
Computational Biology

(BIOSC 1540)

Sep 17, 2024
Lecture 07:
Transcriptomics
Announcements
- A02 will be graded by Sunday
- A03 is due Thursday by 11:59 pm
- The bioinformatics exam is in 16 days
- If you have DRS-approved accommodations, please request an exam time
- We will have a review session the day before the exam
- I will post a list of concepts that will be on the exam
- We will have a poll where you can vote on topics to go over
- Programming+ recitations are Fridays from 2:00 - 3:30 pm in 315 Clapp

After today, you should be able to
1. Define transcriptomics and explain its role in understanding gene expression patterns.
2. Discuss emerging trends in transcriptomics.
3. Compare and contrast transcriptomics and genomics.
4. Explain the principles of RNA-seq technology and its advantages over previous methods.
5. Outline the computational pipeline for RNA-seq data analysis.
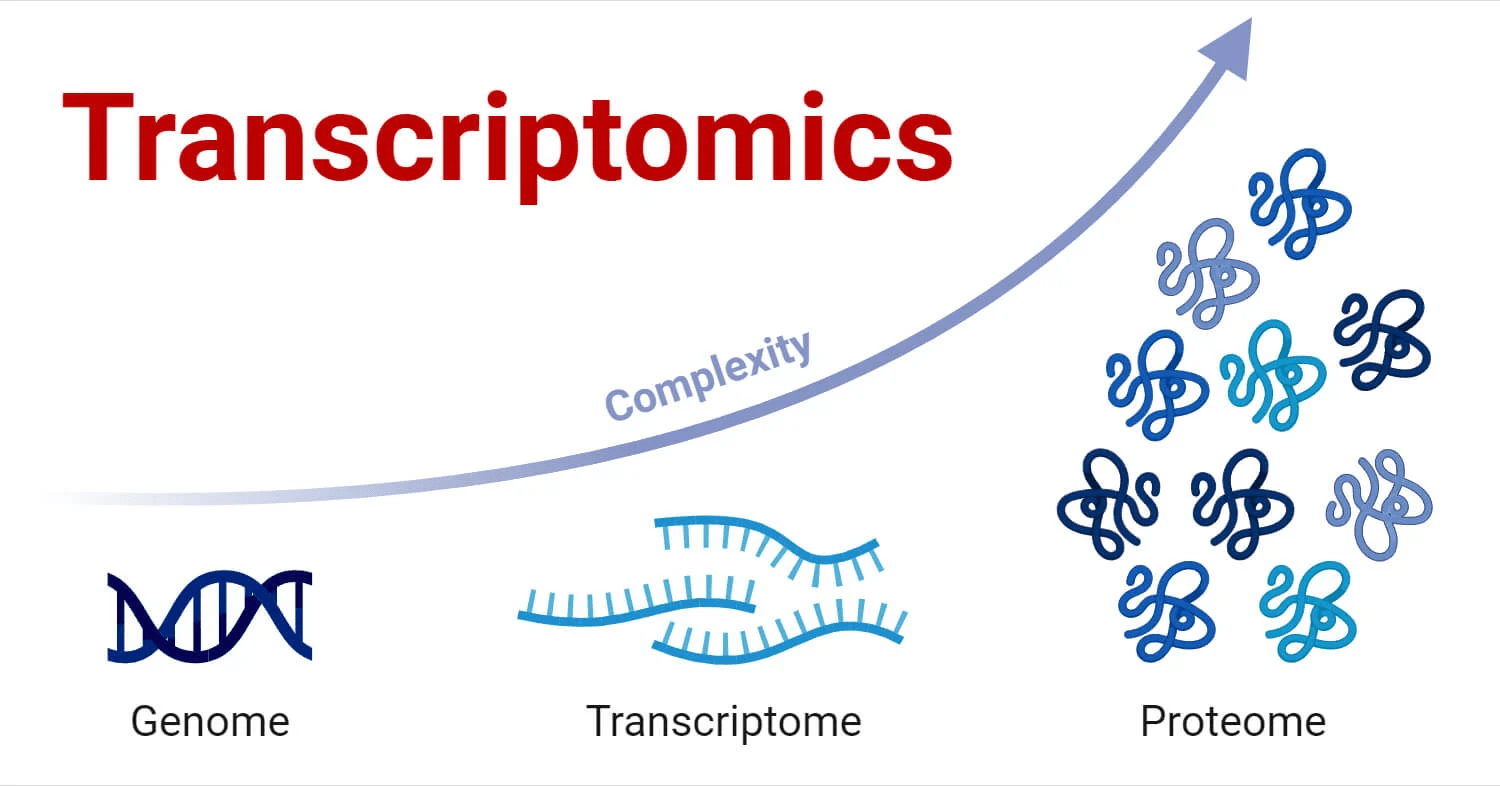
Transcriptomics: A real-time microscope
Transcriptomics allows us to see exactly what genes are active at a given moment
We can see gene expression changes over time
Transcriptomics works with the complete set of RNA transcripts
This includes
mRNA: instructions for protein synthesis
rRNA: forms part of the ribosome structure
tRNA: helps translate the genetic code into proteins
Non-coding RNAs: play regulatory roles in the cell
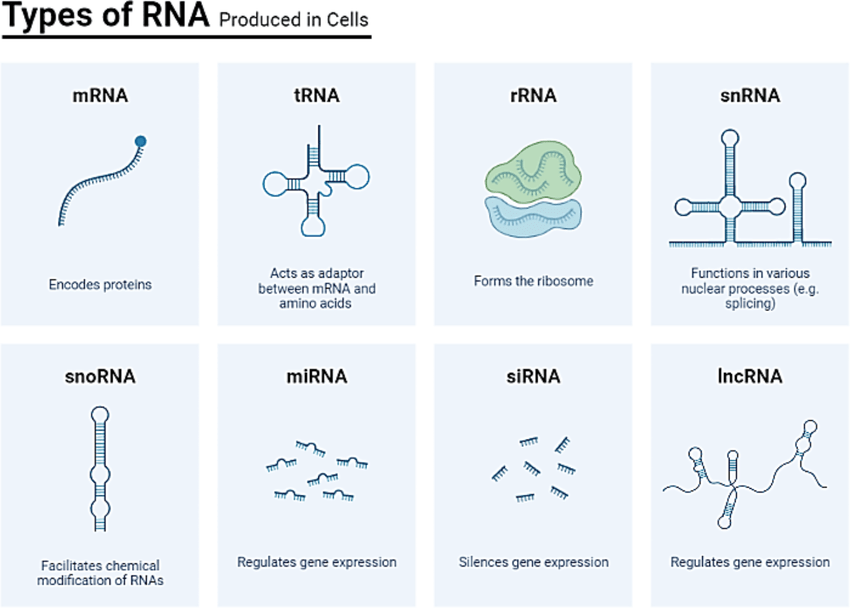



