


Computational Biology
(BIOSC 1540)
Feb 18, 2025
Lecture 07A
RNA Quantification
Foundations
Announcements
- César will provide optional Python recitations on Fridays from 2 - 3 pm (Location TBD).
- Please fill out the Canvas discussion for CBit 06
Assignments
Quizzes
CBytes
CBits
After today, you should have a better understanding of
Quiz 02
Please put away all materials as we distribute the quiz
Fill out the cover page, and do not start yet
Sit with an empty seat between you and your neighbors for the quiz
Quiz ends around 9:51 am
When you are finished, please hold on to your quiz and feel free to doodle, write anything, tell me a joke, etc. on the last page
After today, you should have a better understanding of
The importance of quantification in RNA-seq data analysis
Let's remember the big picture: We want to quantify gene expression differences
Suppose we have isolated a normal and cancerous cell
Cancerous
Normal
We want to identify possible drug targets based on overexpressed genes
We have to quantify the amount of transcripts in our cell(s)
Our transcriptome is the set of mRNA transcripts our cell could have
Let's consider only three transcripts
They have short, medium, and long lengths


We first have to define what possible transcripts our cells have
RNA quantification aims to determine the number of transcripts in my original sample


Normal
Cancerous
Example distribution of mRNA under different conditions
The RNA quantification problem statement
Transcriptome
Reads/Fragments
Given the sequencing reads that were sampled from these transcripts
How many copies of each transcript were in my original sample?
Unknown quantity
Experimental biases and errors
After today, you should have a better understanding of
The relevance of pseudoalignment
One way we could quantify transcripts is to use read mapping (i.e., read alignment)
Transcriptome
Reads/Fragments
We align each read to single transcript using our read mapping algorithms
1
2
4
Number of reads mapped to each transcript
Example: Bowtie 2 uses a FM-index for read mapping
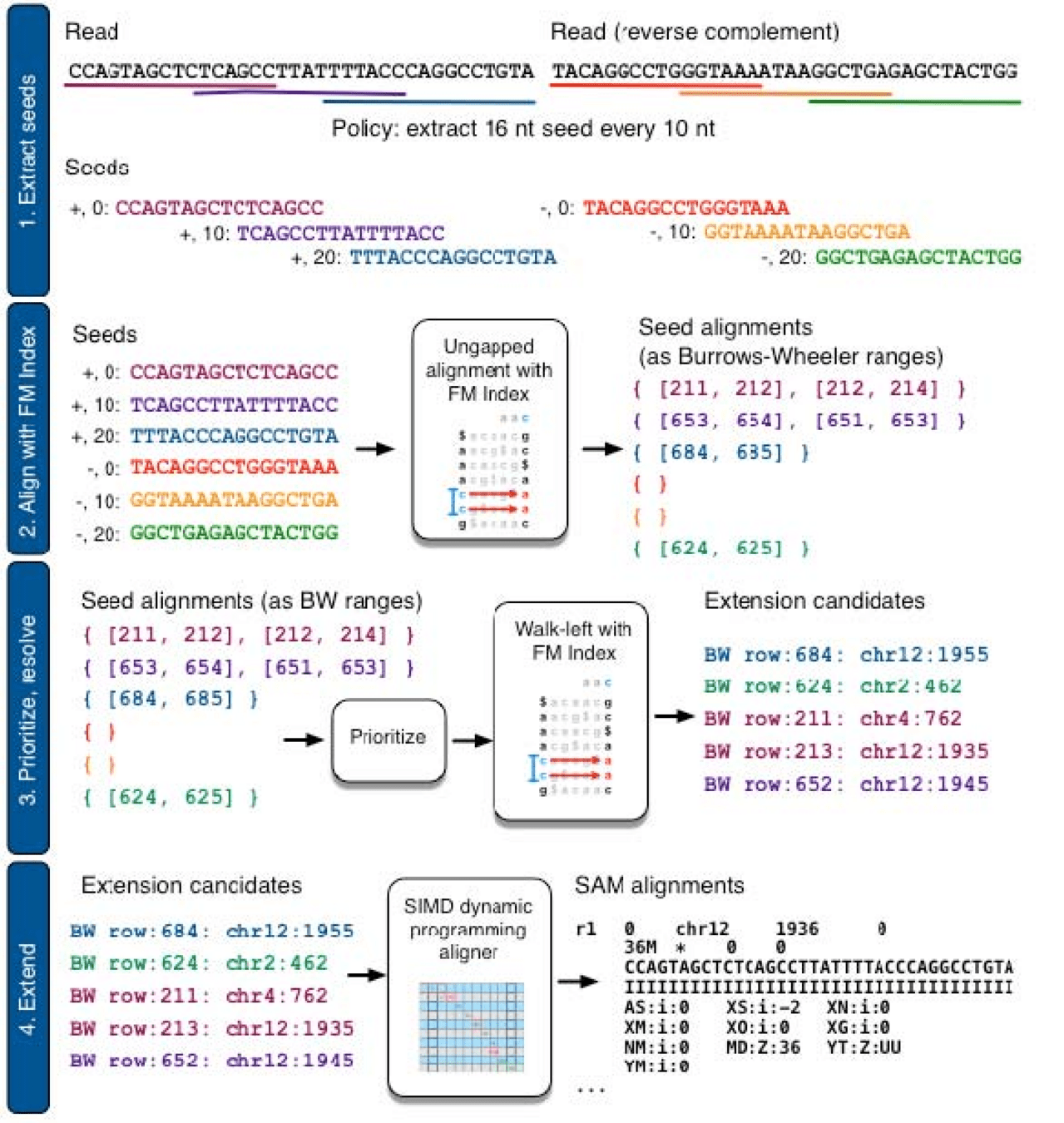
DOI: 10.1038/nmeth.1923
1. Extract k-mer seeds from all reads in our sample

