


Computational Biology
(BIOSC 1540)
Jan 28, 2025
Lecture 04A
Gene prediction
Foundations
Announcements
Assignments
Quizzes
CBytes
ATP until the next reward: 1,783
After today, you should have a better understanding of
Quiz 01
Please put away all materials as we distribute the quiz
Fill out the cover page, and do not start yet
Sit with an empty seat between you and your neighbors for the quiz
Quiz ends at 9:50 am
When you are finished, please hold on to your quiz and feel free to doodle, write anything, tell me a joke, etc. on the last page
After today, you should have a better understanding of
The biological importance of gene prediction and genome annotation
Genome assembly provides the sequence, but gene prediction and annotation assign meaning
In previous lectures, we explored the process of creating contiguous sequences with genome assembly
DNA sequence (i.e., contig)
TACGATCGGATTACGCGTAGGCTAGCTTACGGACTCGATGTACGATCGGATTACG
Gene prediction and genome annotation transform raw sequence data into actionable biological insights, identifying functional elements like genes, regulatory regions, etc.
Gene prediction locates gene-containing regions and functional elements within a genome
Predicted genes
Genes encode proteins, enzymes, and non-coding RNAs essential for cellular function
We often use Hidden Markov Models (HMMs) to statistically predict gene locations
(Topic for L04B)