(And more)
See RNA in action
The genome is relatively static
Allows us to see which annotated genes are actually being used
The transcriptome is constantly changing and captures the cell's response to its environment and internal signals
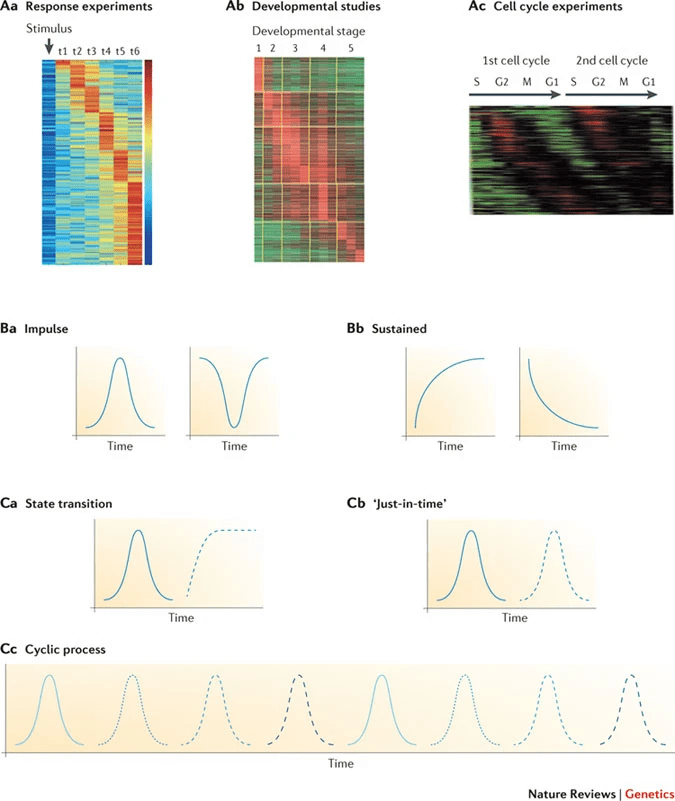
- Cell type: A neuron will have a different gene expression profile than a liver cell
- Developmental stage: The genes active in an embryo differ from those in an adult
- Environmental conditions: Cells respond to stress, nutrients, or pathogens by changing gene expression
This dynamic nature of the transcriptome reflects the functional state of the cell
Transcriptomics is versatile
Developmental biology: Understanding cell differentiation
- Which genes are expressed in a specific cell type or condition?
Disease research: Identifying pathological gene expression patterns
- What are the differences in gene expression between healthy and diseased states?
Drug discovery: Revealing mechanisms of action and side effects
- How does gene expression change over time or in response to stimuli?
Ecology: Studying organism-environment interactions
- How do environmental factors influence gene expression?
