
Computational Biology

(BIOSC 1540)

Oct 8, 2024
Lecture 11:
Structural biology
Announcements
- A03 and A04 will be graded this week
- A05 will be posted on Friday
- We are now entering the world of structural biology (Physical chemistry and Biochemistry)

After today, you should be able to
Categorize atomic interactions and their importance
The atomic world of biology
At the foundation of biological processes lie atoms and their interactions
Atomistic structure determines behavior of biomolecules
Why is Green Fluorescent Protein (GFP) fluorescent, but not the chromophore in solution?
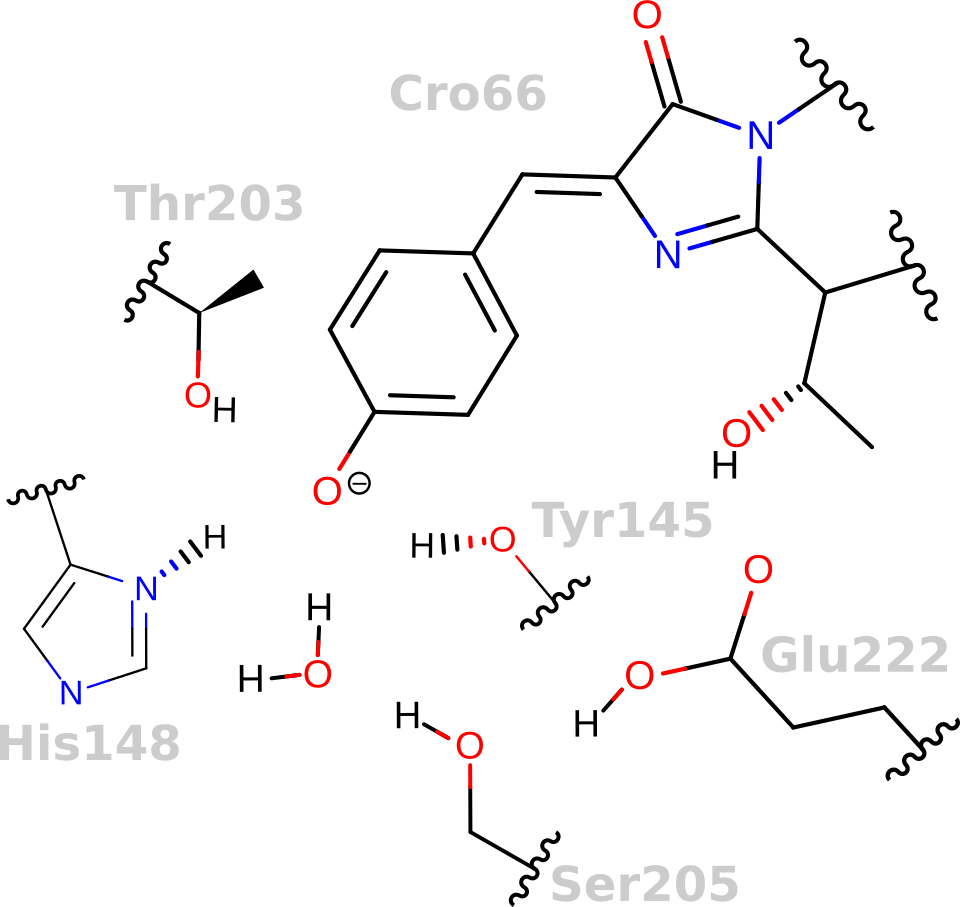
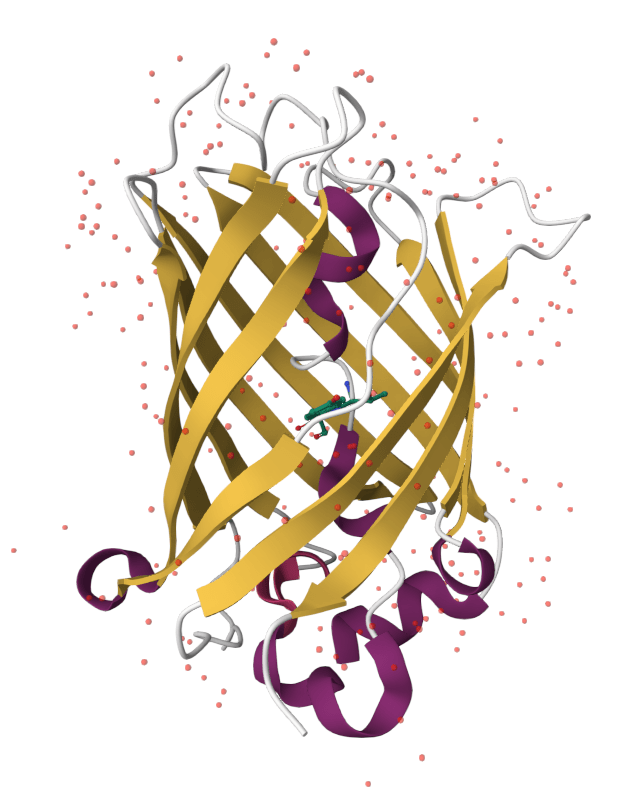
GFP keeps the chromophore planar and facilitates an excited-state proton transfer
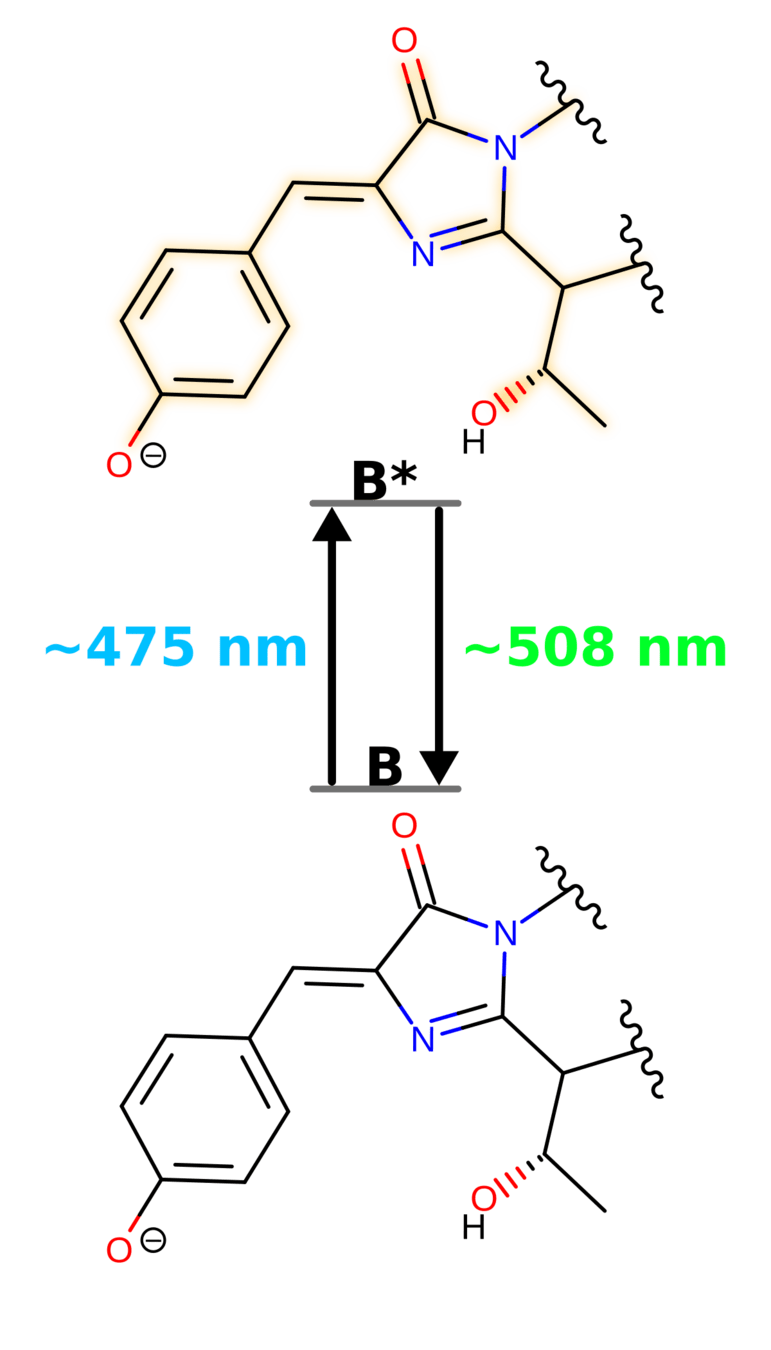
Fluorescent
Not Fluorescent
Two types of atomistic interactions
Covalent
Noncovalent
Covalent bonds: The framework of biomolecules
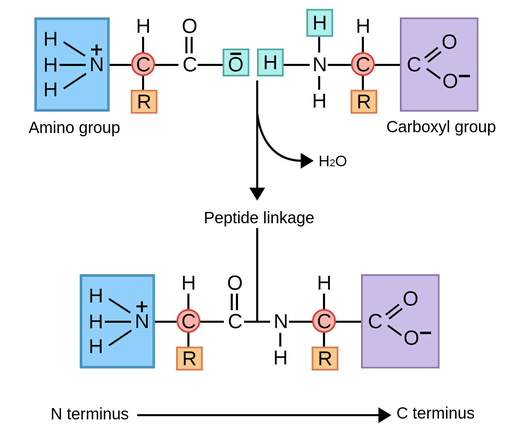
- Peptide bonds covalently link amino acids into polypeptide chains
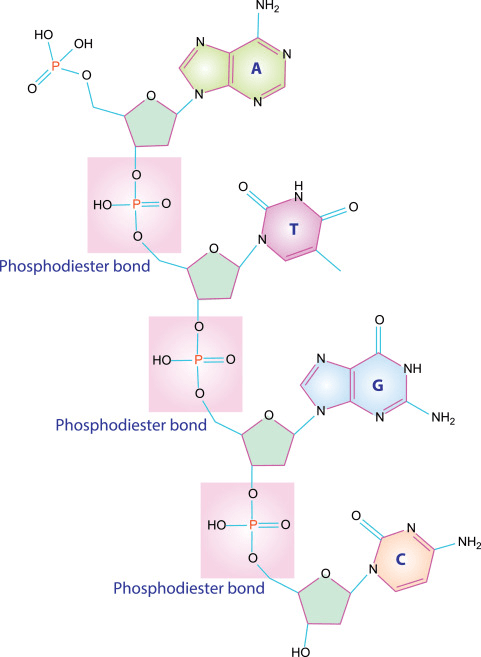
- Phosphodiester bonds form the sugar-phosphate backbone of DNA and RNA
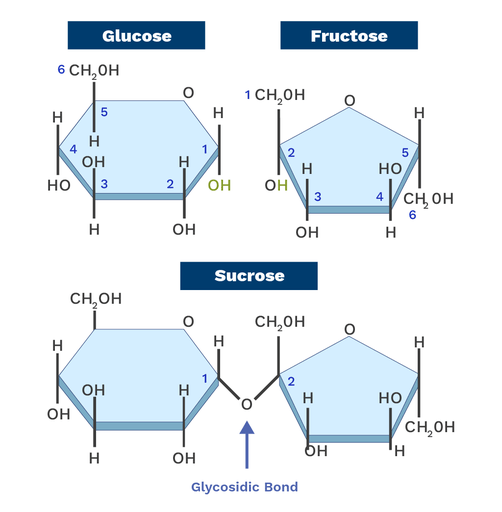
- Glycosidic bonds join monosaccharides to form complex sugars
Covalent bonds are formed when atoms share pairs of electrons that holds molecules together
Relevant characteristics of covalent bonds
