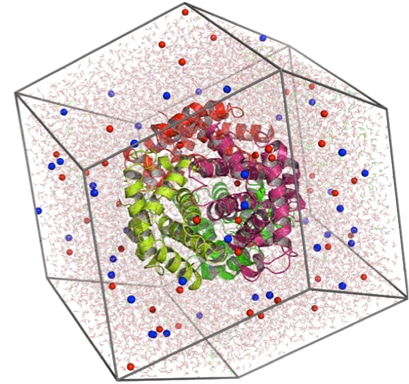
Modeling employs representations that mimic key biological phenomena
Protein-protein binding
Classical force fields
After today, you should be able to
1. Describe the course structure, expectations, and available resources for success.
2. Define computational biology and explain its interdisciplinary nature.
3. Identify key applications and recent advancements.
4. Understand the balance between applications and development.
5. Identify potential career paths and educational opportunities.
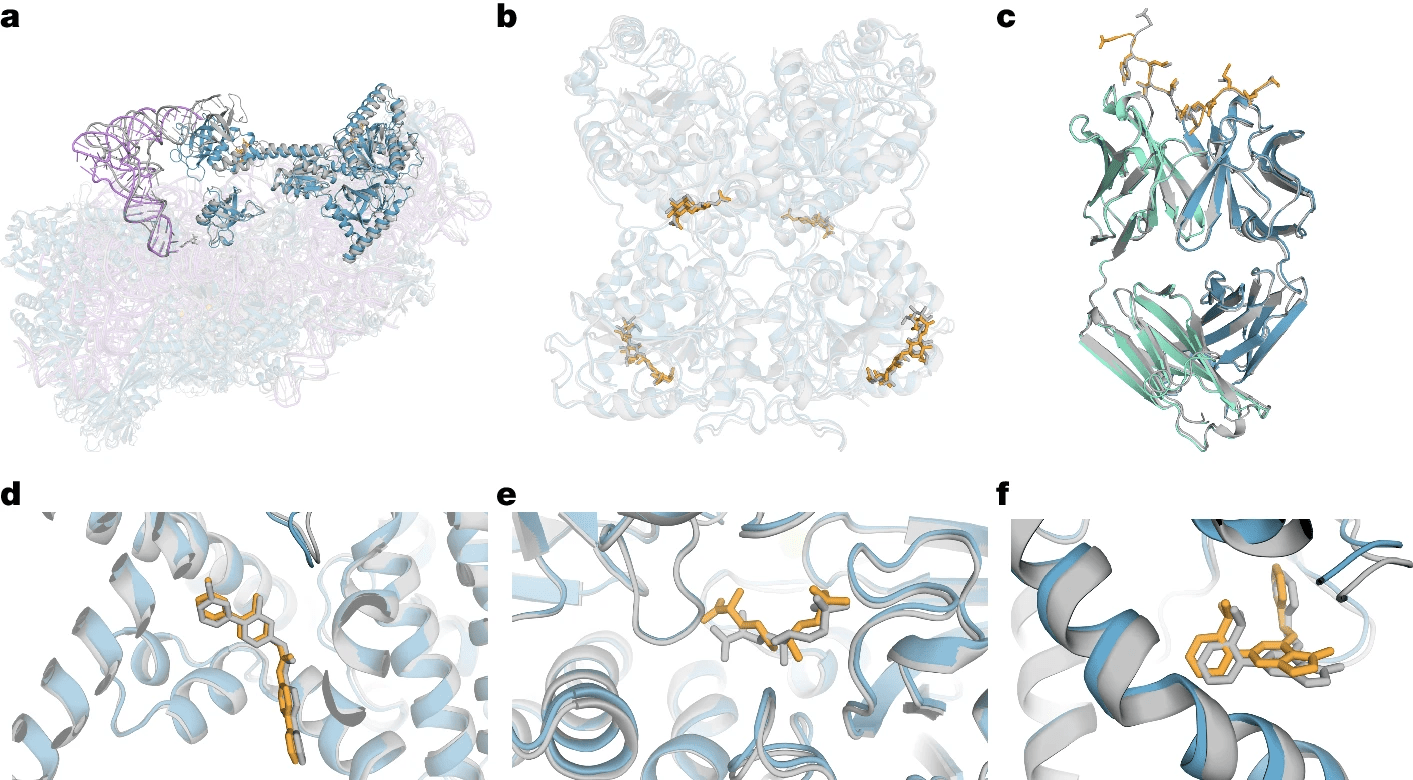
AlphaFold 3
"AlphaFold 3 can predict the joint structure of complexes including proteins, nucleic acids, small molecules, ions, and modified residues."