Transcriptomics reveals alternative splicing and isoforms
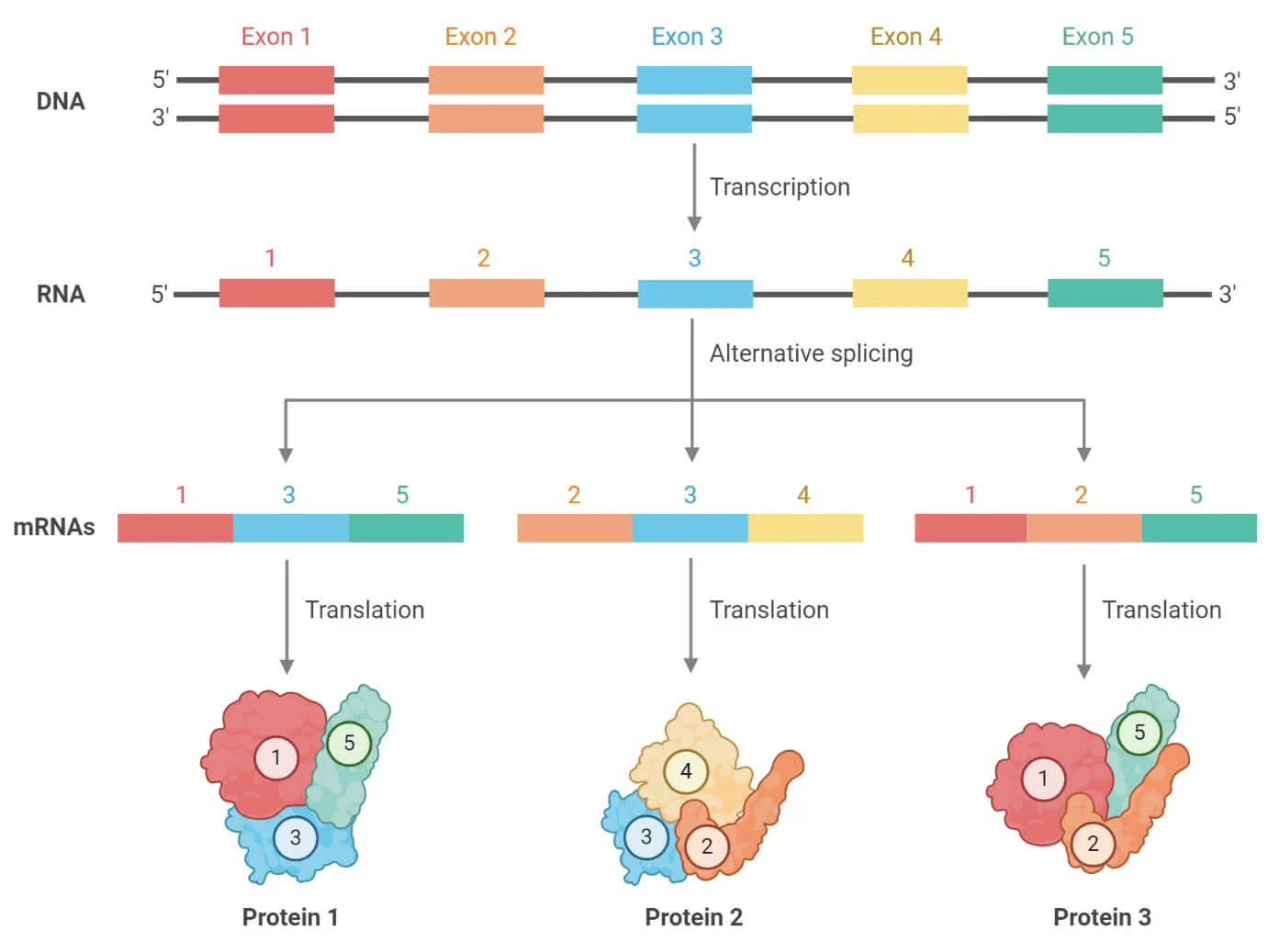
A single gene can produce multiple mRNA transcripts, which we call isoforms
One of the main ways organisms can increase protein diversity without increasing the number of genes
It's estimated that over 90% of human genes undergo alternative splicing
Example: Dscam in Drosophila
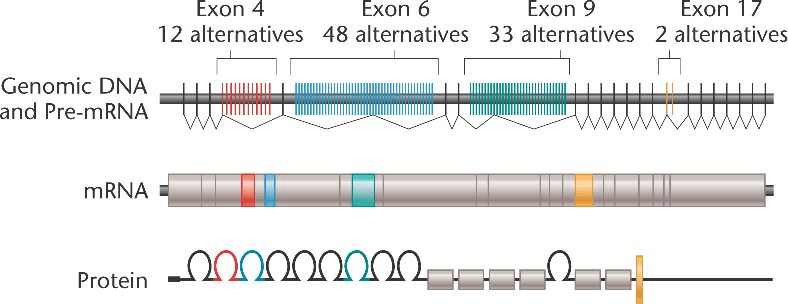
Drosophila melanogaster has over 38,000 isoforms from this one gene
Dscam (Down syndrome cell adhesion molecule) is involved in neural development
After today, you should be able to
1. Define transcriptomics and explain its role in understanding gene expression patterns.
2. Discuss emerging trends in transcriptomics.
3. Compare and contrast transcriptomics and genomics.
4. Explain the principles of RNA-seq technology and its advantages over previous methods.
5. Outline the computational pipeline for RNA-seq data analysis.
Single-cell transcriptomics revolutionizes resolution
- Captures gene expression in individual cells
- Reveals cellular heterogeneity within tissues
- While powerful, data is sparse and noisy


