


Computational Biology
(BIOSC 1540)
Feb 4, 2025
Lecture 05A
Sequence Alignment
Foundations
Announcements
Assignments
Quizzes
CBytes
ATP until the next reward: 1,653
After today, you should have a better understanding of
Why sequence alignment matters
Homology
Homology describes the evolutionary relationship between sequences and is key to understanding biological function and evolution
Homologous sequences share a common ancestor, even if they have diverged over time.
Homology helps transfer knowledge from well-studied genes to newly discovered ones.
Example: The identification of BRCA1 as a breast cancer gene was based on identifying its association with RAD51, which function was known due to its high homology with yeast DNA repair
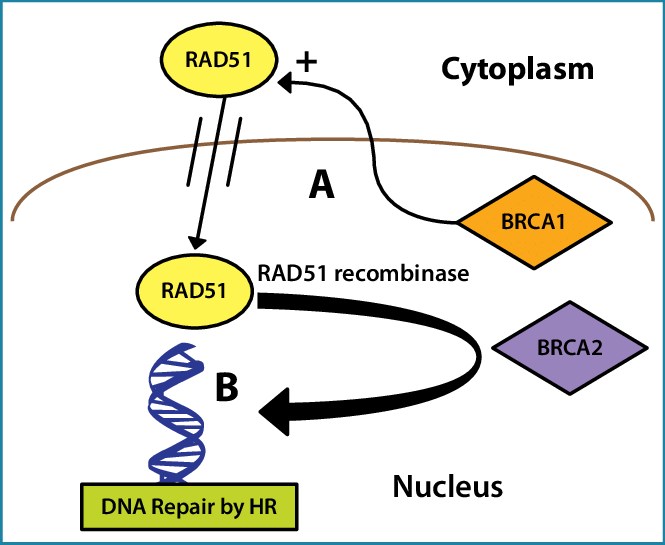
Orthologs are genes in different species that originated from a common ancestor and usually retain the same function
They typically perform the same function across species but may accumulate minor adaptations.
Orthologs arise from speciation events, meaning a single ancestral gene diverges into different species.

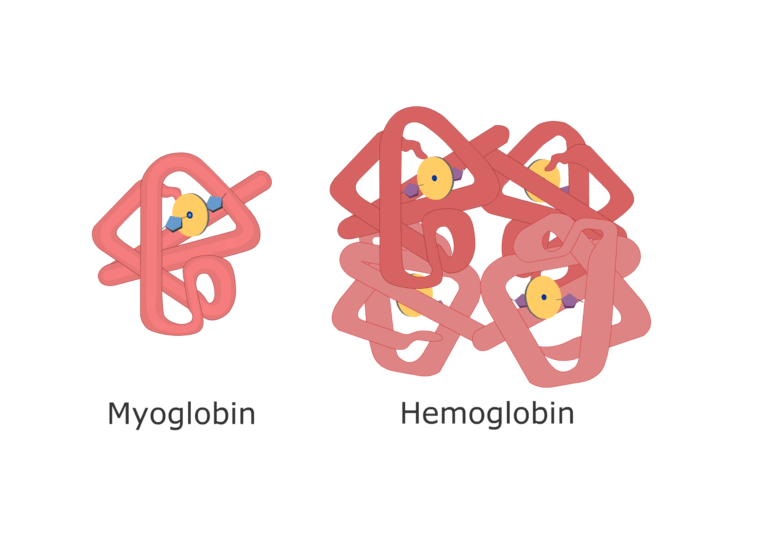
Example: The hemoglobin gene in humans and mice is orthologous, both encoding oxygen-carrying proteins in red blood cells.
Paralogs are genes that arise from duplication within the same genome and may evolve new functions
- Gain a new function (neofunctionalization).
- Specialize in a subset of the original function (subfunctionalization).
- Become a nonfunctional pseudogene.
Paralogs originate from duplication events, which allows one copy to retain the original function while the other copy can
Paralogs drive gene family expansions, leading to specialized and diverse biological functions.

Gene duplication allows new biological functions to emerge while preserving essential roles
Examples of paralog-driven functional diversification:
- Globin family: Myoglobin (muscle oxygen storage) and hemoglobin (blood oxygen transport) evolved from a common ancestor.
- HOX genes: Regulate body plan development, with duplicates specializing in different body regions.
- Opsin genes: Responsible for color vision in vertebrates, arising from multiple duplications.
After today, you should have a better understanding of
Why sequence alignment matters
Homology applications
Functional annotation of genes and proteins relies on identifying homologous sequences
Conserved motifs suggests similar functional roles in different organisms.