- Single Bonds: Allow rotation, contributing to molecular flexibility
- Double/Triple Bonds: Restrict rotation, affecting the rigidity and function of molecules
Strength and stability: Covalent bonds provide the necessary stability for complex biological structures
Directionality: Covalent bonds limit the specific angles and orientations leading to the 3D shapes of biomolecules
Two types of atomistic interactions
Covalent
Noncovalent

Noncovalent Forces: The Dynamic Glue
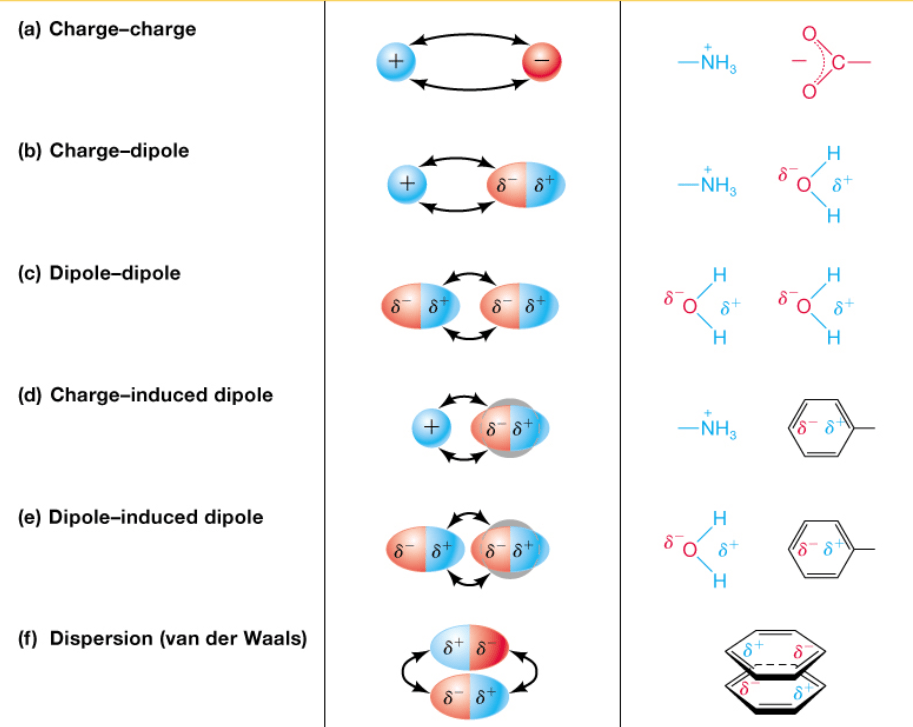

Noncovalent interactions are weaker than covalent bonds and involve electrostatics
We will cover this in a later lecture
Noncovalent interactions drive most of biology
Macromolecular structure
-
Membrane Formation
-
Protein-Protein Interactions
- Base pairing in DNA and RNA
- Protein folding
Molecular recognition
- Enzyme-Substrate Binding
- Antigen-Antibody Interactions
After today, you should be able to
What is structural biology and why is it important?
What is the value of atomistic insight?


The precise arrangement of atoms determines how molecules fold, bind, and perform biological tasks
We cannot exploit what we do not understand
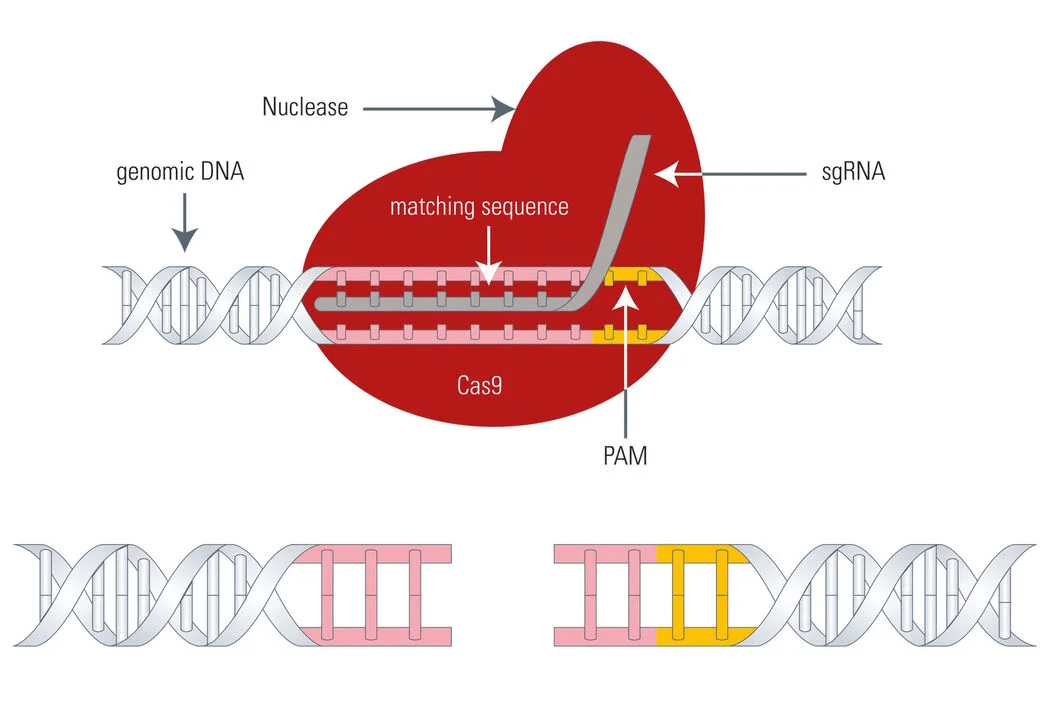
CRISPR-Cas9
COVID-19 treatments
High-throughput sequencing
Innovation and biotechnology depend on molecular understanding
What is structural biology?