Spatial transcriptomics maps gene expression to location
- Preserves spatial information of transcripts within tissue sections
- Reveals how cellular neighborhoods influence gene expression

TopHat questions
Which of the following scenarios would likely benefit most from using single-cell transcriptomics over bulk RNA-seq?

After today, you should be able to
1. Define transcriptomics and explain its role in understanding gene expression patterns.
2. Discuss emerging trends in transcriptomics.
3. Compare and contrast transcriptomics and genomics.
4. Explain the principles of RNA-seq technology and its advantages over previous methods.
5. Outline the computational pipeline for RNA-seq data analysis.
Functional insights
- Reveals which elements are active
- Shows diseases state
- Identifies potential functional elements
- Predicts disease risk
Genomics
Transcriptomics
- Requires one-time sampling
- Reveals evolutionary history
- Captures real-time cellular responses
Temporal insights
After today, you should be able to
1. Define transcriptomics and explain its role in understanding gene expression patterns.
2. Discuss emerging trends in transcriptomics.
3. Compare and contrast transcriptomics and genomics.
4. Explain the principles of RNA-seq technology and its advantages over previous methods.
5. Outline the computational pipeline for RNA-seq data analysis.
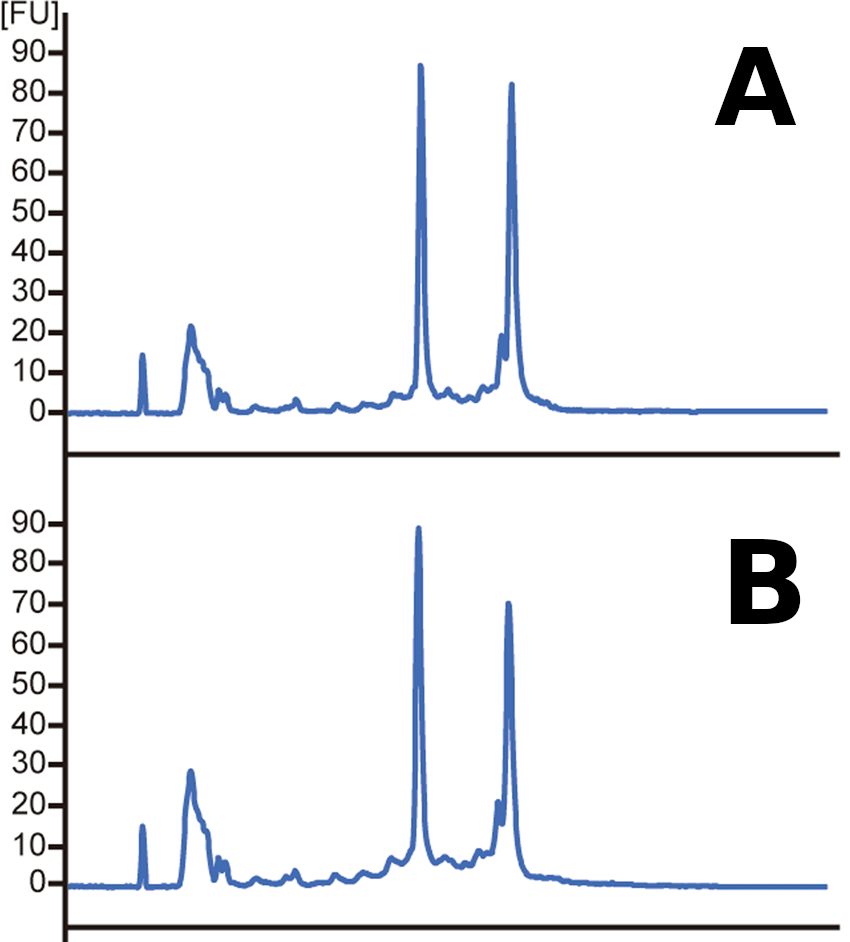
RNA quality is critical for successful sequencing
Assess RNA integrity (RNA Integrity Number)
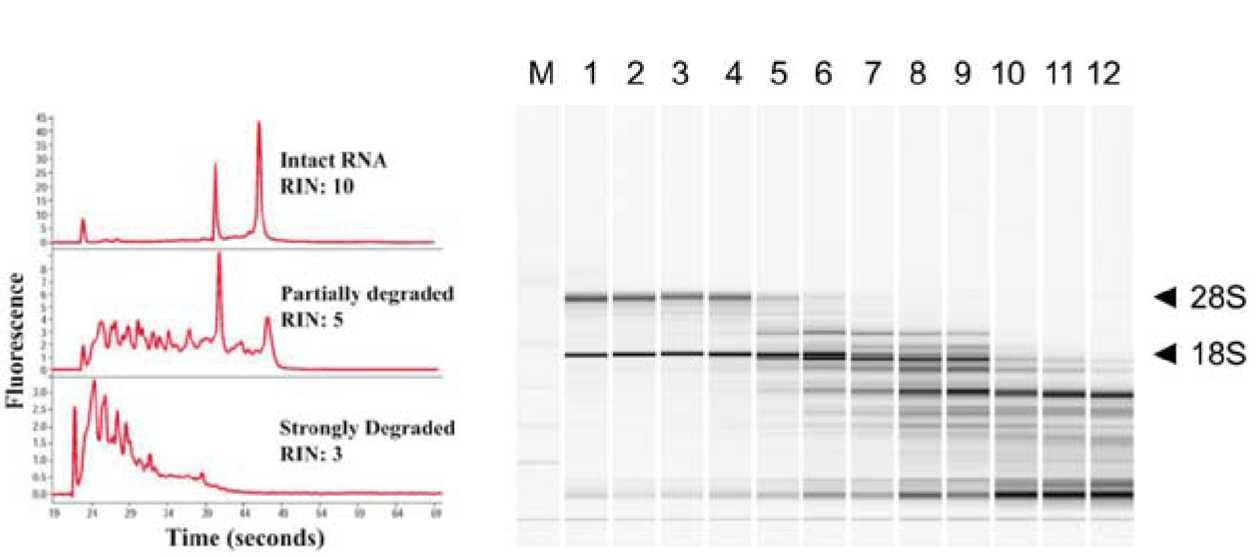
Low RIN
High RIN
- rRNA makes up a large (~85%) of our RNA
- Based on the ratio of 28S and 18S rRNA vs. all RNA
mRNA enrichment focuses sequencing on protein-coding transcripts
Enrichment method affects
- Gene expression measurements
- Detection of non-coding RNAs
- Identification of immature transcripts
Poly(A) selection captures mature mRNAs
How could we filter our sample for only mRNA?