For example, if a newly discovered protein aligns with a known enzyme, it likely shares the same biochemical function.
Homology-based searches (e.g., BLAST) rapidly annotate unknown sequences by comparing them to well-characterized databases.
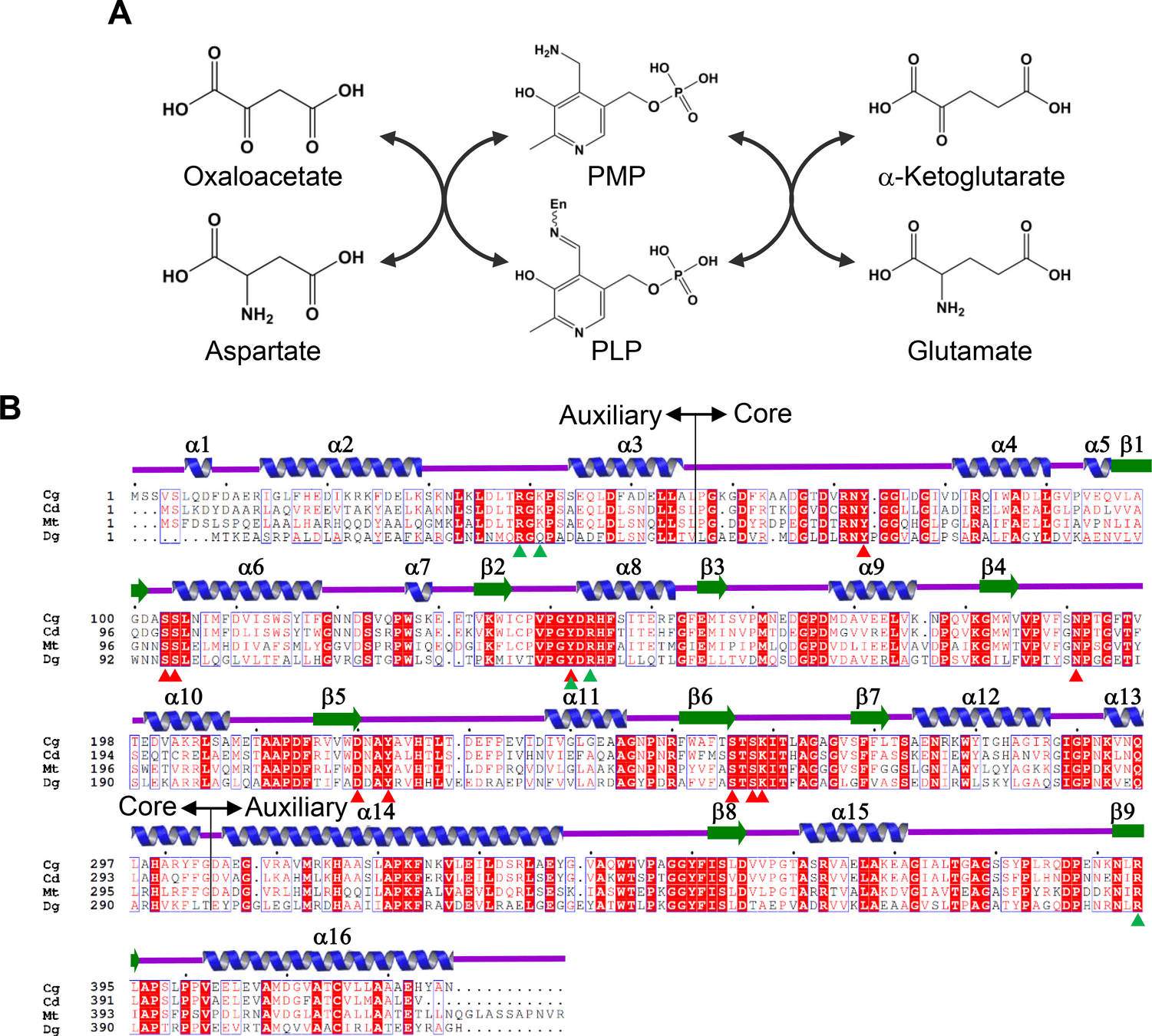

Aspartate aminotransferase (AspAT)
Conserved residues that bind to PLP cofactor are shown with triangles
Protein sequence homology predicts 3D structure and function
Structural homology models unknown proteins based on alignment with known structures.
Highly conserved residues often indicate key structural or catalytic sites.
We will cover this in Lectures 11A and 11B
Protein threading techniques align sequences to structural databases like the Protein Data Bank.
AlphaFold uses sequence alignments as inputs to its deep learning model
Importance in research:
- Used to infer gene function across species (e.g., using model organisms to study human genes).