This is also called "structural annotation"
Genome annotation links gene sequences to biological functions and processes
Annotation assigns putative functions to genes through experimental evidence, similarity to known genes, or ab initio predictions.
Functional annotation helps classify genes into pathways (e.g., KEGG), ontologies (e.g., GO terms), and systems (e.g., metabolic networks).
Gene prediction and annotation provides the starting point for downstream analyses and discoveries
All downstream (i.e., after) analyses are often gene-specific, so any errors here will propogate
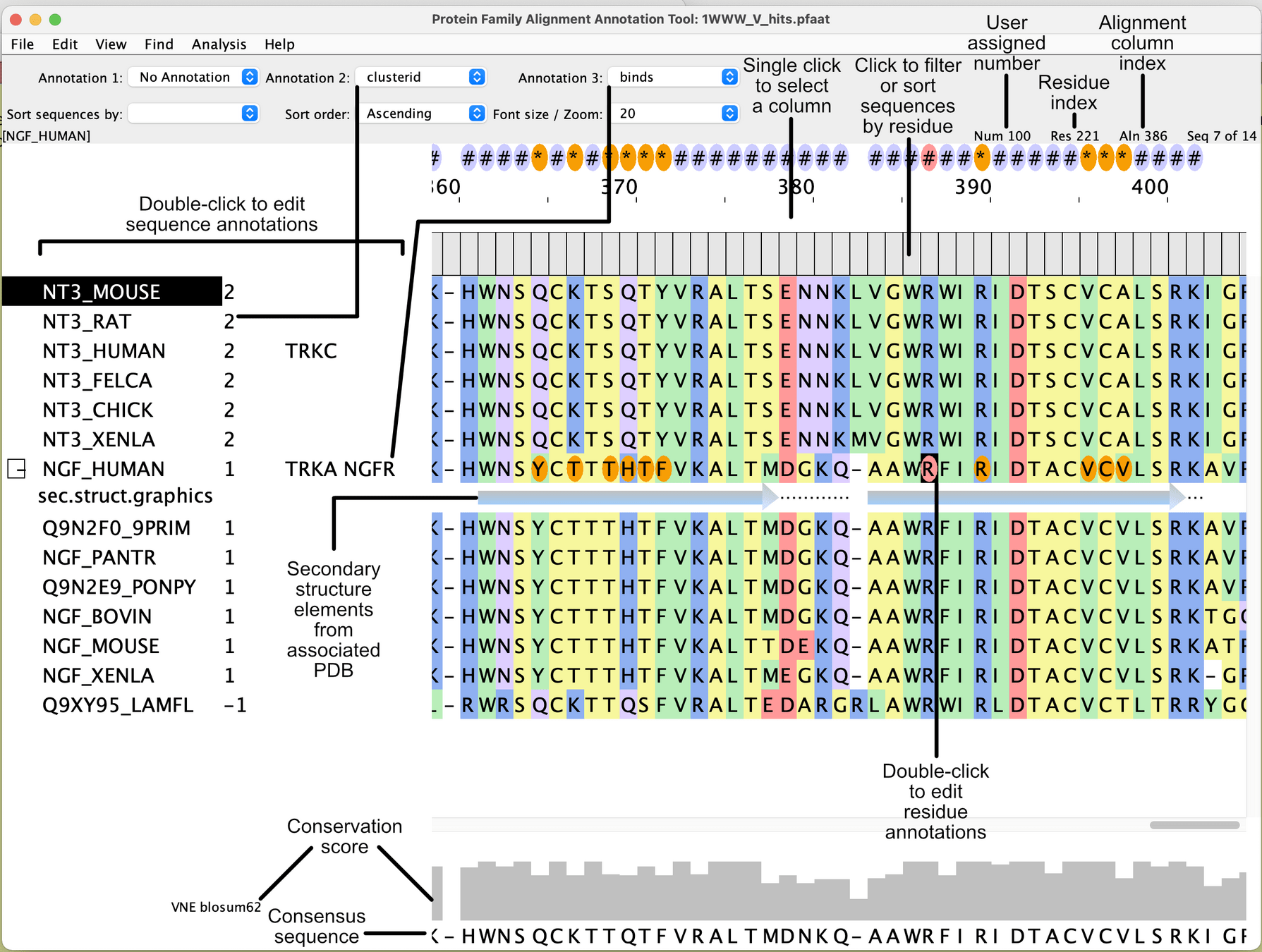
After today, you should have a better understanding of
Key differences and challenges of prokaryotic and eukaryotic gene prediction
Prokaryotes
Prokaryotic genomes are relatively straightforward due to their compact structure
Most genes are readily identifiable by open reading frame (ORF) detection.
Most genes are organized in operons—clusters of co-transcribed genes under a single promoter.
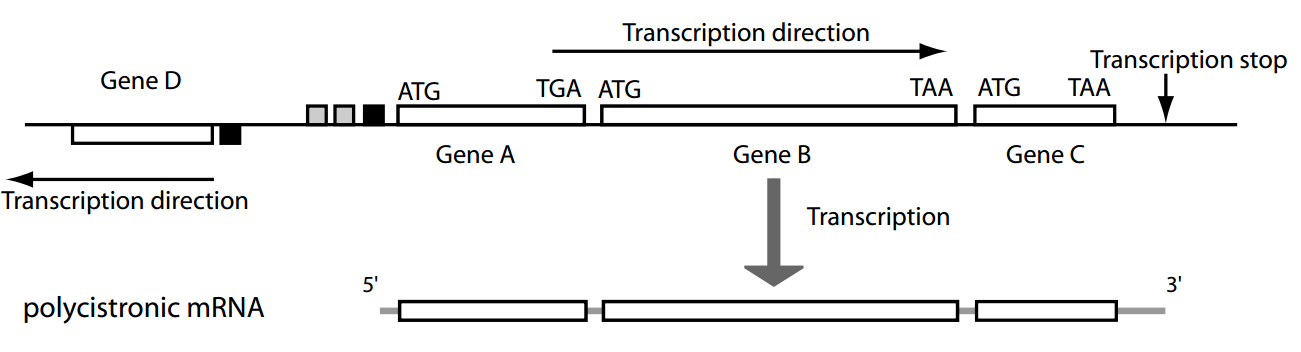
Promoters
Regulatory sequences
Operon
Polycistronic: coding sequences for two or more polypeptide chains that are transcribed in succession from the same promoter
While prokaryotic genomes are simpler, challenges still exist
Horizontal gene transfer: Foreign genes may lack organism-specific sequence patterns, complicating detection.