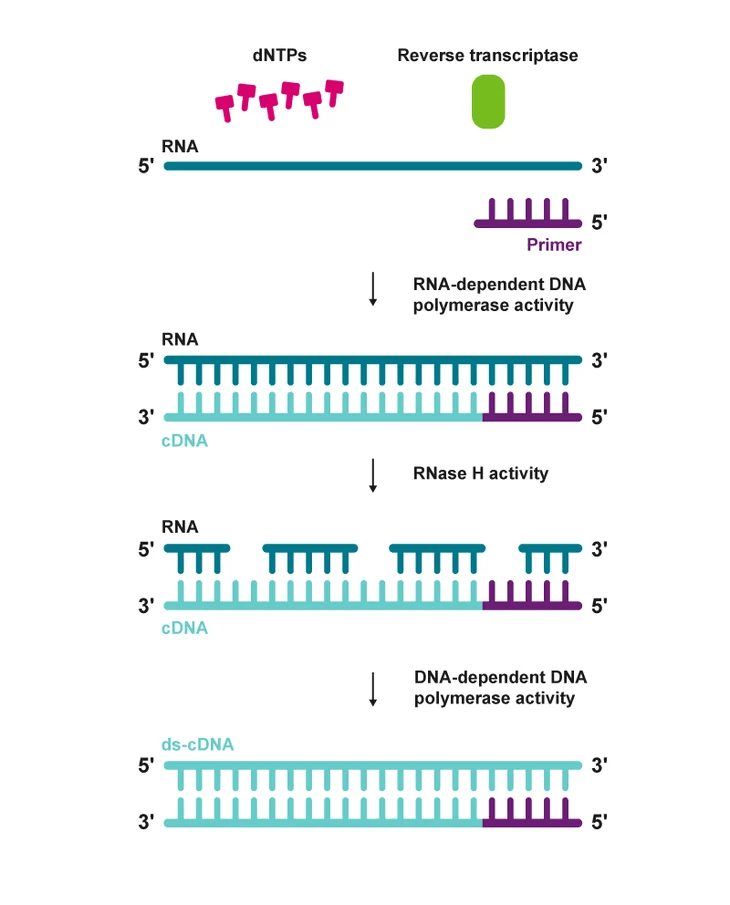
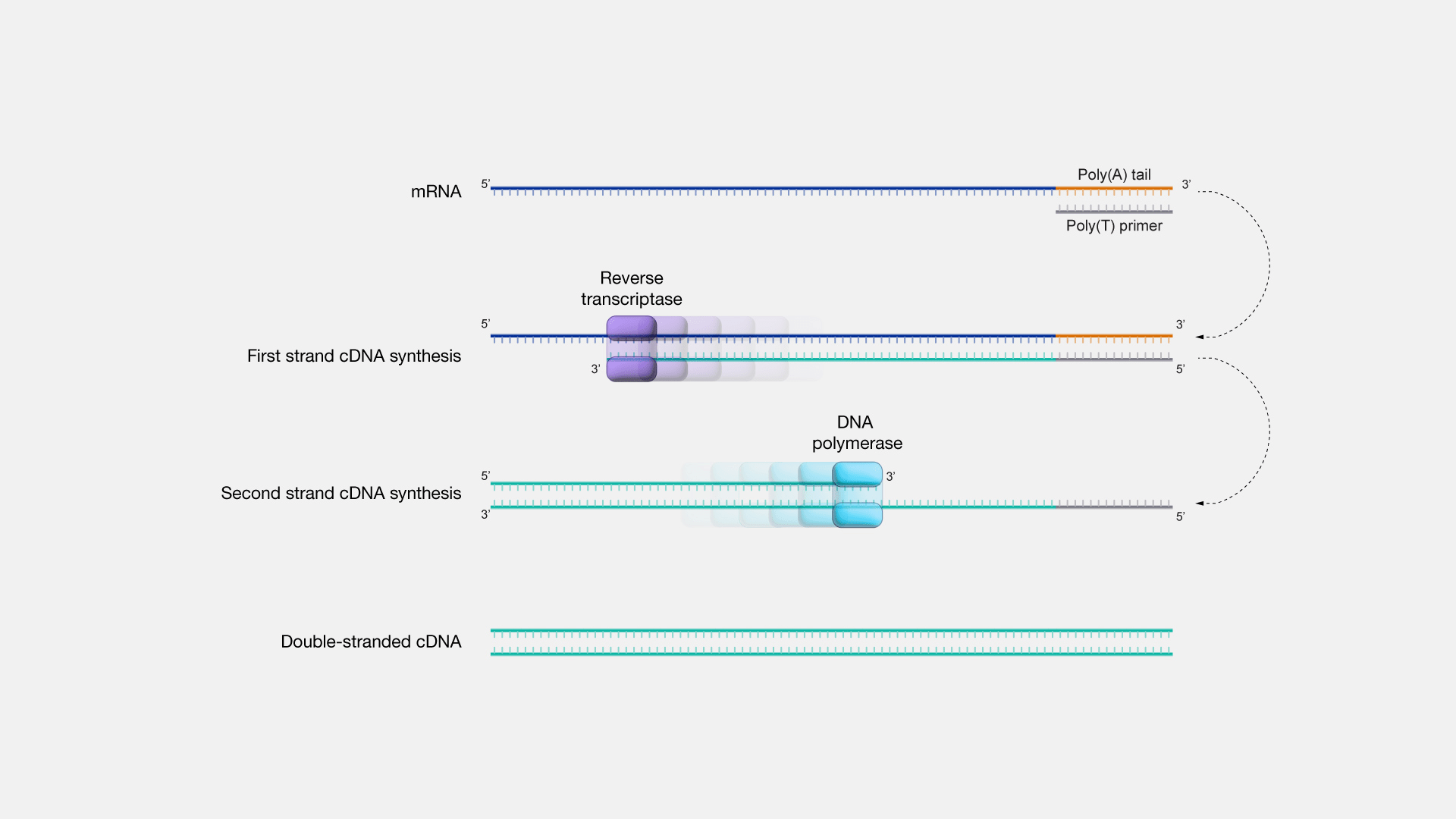
Reverse transcription introduces unique challenges
RNA is converted to cDNA using reverse transcriptase
- Random or oligo(dT) primers influence transcript representation
- Second-strand synthesis method can preserve strand information
Once upon a time, we had microarrays
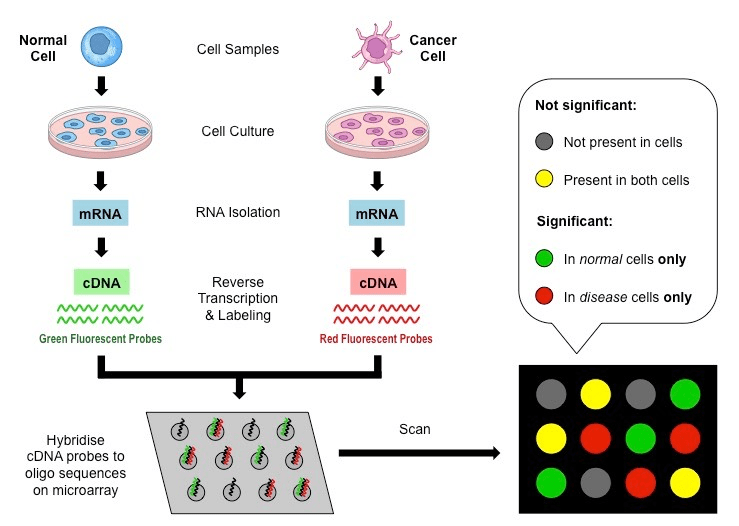