2. Use FM-index of transcriptome to search for k-mer matches

3. Extend alignments starting from k-mer seeds
These approaches are extremely slow (we have 30 to 60 million reads)
Suppose someone took library books (transcripts) and then shredded them (reads)

We would need to find which book each piece came from, but also which page
This would take a long time
Relating it back: Alignment-based methods need to determine the read's exact position in the transcript
Alignment-based methods are computationally expensive
Why?
We want to tape these books back together by using another library's copy as a reference
Strategy: Identify which transcripts are compatible with the read, skipping the precise location (i.e., alignment) step
Pseudoalignment finds which transcript reads came from but not the exact position
Instead of having to find the exact page each piece is from, suppose we just need to figure out which book

This is all we need to know for RNA quantification: Where did this read come from?
Alignment
Pseudoalignment
Specifies where exactly in the transcript this read came from
(e.g., at position 478)
Specifies that it came somewhere from this transcript (i.e., compatible)
Reads
Transcripts
Bypassing alignment accelerates quantification, but how can we do this?
Comparison
After today, you should have a better understanding of
Understand the use of generative models
Let's understand our problem
Initial sample
We can only use reads to quantity our initial mRNA sample
Has some number of transcripts
Fragments
Reads
After PCR amplification and fragmentation
Sequencing with errors
We can use a generative model to back-calculate transcript quantities from our reads
Generative model: A statistical model that explains how the observed data are generated from the underlying system
Defines a computational framework that produces sequencing reads from a transcriptome
Let's walk through a conceptual example
Exploring generative modeling with a bag of marbles
Suppose we take some number of green, blue, and red marbles (i.e., transcripts)
We put the marbles into a bag and crush them (i.e., PCR amplification and fragmentation)
Exploring generative modeling with a bag of marbles
We then take a handful of marble fragments (i.e., reads)

How would you estimate the distribution of marbles in the bag?
and then determine their color (i.e., map)
Suppose our marbles have different characteristics
Red marbles are twice the size
These differences are called biases
Blue marbles break easily
How would you adjust your approach?

Generative models estimate parameters that explain our observed experimental results
1. Guess some number of marbles
0. Determine/learn bias probabilities of sampling that specify our framework
2. Randomly sample n fragments
3. Compare to our original distribution
- The probability of grabbing a blue fragment is 1.4x the green probability
- The probability of grabbing a red fragment is 2.0x the green probability
If our simulated distribution closely matches our observed distribution, we have a likely estimate of the original (hidden) distribution
Does not match
We need to maximize the probability that our generative model and parameters explain our observations
1. Guess some number of marbles
2. Randomly sample n fragments
We keep trying new marble combinations until our generated fragments look very similar to our observed fragments
This is conceptually similar to how RNA quantification methods work (e.g., Salmon)
After today, you should have a better understanding of
Python practice
Before the next class, you should
Lecture 07B:
Quantification -
Methodology
Lecture 07A:
Quantification -
Foundations
Today
Thursday
Wait, what about sequencing depth?
Longer transcripts will have more reads
Read per kilobase (RPK) corrects this experimental bias through normalization by gene length
(Length is usually just the exons)
RPK example
Reads per kilobase of transcript per million reads mapped
Transcripts per million
Learning bias parameters
For the first 5,000,000 observations we learn these defined bias parameters
Probability of generating fragment j from transcript i
Probability of drawing a fragment of the inferred length given t
Probability of fragment starting at position p on t
Probability of obtaining a fragment with the given orientation
Probability of alignment of fragment j given these mapping and transcript conditions
Spliced Transcripts Alignment to a Reference (STAR)
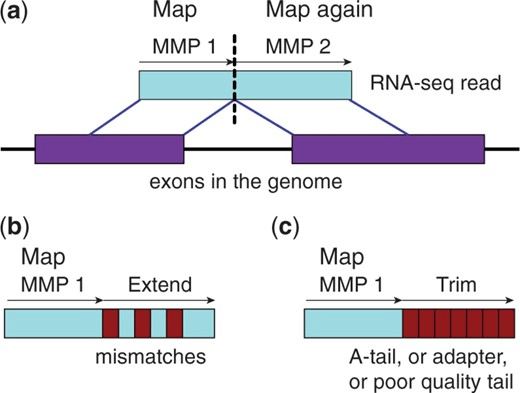
Maximum Mappable Prefix (MMP) approach for fast, accurate spliced alignments
Finds prefix that perfectly matches reference then repeats for unmatched regions
This automatically detects junctions instead of relying on databases
Probability of alignment fragment to transcript
Represents the probability of the alignment starting at a particular position or state sos_o
The probability of the fragment "transitioning to another state"
Essentially, how likely does this fragment align somewhere else
For fun: Hidden Markov Models
We have to normalize our transcriptome before making comparisons
2
3
4
We can use transcripts fraction
Ratios are sensitive to total amount


Normal
Cancerous
Because the cancer cell is transcribing more overall, we still get changes across the board
Scaling data to "parts per million"
Real data has more than three transcripts and ratios are substantially smaller (e.g., 0.000001)
This can make computations and communications challenging, so we often scale everything to a million to use unsigned integers
| Transcript | Normal | Cancerous |
|---|---|---|
| 1 | 222,222 | 266,666 |
| 2 | 333,333 | 400,000 |
| 3 | 444,444 | 333,333 |
| Total | 1,000,000 | 1,000,000 |
Small floats require high precision (i.e., float64) and thus memory