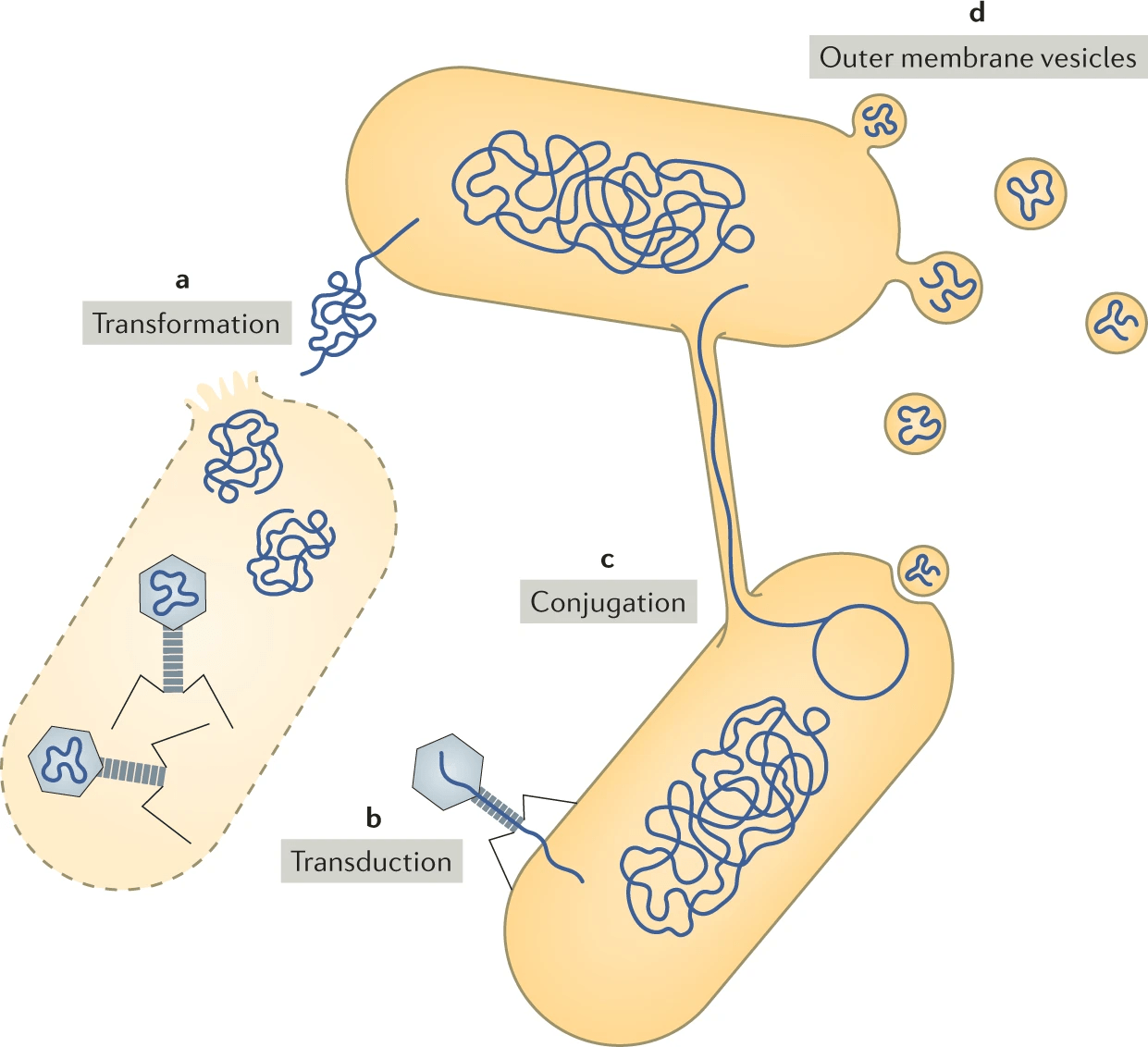
Short genes: Genes shorter than 150 bp are harder to distinguish from random ORFs.