(Now obsolete)
Microarrays have some caveats
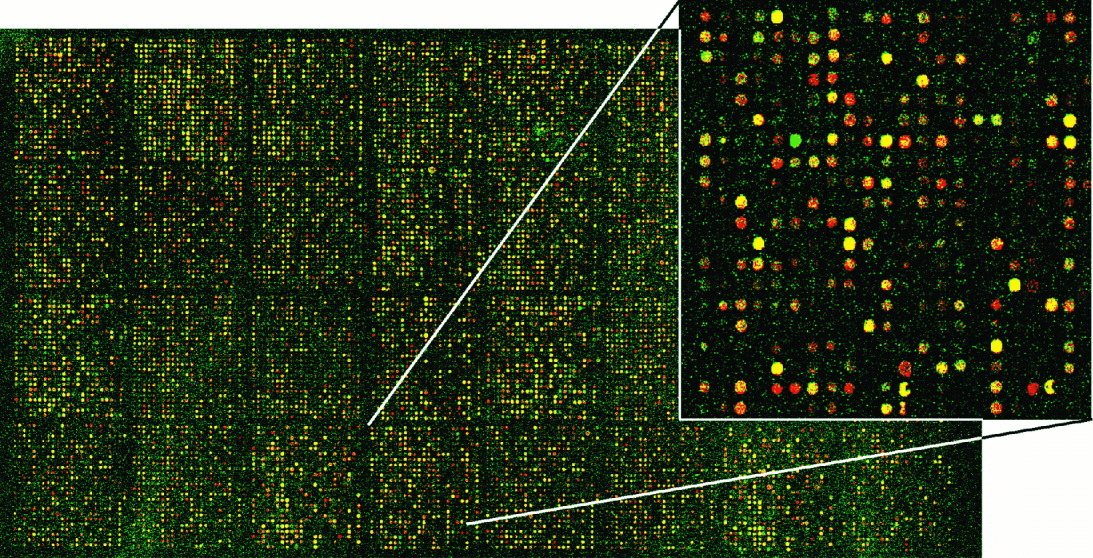
- Limited to known sequences: Can only detect pre-defined sequences
- Cross-hybridization: Similar sequences may cause false positives
- Limited dynamic range: May miss very low or high abundance transcripts
- Normalization challenges: Complex process, potential for bias