- Enable comparative genomics and evolutionary studies.
- Help in drug discovery, as conserved drug targets can be tested in different organisms.
- Knockout and mutation studies in animals help determine gene function in humans.
- Evolutionarily conserved pathways (e.g., DNA repair, metabolism) can be studied using orthologs.
Homologous genes allow functional studies using model organisms to understand human biology
After today, you should have a better understanding of
Conceptual interpretation of alignment results
Match and mismatches
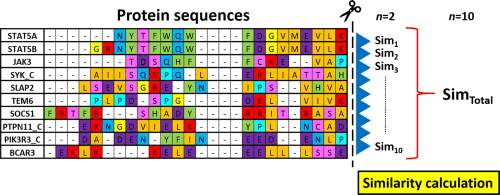
Alignment algorithms compare sequences to identify conservation, mutations, and functional domains.
Alignment patterns reflect evolutionary events, including mutations, conservation, and sequence divergence
Alignment results provide insight into sequence similarity, evolutionary relationships, and functional conservation
Identical residues at aligned positions suggest evolutionary conservation and functional stability
Highly conserved sequences often correspond to:
- Protein active sites (e.g., catalytic residues in enzymes).
- DNA regulatory elements (e.g., promoters, enhancers).
- RNA structural motifs (e.g., ribosomal RNA stems and loops).
Matches (|) indicate strong evolutionary constraints, meaning the sequence is critical for function.
CGACGATTCTATAGTCTAACATGCGAGCGTGACGAATAAAAGATCTCGCG
||||||||||||||||||||||||||||||||||||||||||||||||||
CGACGATTCTATAGTCTAACATGCGAGCGTGACGAATAAAAGATCTCGCGSequence mismatches ( ) highlight mutations that contribute to genetic variation and adaptation
Point mutations can be neutral, beneficial, or deleterious.
- Synonymous mutations that do not alter the protein sequence.
- Nonsynonymous mutations that may change protein function.
Mismatches occur when different residues are aligned
CGCGATTCTATAGTCTAACATGCGAGCGTGGAAAAAAGATCTCGCG
|| |||||||| | | | | | ||| ||| |||
CGACGATCTATAGTAACATGCGAGCGTGACGAATAAAAGATCTGCGAfter today, you should have a better understanding of
Conceptual interpretation of alignment results
Insertions and deletions
Insertions (–) introduce new genetic material, impacting protein structure and genome evolution
Functional and evolutionary impact:
- Short insertions in proteins modify binding sites or enzyme activity.
- Insertions in DNA regulatory regions affect gene expression patterns.
Causes of insertions:
- Gene duplications leading to new protein functions.
- Insertion of transposable elements modifying gene regulation.
- Microindels affecting protein structure and function.
Gaps are used to indicate insertions
CGACGATTCTATAGTC-------------TGACGAATAAAAGATCTCGCG
|||||||||||||||| |||||||||||||||||||||
CGACGATTCTATAGTCTAACATGCGAGCGTGACGAATAAAAGATCTCGCGDeletions (–) remove genetic material, leading to functional changes or species divergence
Causes of deletions:
- Loss of nonessential genes in parasitic or symbiotic organisms.
- Regulatory deletions affecting developmental pathways.
- Frameshift deletions that drastically alter protein coding.
Functional and evolutionary consequences:
- Can disable genes, leading to loss of function.
- Can optimize metabolic efficiency, as seen in endosymbiotic bacteria with streamlined genomes.
Gaps are used to indicate deletions
CGACGATTCTATAGTCTAACATGCGAGCGTGACGAATAAAAGATCTCGCG
|||||||||||||||| |||||||||||||||||||||
CGACGATTCTATAGTC-------------TGACGAATAAAAGATCTCGCGSmall insertions and deletions (indels) are a major cause of genetic disorders
- Cancer-related genes (e.g., TP53, BRCA1).
- Neurological disorders (e.g., Huntington’s disease, caused by repeat expansions).
- Metabolic disorders (e.g., cystic fibrosis, due to a 3-base deletion in CFTR).
Indels play a major role in speciation by modifying gene structure and expression.
Indels can disrupt coding sequences or regulatory elements, leading to disease.
Frameshift mutations caused by small indels result in completely altered protein sequences.
After today, you should have a better understanding of
Conceptual interpretation of alignment results
Alignment scores
Alignment scores measure sequence similarity by rewarding matches and penalizing mismatches and gaps
Alignment algorithms assign numerical scores to quantify how well two sequences align.