After today, you should have a better understanding of
Key differences and challenges of prokaryotic and eukaryotic gene prediction
Eukaryotes
Eukaryotic genomes are more complex due to non-coding regions and regulatory sequences
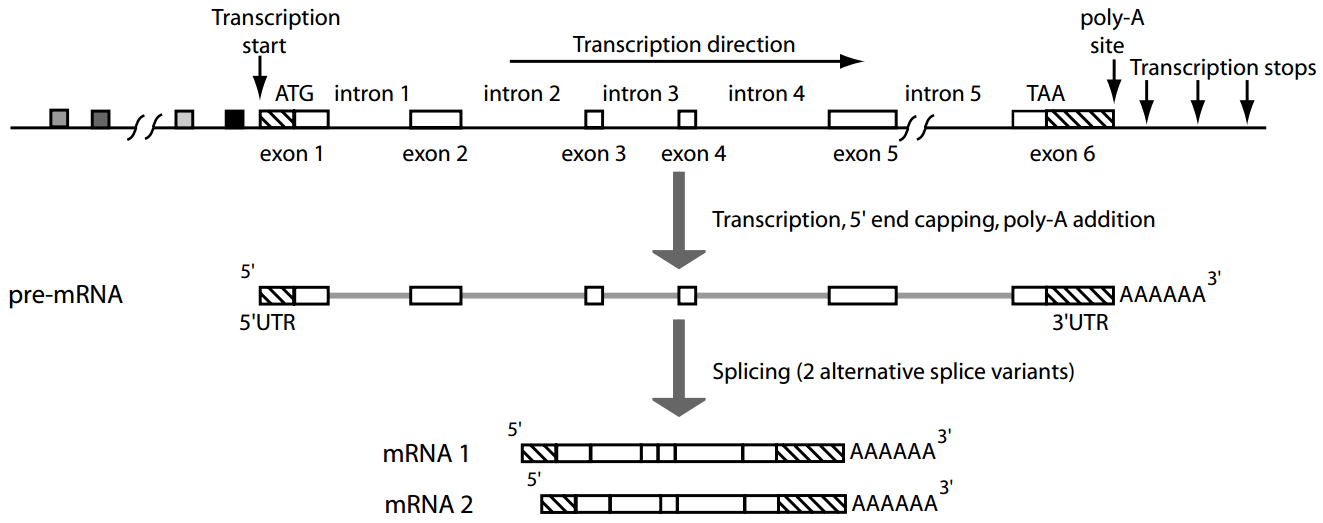
Genes contain introns (non-coding regions) and exons (coding regions), requiring splicing for expression
Intergenic (i.e., between genes) regions are large and often contain regulatory elements (e.g., enhancers, silencers).
Eukaryotic gene prediction faces additional challenges due to complexity
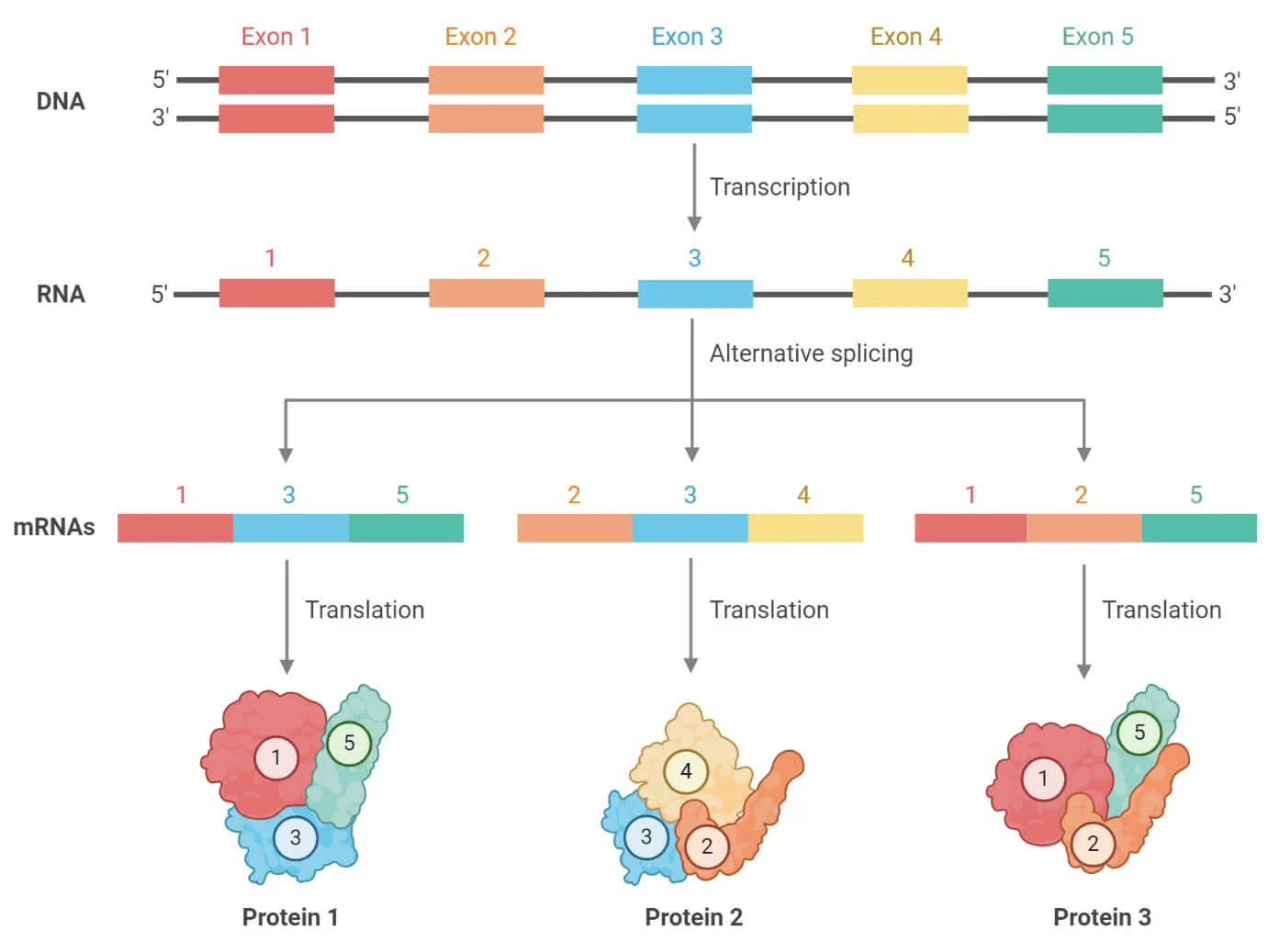
Eukaryotic genes undergo splicing which will remove introns and then join exons to form mature mRNA
Gene prediction has to predict intron boundaries, which are often much longer than exons and not always consistent
Furthermore, eukaryotes use alternative splicing to join different exons of the same gene to form multiple different proteins
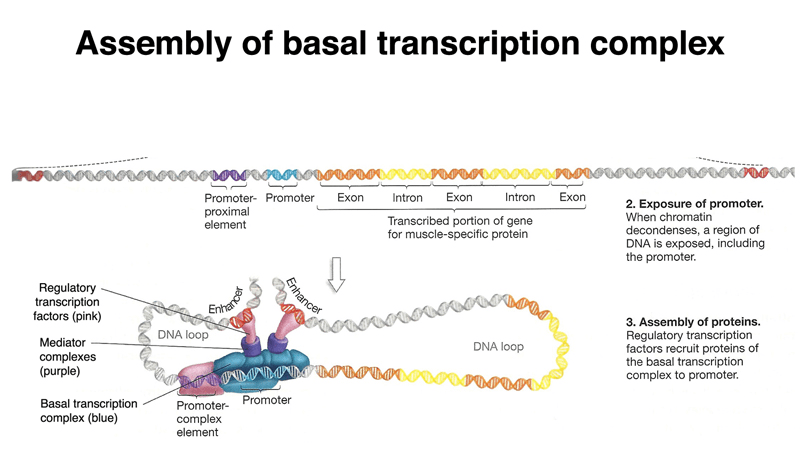
Regulatory elements are critical for expression but are hard to predict
Promoters, enhancers, and silencers regulate transcription but are often far from the gene they control.
These elements lack a universal sequence pattern, making them difficult to identify.
Eukaryotic gene prediction faces additional challenges due to complexity
Repetitive sequences: Large portions of eukaryotic genomes are repetitive, often confusing prediction algorithms.
AGCTGATC
TTAGCCGA
CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT
CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT
CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT
CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT
CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT
CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT CGAT
After today, you should have a better understanding of
The principles behind ab initio and homology-based gene prediction approaches
Ab initio
Ab initio gene prediction identifies genes based on intrinsic sequence features
Relies on patterns like:
- Start and stop codons.
- Coding sequence biases
(e.g., codon usage, GC content). - Splice sites and promoter regions (in eukaryotes).
Detects genes without requiring prior knowledge or reference sequences.