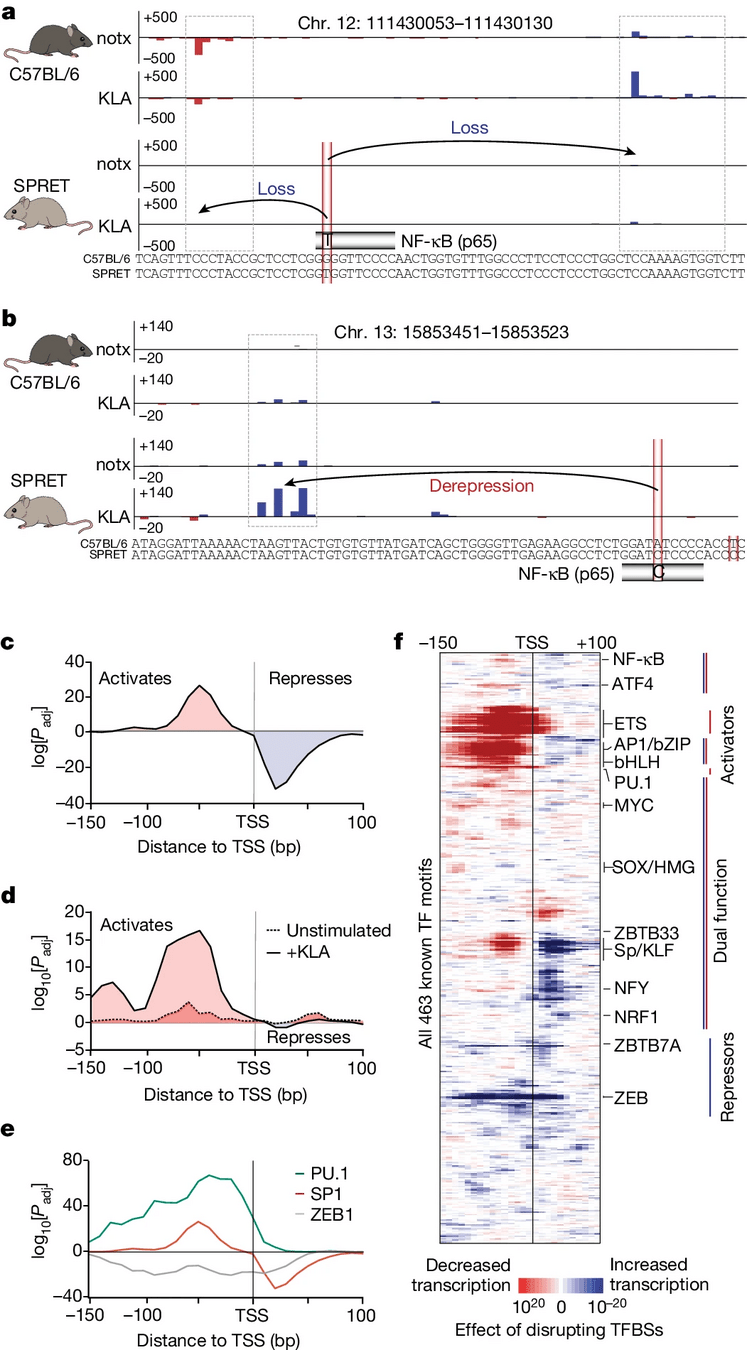
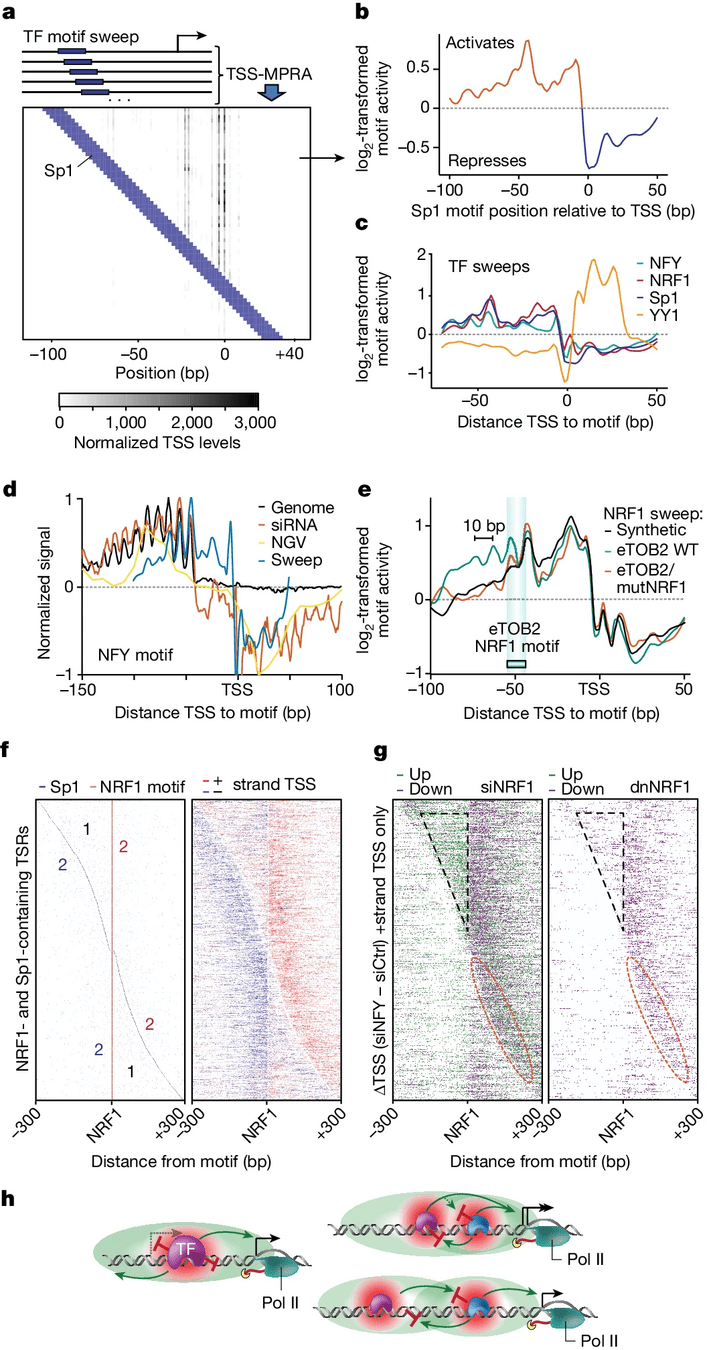
HOMER2
"We show that the effect of transcription factor binding on transcription initiation is position dependent."
Miniprot: protein-genome aligner
"Miniprot [...] is tens of times faster than existing tools while achieving comparable accuracy on real data."
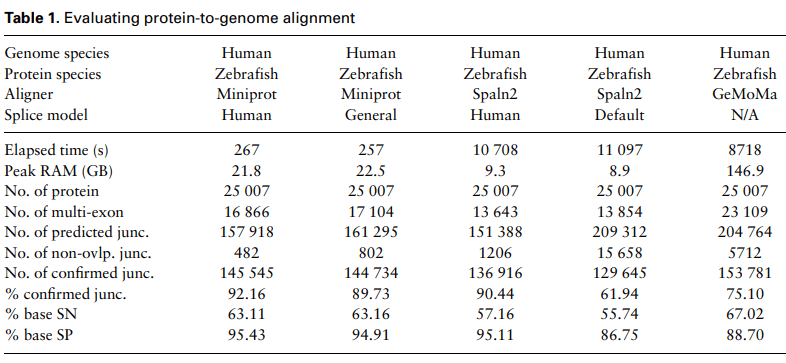
Why would we use protein-genome instead of genome-genome mapping?