Structural biology determines the 3D shapes of biological macromolecules and how these shapes relate to function
Why study structure?
- Proteins and nucleic acids adopt specific shapes crucial for their biological roles.
- Example: The shape of an enzyme’s active site determines how it binds substrates and catalyzes reactions.
Primary Goal: To understand how molecular machines in cells work by deciphering their atomic arrangements.
Primary structure
The primary structure of a protein is the linear sequence of amino acids, held together by covalent peptide bonds
The primary structure is crucial because it dictates how the protein will fold into higher-order structures
The primary structure alone does not reveal the protein's functional form or activity
While the primary sequence is critical, the folding process may also depend on cellular factors (e.g., chaperones)
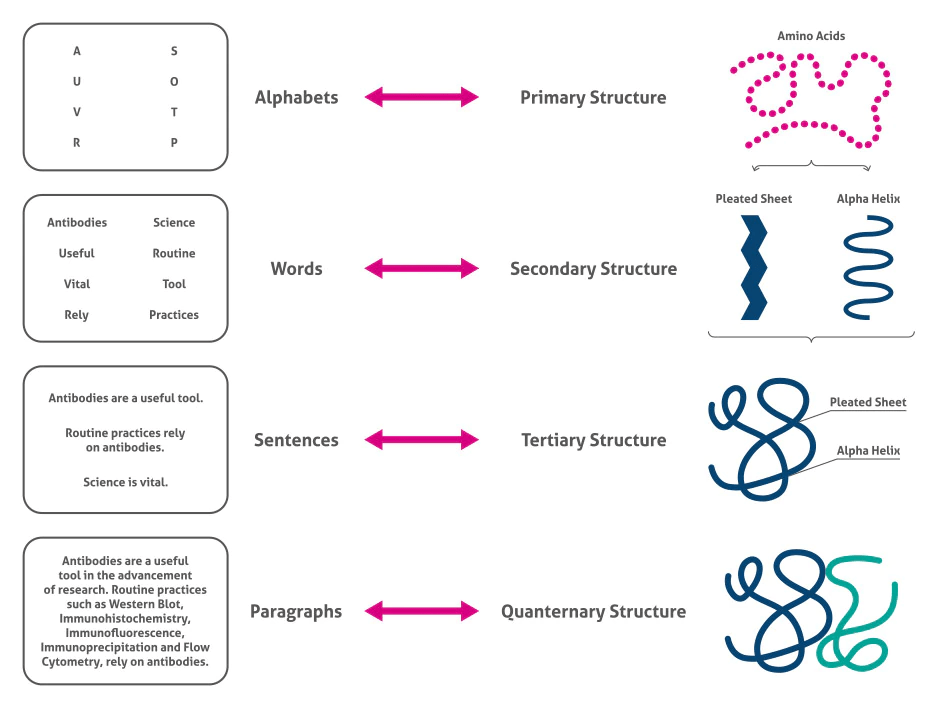
Secondary structure
The secondary structure refers to local conformations of the polypeptide chain, stabilized primarily by hydrogen bonds
Secondary structures can undergo local fluctuations—alpha helices can unwind, and beta-sheets can twist—adding to functional flexibility

These structural motifs are critical for certain functions
- For example, DNA-binding domains often contain alpha-helices
While alpha-helices and beta-sheets dominate, other structural motifs (e.g., 310 helices) are less common and sometimes overlooked
Tertiary Structure
The tertiary structure refers to the complete 3D shape of a single polypeptide chain
Tertiary structures reveal active sites or binding pockets where catalysis or molecular interactions occur

Predicting how a sequence folds into its tertiary structure is complex, even with knowledge of secondary structures
After today, you should be able to
Explain the fundamentals of electron density
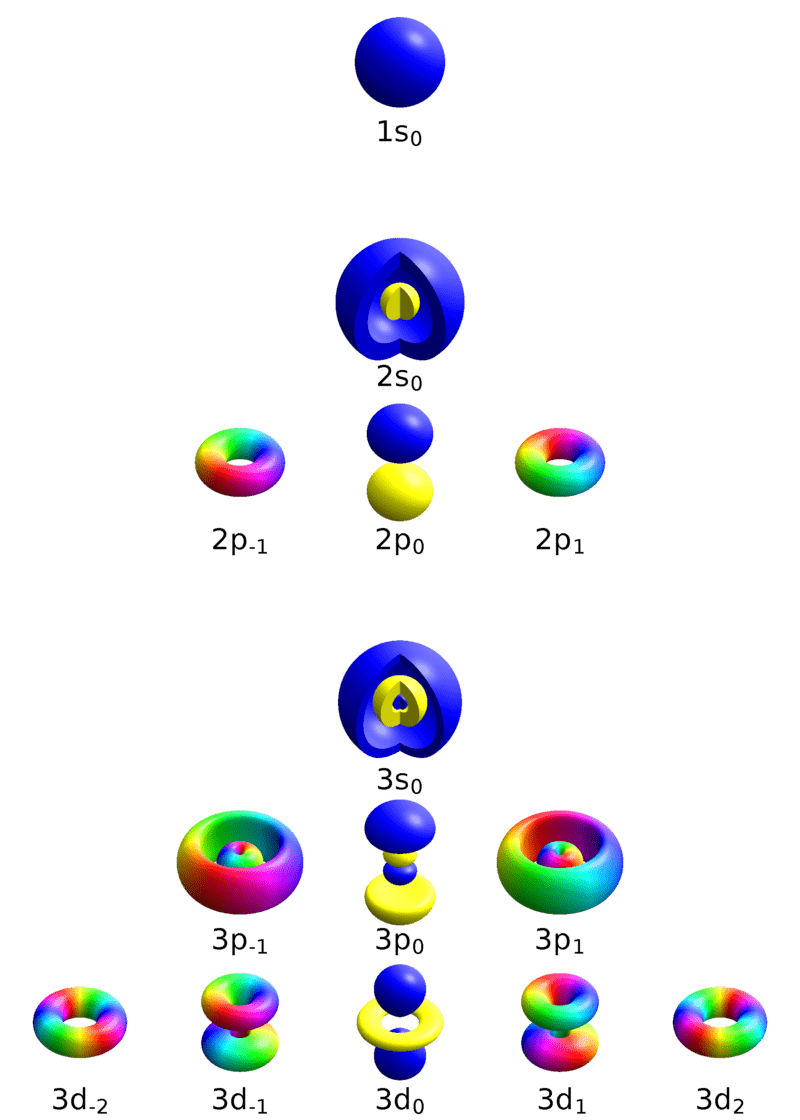
Particle behaviors are determined by quantum numbers
Principle quantum number
1
2
3
Orbital quantum number
0
-1
0
1
-1
0
1
-2
2
Magnetic quantum number
1
1
2
3
2
1
(You don't need to know what these mean)
An electron at (n, l, m) will have a specific energy level and characteristics
Each atom contributes electrons to the molecule
Benzene has . . .