We use HMMs to detect these
Ab initio methods are powerful but limited by genome complexity
Eukaryotes:
- Accurate prediction requires identifying introns, exons, and splice sites.
- Alternative splicing and non-coding regions can confound predictions.
Prokaryotes: Compact genomes make ORF detection easier, but short genes and overlapping genes can still pose challenges.
False positives and false negatives are common, especially in large, complex genomes.
After today, you should have a better understanding of
The principles behind ab initio and homology-based gene prediction approaches
Homology
Homology-based gene prediction identifies genes by comparing sequences to known databases
Tools often use sequence alignment methods (e.g., BLAST, HMMER) to detect homologous genes.
Searches for regions of similarity between the query genome and annotated sequences in databases.
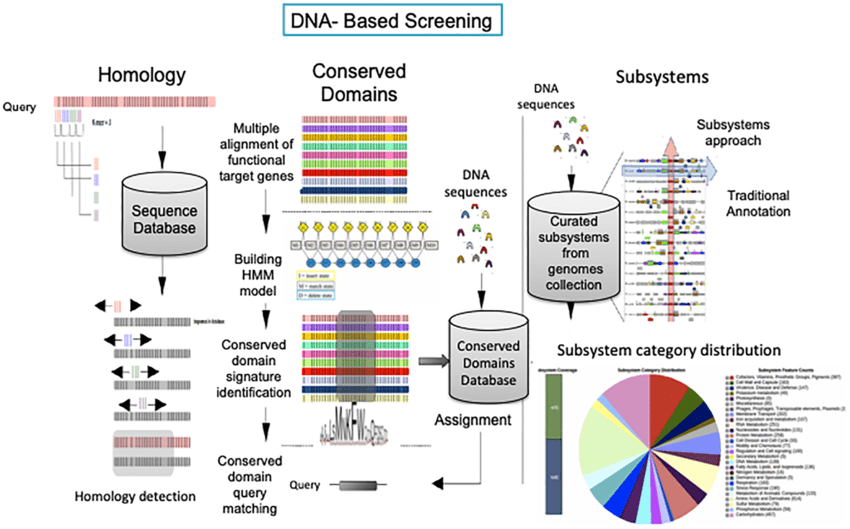
Assumes genes are evolutionarily conserved across species.
Homology-based methods depend on accurate and complete reference data
Advantages: High accuracy for conserved genes with reliable reference sequences.
Limitations:
- Cannot predict novel genes or those without significant similarity to database entries.
- Errors in reference annotations propagate into predictions.
- Divergence and mutation can obscure homology signals.
Combining ab initio and homology-based methods improves gene prediction accuracy
Ab initio methods can detect novel genes, filling gaps left by homology-based methods.
Integrated pipelines (e.g., Prokka, AUGUSTUS) use both approaches to produce more reliable results.
Homology-based methods provide functional validation for predictions from ab initio.
After today, you should have a better understanding of
Practical examples of gene prediction tools and how to interpret their outputs
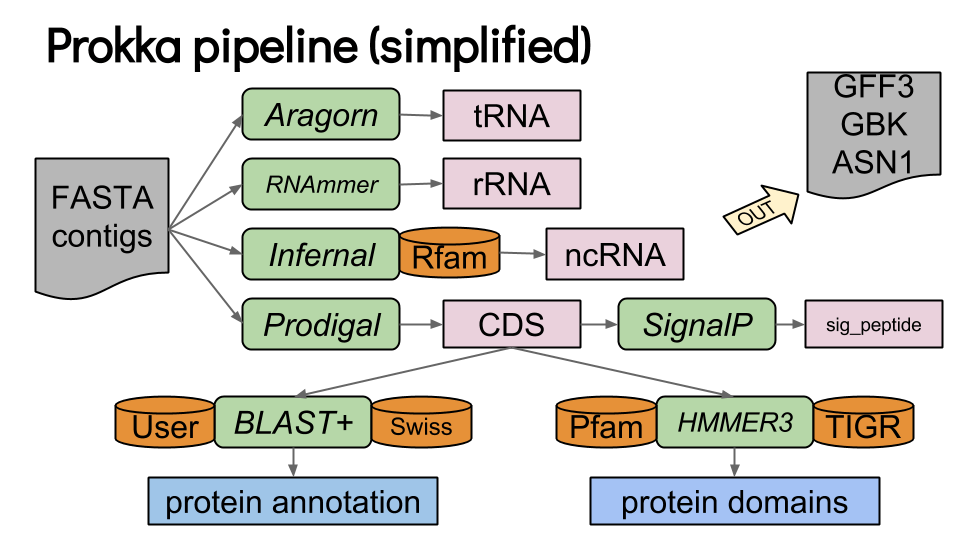
Prokka
Gene prediction tools apply computational principles to real-world problems
Selecting the right tool depends on the organism, genome complexity, and research goals.