TopHat: 173423
A. Protein-genome mapping is more sensitive for detecting distant homologs
B. Genome-genome mapping is too slow for large-scale comparisons
C. Protein-genome mapping allows for the detection of RNA editing events
D. Genome-genome mapping cannot handle intron-exon structures
(Not for points)
After today, you should be able to
1. Describe the course structure, expectations, and available resources for success.
2. Define computational biology and explain its interdisciplinary nature.
3. Identify key applications and recent advancements.
4. Understand the balance between applications and development.
5. Identify potential career paths and educational opportunities.
Computational Biology is broad
Data science
Computer science
Biology
Physics/
Engineering
Chemistry/
Biochemistry
Mathematics
You can tailor your career to these interests
We will touch on all of these topics in this course
Method development or applying tools?
Computer science
Biology
Developing
Applying
Typically, it is harder to pick up after the fact
(a different way of thinking)
Many, many, many specalities
Both separately are pretty saturated
After today, you should be able to
1. Describe the course structure, expectations, and available resources for success.
2. Define computational biology and explain its interdisciplinary nature.
3. Identify key applications and recent advancements.
4. Understand the balance between applications and development.
5. Identify potential career paths and educational opportunities.
Bioinformatics Scientist
Description: Develops software tools and approaches for analyzing biological data, particularly genomic and proteomic data.
Expected Salary: $80,000 - $130,000
Qualifications:
- PhD in Bioinformatics, Computational Biology, or related field
- Strong programming skills (Python, R, C++)
Example companies: UPMC, Illumina, 23andMe, Genentech, Regeneron Pharmaceuticals, Broad Institute
Computational Biologist
Description: Applies computational methods to study biological systems, often focusing on modeling complex biological processes.
Expected Salary: $75,000 - $135,000
Qualifications:
- PhD in Computational Biology, Systems Biology
- Expertise in mathematical modeling and simulation
- Strong programming and data analysis skills
Example companies: Moderna, Vertex Pharmaceuticals, Biogen, Allen Institute for Brain Science, Flatiron Health
Biostatistician
Description: Applies statistical methods to analyze biological and health-related data, often in clinical trials or epidemiological studies.
Expected Salary: $72,000 - $119,000
Qualifications:
- Master's or PhD in Biostatistics or related field
- Strong background in statistics and mathematical modeling
- Proficiency in statistical software (R, SAS, STATA)
Example companies: Pfizer, Merck, Johnson & Johnson, IQVIA, Fred Hutchinson Cancer Research Center
Molecular Modeler
Description: Uses computational methods to model and simulate molecular structures and interactions, often in drug discovery.
Expected Salary: $85,000 - $140,000
Qualifications:
- PhD in Computational Chemistry, Biophysics, or related field
- Experience with molecular dynamics simulations
- Knowledge of drug design principles
Example companies: Schrödinger, Novartis, GlaxoSmithKline (GSK), Atomwise, Dassault Systèmes BIOVIA
If these careers sound interesting, a PhD should be on your radar
Note: There tend to be more jobs in bioinformatics than simulation and modeling
Okay, but what about a Bachelor's degree?
Challenging for computational biology jobs, but other options are available
Focus on one half of your major
I'm unfamiliar with options here (your advisors are well-versed)
Computer Science
Biology
Software engineer, data science, machine learning, web development
To be honest: Engineering degrees give the highest chance for a well-paying job after graduation
What will help you prepare for
Everyone applying for the same positions has a college degree
Distinguish yourself with extracirriculars
Employers and graduate schools do not care about the classes you took, they care about what you can do
?
How to do this?
Show and tell
Show what you can do
Contribute to open-source projects

Hackathons and competitions


Show what you can do
Your marketable skills are learned outside the classroom
Computer science: Python, GitHub, machine learning
Graphic design: Illustrator/Inkscape, Photoshop/Gimp, Blender
Communication: Writing and presenting
Classes give foundational knowledge to learn hands-on skills in research and internships
Computational biology: You will get a small taste of this in classes; you need some research or project experience
Before the next class, you should
- Check that you are subscribed to Canvas notifications
- Make an account on the following sites (for the project):
Lecture 02:
Reference genome assembly
Lecture 01:
Introduction to computational biology
Today
Thursday