Six carbon atoms with 1s2 2s2 2p2
Six hydrogen atoms with 1s
located at the center of each atom's position












Electrons "mix" into molecular orbitals to a specific energy level
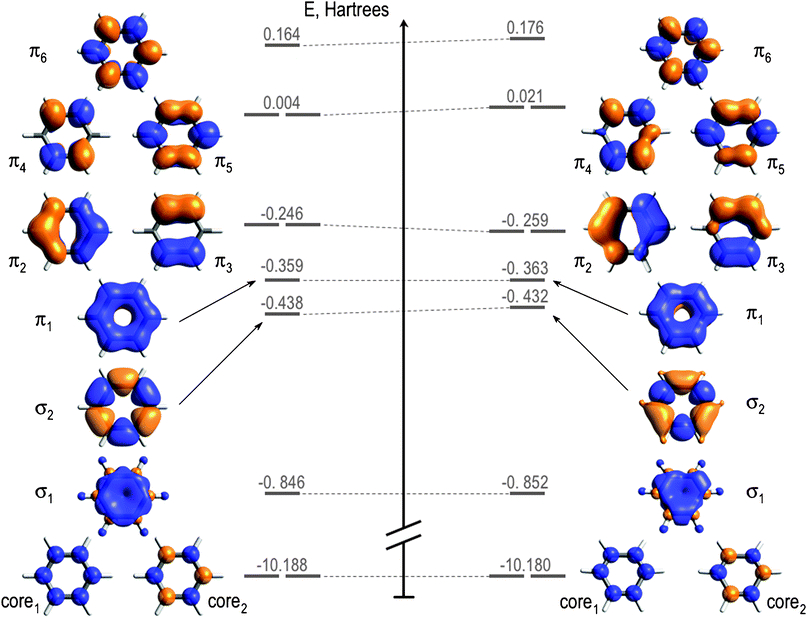
Particles (e.g., electrons and photons) can interact with these molecular orbials
D6h structure
D3h structure
These molecular orbitals determine behavior
Changing the positions (or symmetry) change molecular orbitals
Result: An electron density distribution unique to that structure
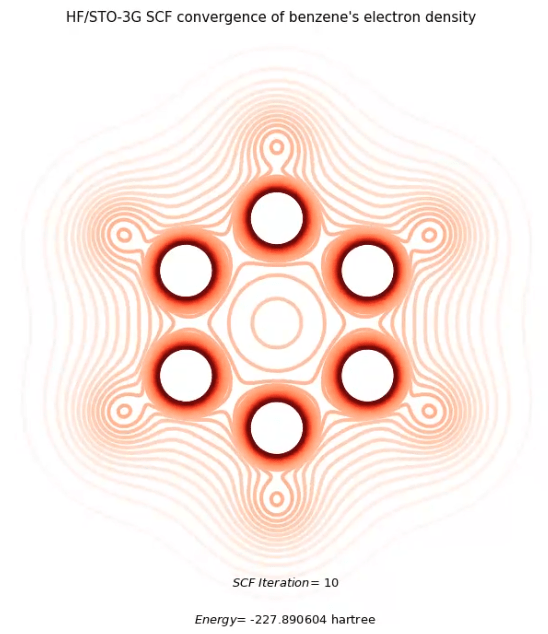
All experimental techniques are based on probes interacting with molecule's electron density to reveal structural information
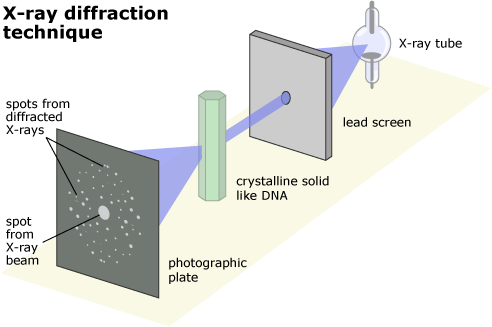
- X-ray Crystallography: How a crystal of molecules diffracts X-rays
- NMR Spectroscopy: How atomic nuclei interact with magnetic fields and radiofrequency pulses
- Cryo-Electron Microscopy: How molecules scatter electron beams
Electron density of benzene
After today, you should be able to
Communicate the basics of X-ray crystallization
Fundamentals of X-ray Crystallography
Basic Principle: Photons scatter when they interact with atoms
The scattered X-rays form a diffraction pattern unique to the crystal
Probe: Photon (carrier of electromagnetic radiation)
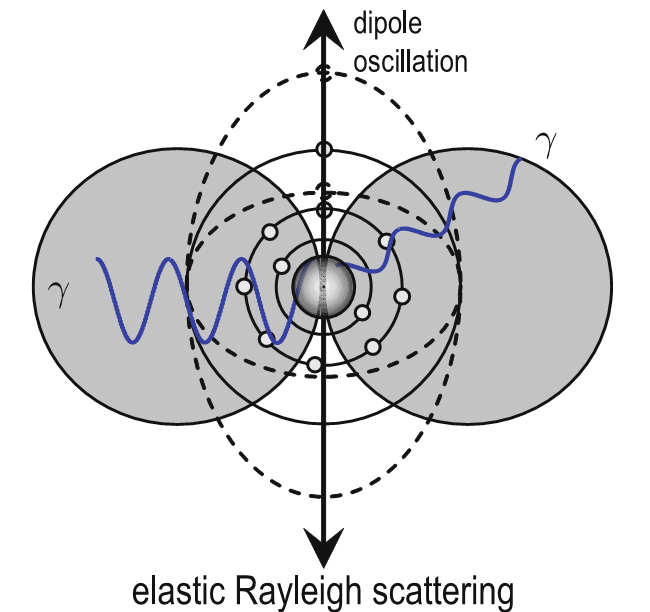
X-rays undergo elastic scattering by electrons
- Incident photon induces an oscillating dipole by distorting the electron density (Rayleigh)
- An oscillating dipole acts as an electromagnetic source and re-emits photons at the same wavelength in all directions
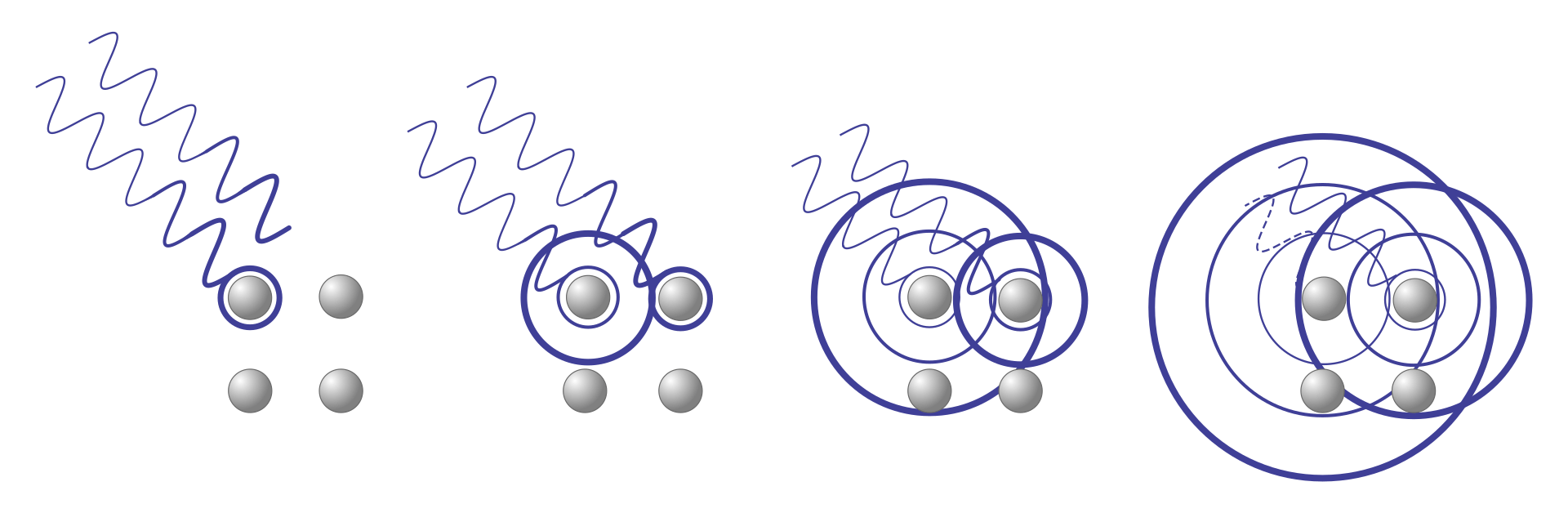
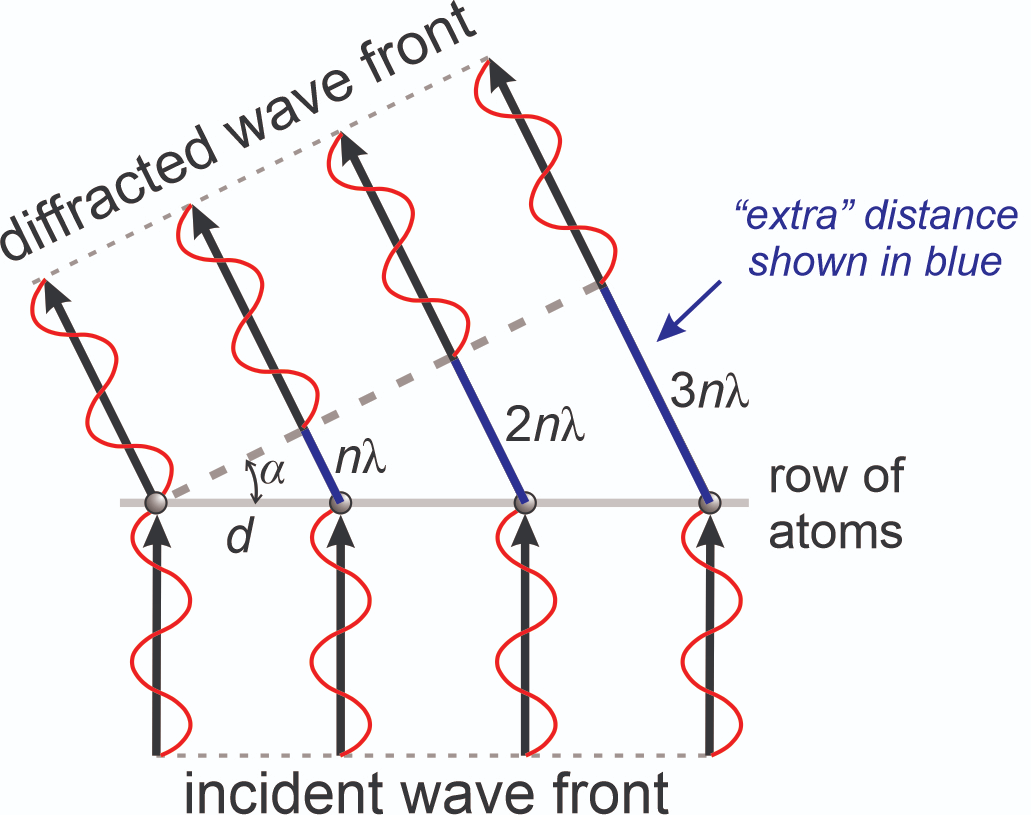
What happens when two waves overlap?