RNA sequencing changed the game
Now we just sequence the cDNA
- RNA-seq doesn't require prior knowledge of sequences
- Enables discovery of novel transcripts and isoforms
- Provides absolute quantification rather than relative concentration
Advantages

TopHat questions
What is the primary advantage of RNA-seq over microarray technology?

Which sample has a higher RIN?
After today, you should be able to
1. Define transcriptomics and explain its role in understanding gene expression patterns.
2. Discuss emerging trends in transcriptomics.
3. Compare and contrast transcriptomics and genomics.
4. Explain the principles of RNA-seq technology and its advantages over previous methods.
5. Outline the computational pipeline for RNA-seq data analysis.
Read Alignment:
Mapping Transcripts to the Genome
- Consideration of splice junctions and gene isoforms
- Needs to account for known and novel splice sites
- Requires specialized alignment algorithms (e.g., STAR, HISAT2)
- Critical for accurate transcript reconstruction and quantification

Quantification:
Measuring Gene Expression Levels
- Counting aligned reads with HTSeq or featureCounts
- Transcript-level quantification with Salmon or Kallisto
- Normalization methods: For example, TPM (transcripts per million)
- Distinguishing between different isoforms of the same gene
- Requires probabilistic models for read assignment

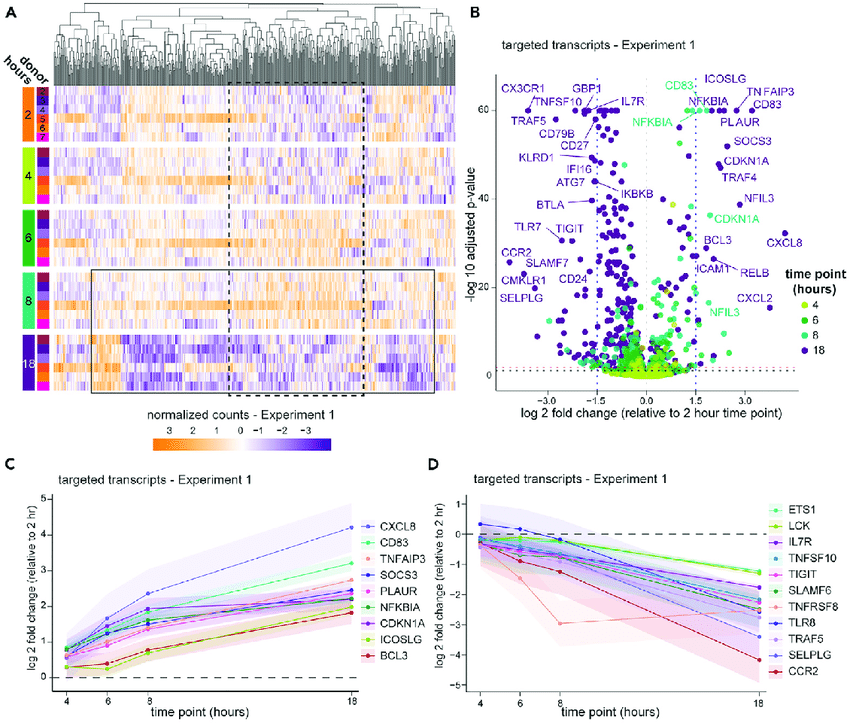
Differential Expression Analysis:
Identifying Key Genes
- Compares gene expression levels
- Statistical testing with DESeq2 or edgeR
- Visualization of results (volcano plots)
- Clustering of differentially expressed genes
- Results in lists of up- and down-regulated genes

High confidence that expression levels changed in these genes
Dimensionality Reduction:
Visualizing Complex Data
- Reduces high-dimensional data to 2D or 3D for visualization
- Reveals patterns and clustering in the data
- Techniques include PCA, t-SNE, and UMAP
- This practice is widely used, but extreme caution needs to be used and is not generally recommended

Group cells and assign types based on gene expression data
Before the next class, you should
Lecture 08:
Read mapping
Lecture 07:
Transcriptomics
Today
Thursday