Prokka is a popular tool for prokaryotic genome annotation
Outputs:
- GenBank files for visualization.
- FASTA files of predicted genes/proteins.
- Summary statistics of genome features.
Combines ab initio and homology-based methods for prokaryotic genomes.
Annotates coding sequences, tRNAs, rRNAs, and regulatory regions.

Prokka provides an efficient workflow for bacterial genome annotation
Inputs: Assembled genome in FASTA format.

Outputs:
- A list of coding sequences (CDSs) with predicted functions.
- Identification of antibiotic resistance genes (e.g., beta-lactamases).
Prokka output files example

>ECNNONJI_02637 Dihydrofolate reductase
MTLSILVAHDLQRVIGFENQLPWHLPNDLKHVKKLSTGHTLVMGRKTFESIGKPLPNRRN
VVLTSDTSFNVEGVDVIHSIEDIYQLPGHVFIFGGQTLFEEMIDKVDDMYITVIEGKFRG
DTFFPPYTFEDWEVASSVEGKLDEKNTIPHTFLHLIRKKAfter today, you should have a better understanding of
Practical examples of gene prediction tools and how to interpret their outputs
AGUSTUS
AUGUSTUS excels at predicting genes in eukaryotic genomes
Outputs:
- Predicted gene structures, including exons, introns, and UTRs.
- GFF3 files for integration with genome browsers.
Focuses on ab initio gene prediction but integrates hints like RNA-seq data for improved accuracy
Suitable for genomes with limited or no reference annotations


After today, you should have a better understanding of
Practical examples of gene prediction tools and how to interpret their outputs
HMMER uses Hidden Markov Models (HMMs) for detecting homologous genes
Outputs:
- Alignment scores for detected homologs.
- Functional annotations from database hits.
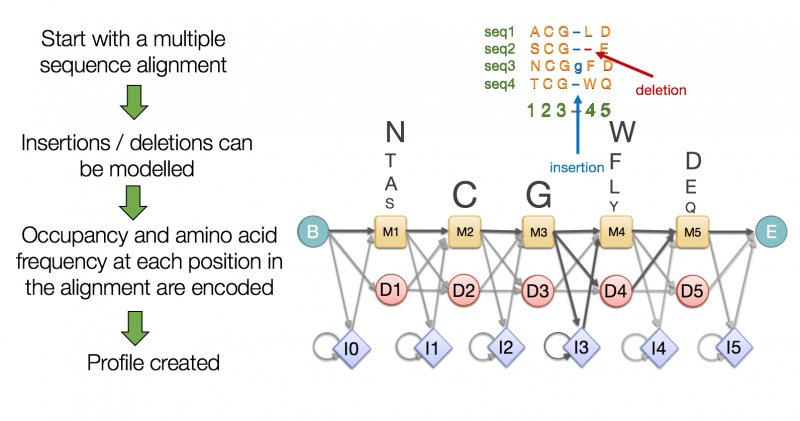
Aligns query sequences to profiles of known genes/proteins in curated databases like Pfam.
Identifies genes based on conserved domains or motifs.
Interpreting outputs requires understanding key metrics and visualizations
- Gene locations: Coordinates of start and stop codons or exon-intron boundaries.
- Scores: Confidence values for predictions, such as e-values in HMMER or reliability scores in AUGUSTUS.
- Functional annotations: Gene ontology (GO) terms, protein domains, or pathway mappings.
Before the next class, you should
Lecture 04B:
Gene prediction -
Methodology
Lecture 04A:
Gene prediction -
Foundations
Today
Thursday