Constructive interference is needed to amplify signal for detectors
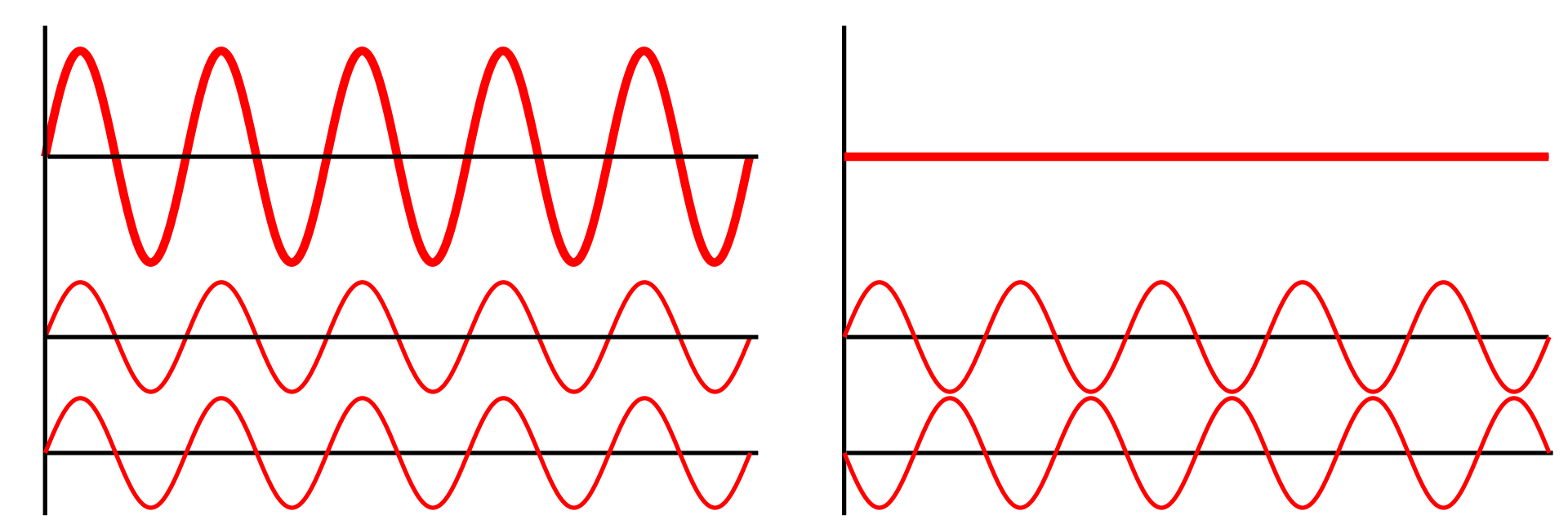



If wavelengths are similar and in phase, they constructively interfere
If waves are out of phase, they deconstructively interfere
Constructive interference leads to distinct patterns
If wavelengths are similar and in phase, they constructively interfere and form spots based on atom type and distance
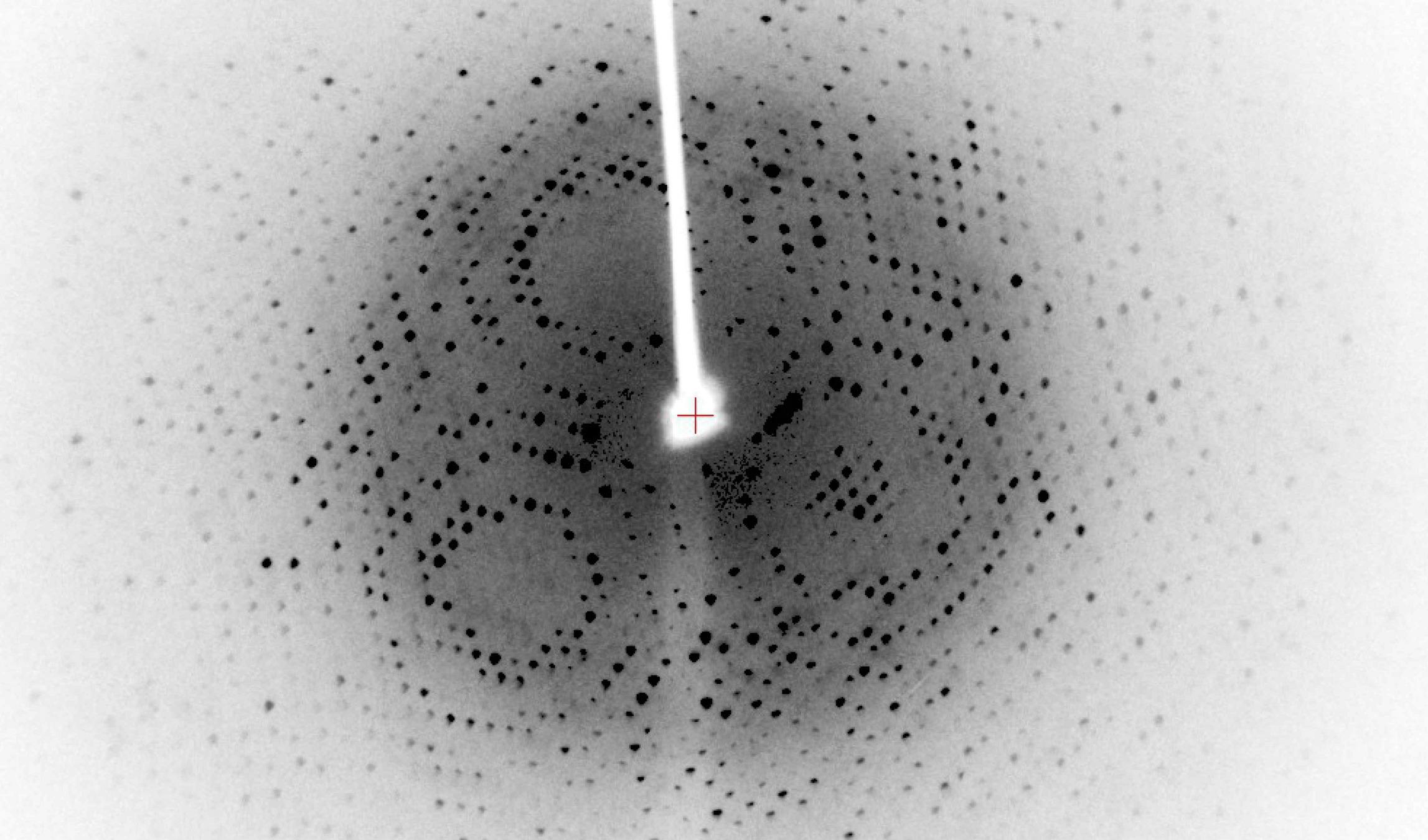
The diffraction pattern
The spots on the detector represent the reflections of the scattered X-rays
- Intensity of the spots reflects the electron density in the crystal
- Position and angle: The position of the spots corresponds to the geometry
The diffraction pattern does not directly show the atomic positions, but provides the data needed to infer the electron density

Building the electron density map
The 3D electron density map reveals the distribution of electrons in the crystal, indicating where atoms are located
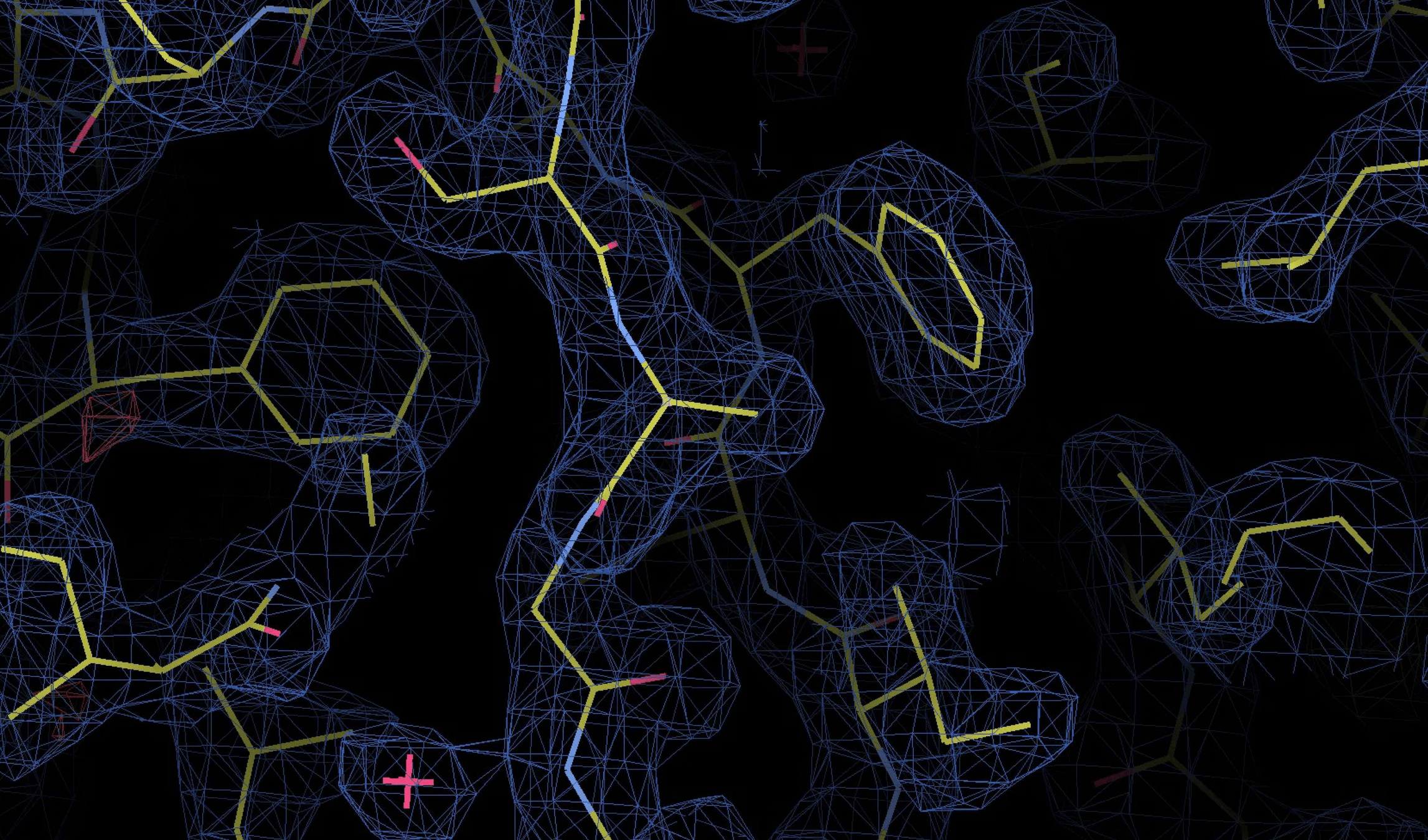
The electron density map is interpreted by fitting atomic models (e.g., amino acids for proteins) into the density
Low-resolution data make it difficult to assign atomic positions precisely, leading to uncertainty in the model
Why do we need crystals?
Molecules
Crystals
Crystals have the same repeating unit cell, which amplifies our signals