Matches receive positive scores (e.g., +1 or +2)
Mismatches receive negative scores (e.g., -1 or -2)
- Ensures meaningful evolutionary comparisons.
- This reflects that large insertions/deletions are less common than point mutations.
Gaps are heavily penalized (e.g., -2, or -3).
- Lower penalties for conservative substitutions (e.g., leucine to isoleucine).
- Higher penalties for radical substitutions (e.g., leucine to arginine, which changes charge and structure).
- Higher values are assigned to matches in functionally critical regions.
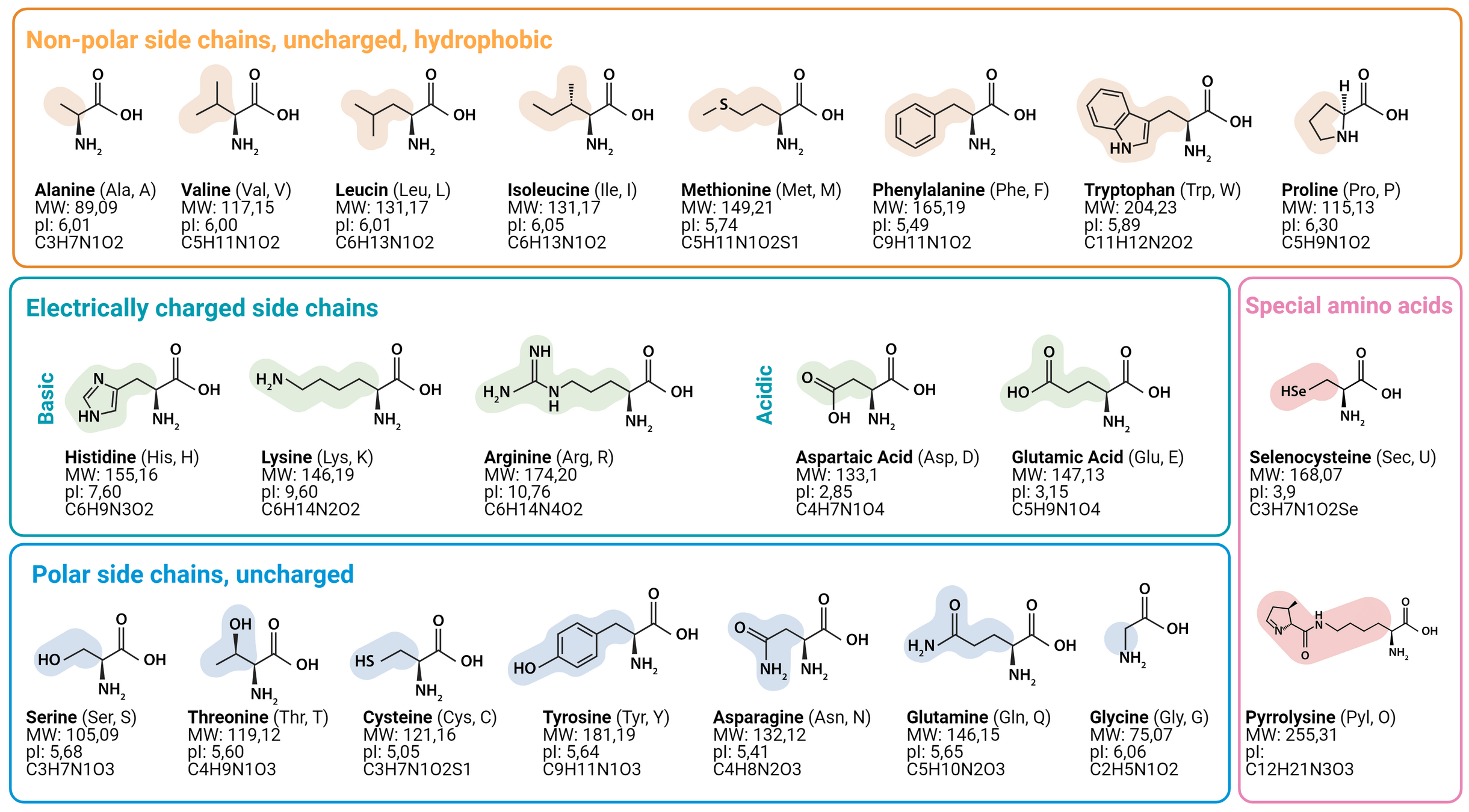
Substitution matrices model evolutionary relationships by assigning biologically meaningful scores to amino acid replacements
Not all mutations are equally likely—some substitutions occur more frequently due to biochemical properties.
Impact on alignment quality:
- Helps distinguish true homology from random similarity.
- Improves evolutionary modeling.