If in solution, particles would be
- Too sparse to diffract
- Moving and diffraction pattern would constantly change
What actual protein crystals look like
After today, you should be able to
Find and analyze protein structures in the Protein Data Bank (PDB)
UniProt is a protein information database
Let's find information about our project's drug target: Dihydrofolate reductase
UniProt is a comprehensive database to access curated data about protein structures, functions, sequences, and annotations.
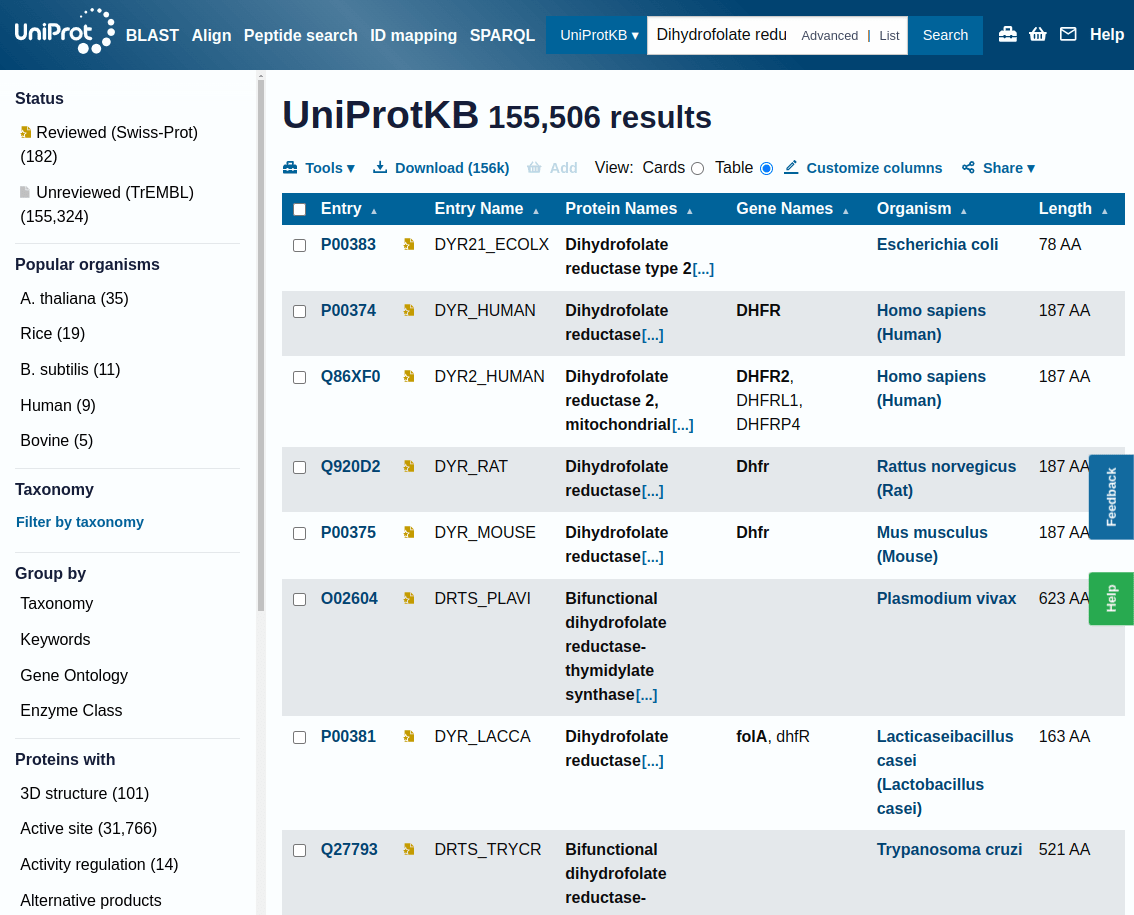
This page shows the results of a search in UniProtKB for a specific protein, in this case, "Dihydrofolate reductase"
On the left side, you have multiple filters to narrow your search results:
-
Reviewed (Swiss-Prot): Experts manually curated and verified these entries, ensuring high accuracy
-
Unreviewed (TrEMBL): These entries are automatically generated and have not been manually reviewed
Each row in the table represents a different protein entry
Entry ID: A unique identifier for the protein (e.g., P00383). You can click on this ID for detailed information about the protein
Protein Data Bank contains structures
After today, you should be able to
Compare and contrast Cryo-EM to X-ray crystallization
Why Cryo-EM?
In Cryo-EM, a beam of high-energy electrons is used instead of photons
Why Electrons?
- Electrons have a much shorter wavelength (~0.02 Å at 300 keV) than photons
- Light elements which scatter electrons more effectively than X-rays
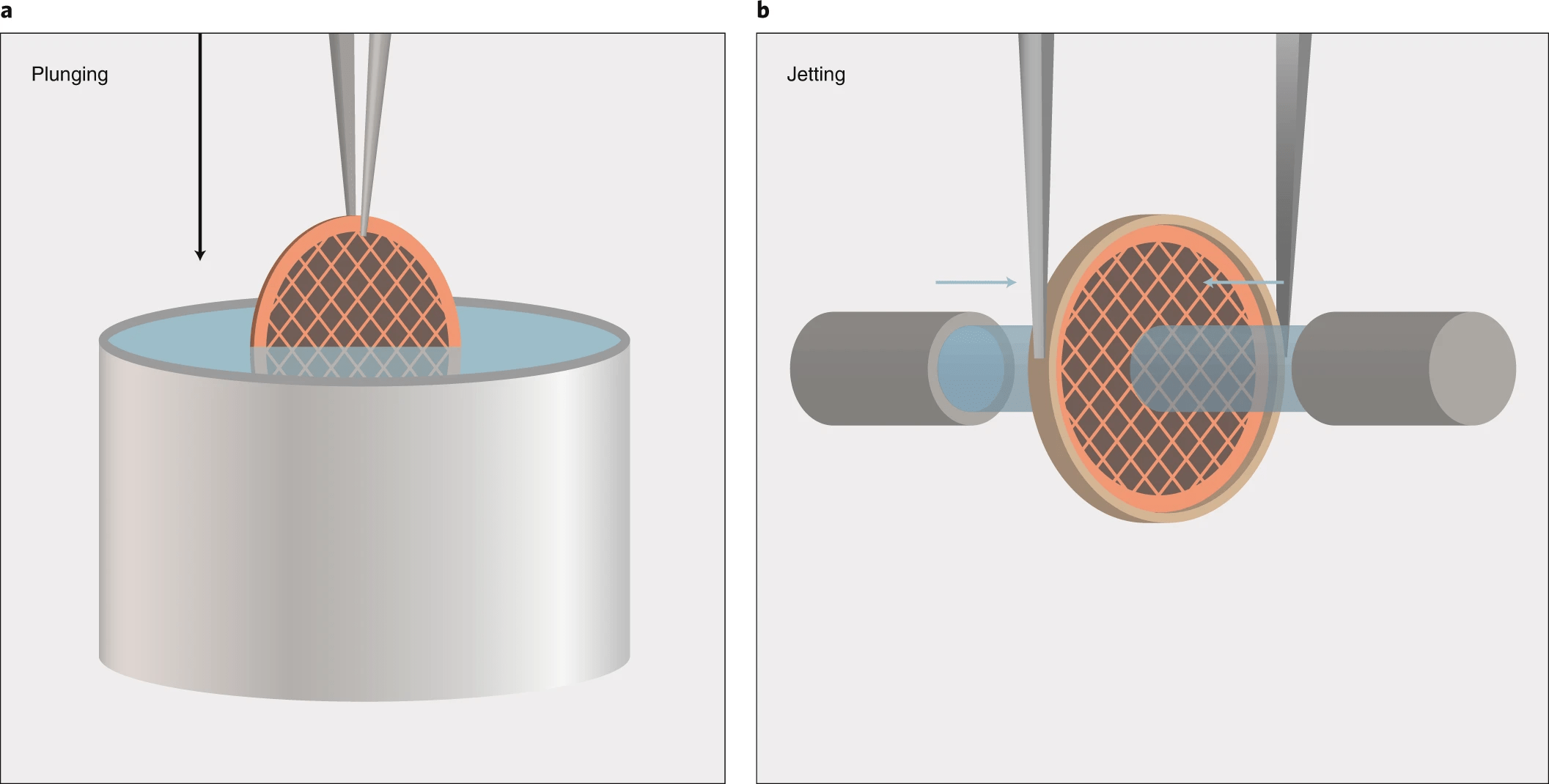
No crystals: The sample is rapidly frozen in vitreous ice to preserve its native structure
- By freezing the sample, biological molecules are imaged in their native hydrated state
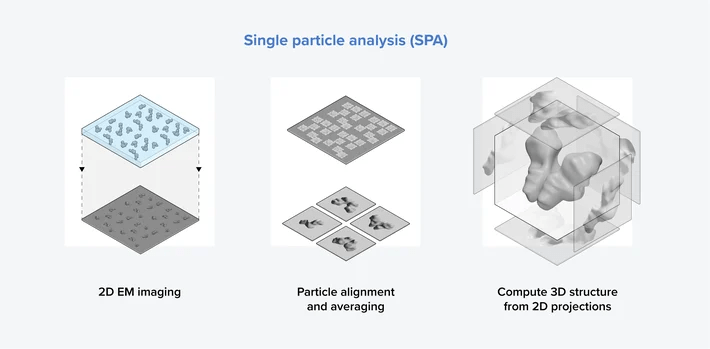
Single Particle Analysis (SPA)
Single Particle Analysis is the main Cryo-EM technique used to determine the 3D structures of individual macromolecules
- Millions of images of individual particles are collected from a thin layer
- Particles are computationally aligned and classified into different orientations