- Adjusts mismatch penalties based on real-world observations.
Substitution matrices assign different scores to different amino acid replacements based on their evolutionary likelihood.
Two widely used matrices are PAM (Point Accepted Mutation) matrices and BLOSUM (Blocks Substitution Matrix)
More frequent substitutions have lower penalties, while rare substitutions are penalized more heavily
Physicochemical properties influence substitution likelihood:
- Hydrophobic residues often replace other hydrophobic residues.
- Charged residues tend to substitute with others of similar charge.
After today, you should have a better understanding of
Conceptual interpretation of alignment results
E-values
E-values measure the likelihood that an alignment occurs by chance, helping assess biological relevance
E-value (Expectation Value): Number of expected random matches in a database search.
Lower E-value = Higher significance (e.g., E = 0.001 means only 1 in 1,000 alignments is due to chance).
Database size affects E-value: Larger databases increase the probability of chance alignments.
After today, you should have a better understanding of
Pairwise versus multiple sequence alignment
Pairwise sequence alignment is the fundamental method for comparing two biological sequences
Methods like global and local alignment provide different perspectives on sequence similarity.
Pairwise alignment finds the optimal arrangement of two sequences to maximize similarity and minimize differences.
Query 1 ATGACTTTATCCATTCTAGTTGCACATGACTTGCAACGAGTAATTGGTTTTGAAAATCAA 60
Sbjct 2555705 .....A.....A..AA.T..C..T..C..TAAA...A....C.....G.ACC........ 2555646
Query 61 TTACCTTGGCATCTACCAAATGATTTGAAGCATGTTAAAAAATTATCAACTGGTCATACT 120
Sbjct 2555645 ...........CT.............A......A.....C..C.GA.C.....GA....A 2555586
Query 121 TTAGTAATGGGTCGTAAGACATTTGAATCGATTGGTAAACCACTACCGAATCGTCGAAAT 180
Sbjct 2555585 C.T.......CA..G..A..T...A.T..T..A..G..G...T.G..A...A.A..T..C 2555526
Query 181 GTTGTACTTACTTC---AGATACAAGTTTCAACGTAGAGGGCGTTGATGTAATTCATTCT 237
Sbjct 2555525 ..C.....C...AACCA..C.T..--.....C.A.GA....-..A.....T..AA.C... 2555469
Query 238 ATTGAAGATATTTATCAACTACCGGGCCATGTTTTTATATTTGGAGGGCAAACATTATTT 297
Sbjct 2555468 C....T..A...A.AG.GT..T.T..T....................A.....G....AC 2555409
Query 298 GAAGAAATGATTGATAAAGTGGACGACATGTATATTACTGTTATTGAAGGTAAATTTCGT 357
Sbjct 2555408 ....C.........CC.G..A..T..T........C..A..A..A..T..A..G....AA 2555349
Query 358 GGTGATACGTTCTTTCCACCTTATACATTTGAAGACTGGGAAGTTGCCTCTTCAGTTGAA 417
Sbjct 2555348 ..A..C..A...........A..C.....C...A..........C.AA........A... 2555289
Query 418 GGTAAACTAGATGAGAAAAATACAATTCCACATACCTTTCTACATTTAATTCGTAAAAAA 477
Sbjct 2555288 ...C..........A........T..A..G.....A..CT........G.G....G.... 2555229While pairwise alignment is effective for comparing two sequences, it has limitations for analyzing multiple sequences
Strengths:
- Computationally efficient for two sequences.
- Provides a direct, detailed comparison.
- Useful for identifying single mutations or evolutionary changes.
Example: A pairwise comparison of hemoglobin genes between humans and chimpanzees provides insight into species divergence but does not reveal broader evolutionary trends across mammals.
Limitations:
- Cannot reveal conserved regions across multiple species.
- Cannot model evolutionary relationships between many sequences.
- Performance and accuracy decline when extended to multiple sequences
MSA extends pairwise alignment to multiple sequences, enabling more powerful biological interpretations
MSA aligns three or more sequences to reveal conserved motifs, functional domains, and evolutionary relationships.
Unlike pairwise alignment, MSA considers multiple substitutions, insertions, and deletions across species.
Example: ClustalW and MUSCLE generate MSAs to compare entire protein families

Example: MSA of SARS-CoV-2 spike proteins identifies conserved regions for vaccine development

Sequence alignment identified key conserved residues that are often epitopes
Residues in orange are stem helix epitope region
Before the next class, you should
- Start P01D (due Feb 10)
Lecture 05B:
Sequence alignment -
Methodology
Lecture 05A:
Sequence alignment -
Foundations
Today
Thursday