After today, you should be able to
Communicate the challenges of disorder
Challenge of flexibility and disorder in biomolecules
Molecules are not static
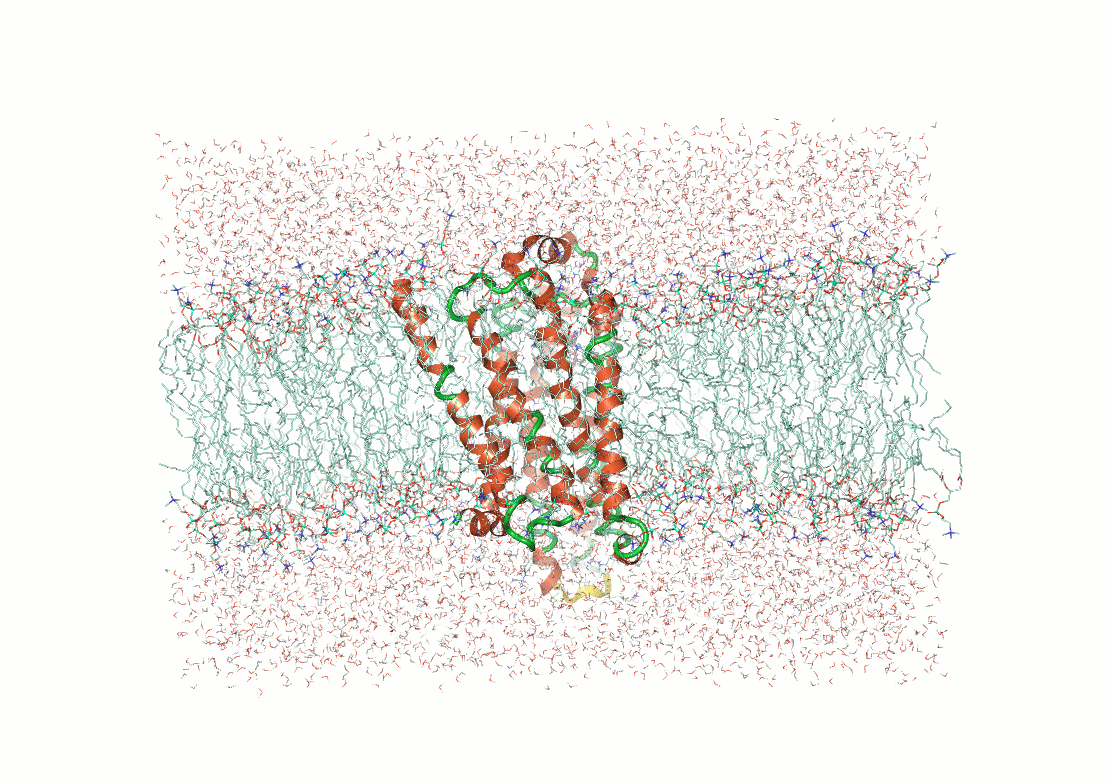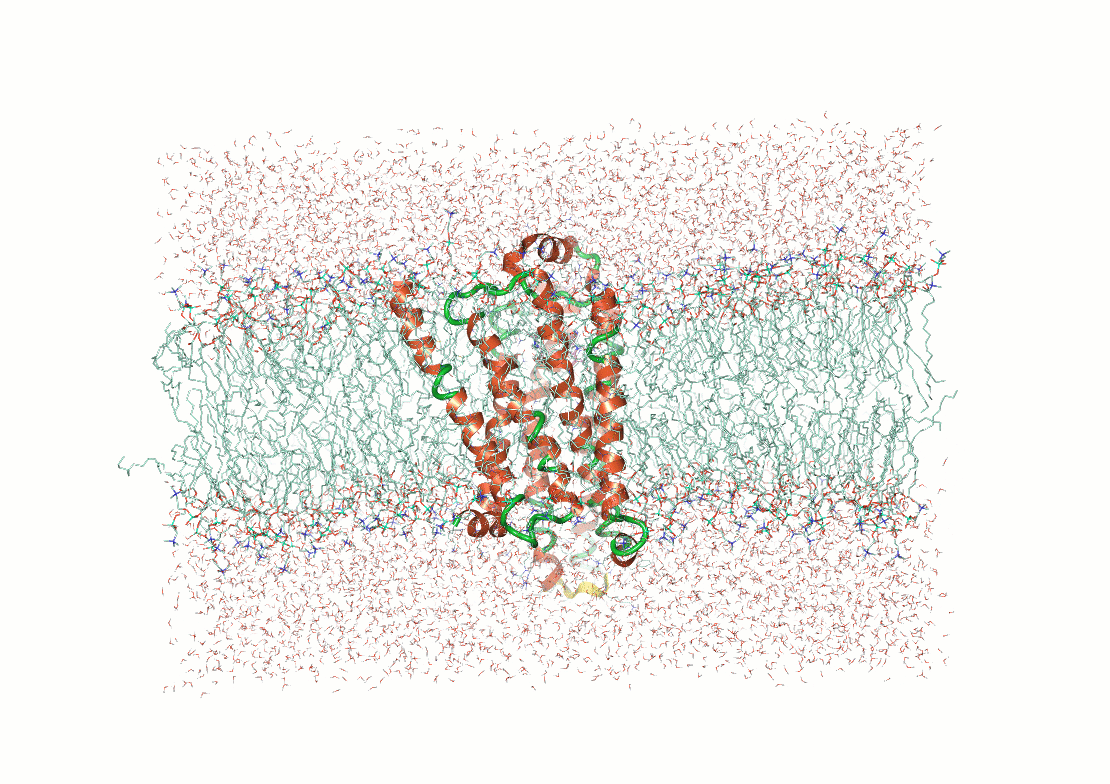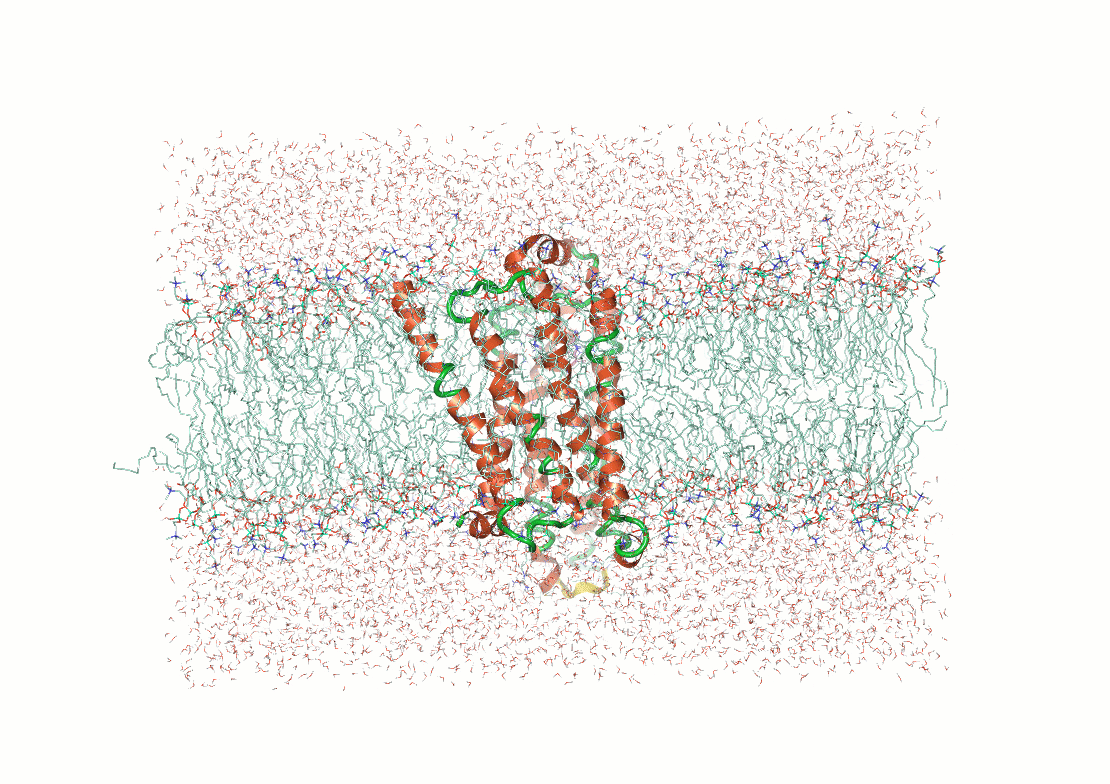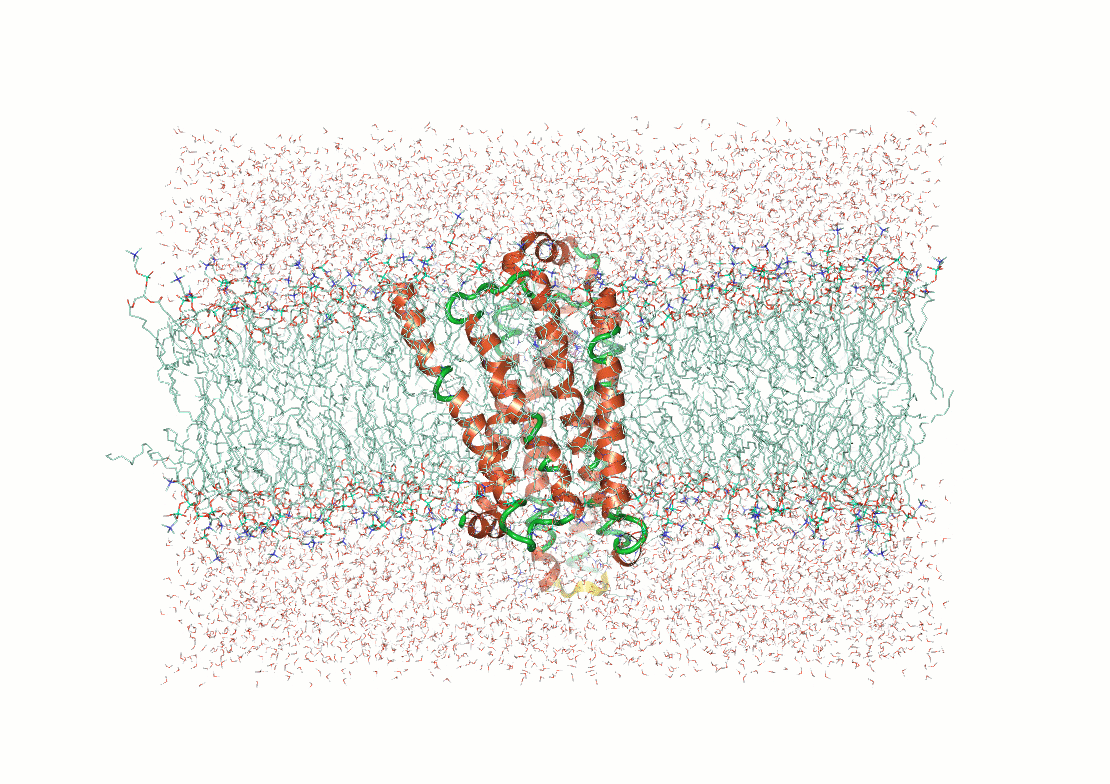
Example: The p53 tumor suppressor protein has flexible regions critical for its regulation and binding interactions
Proteins often exhibit flexibility, disordered regions, and multiple conformations
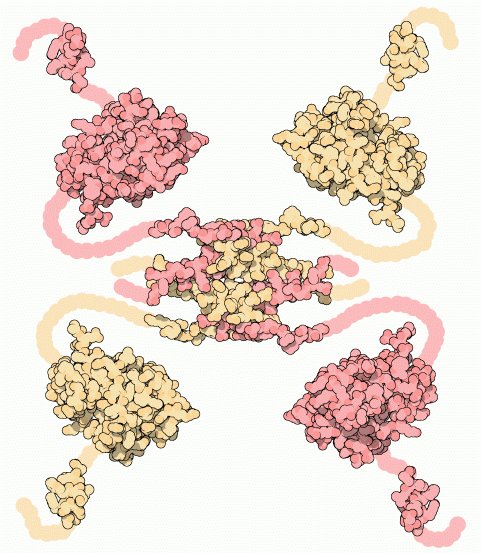
Why It Matters: Structural techniques often require ordered or stable configurations
Challenges in X-ray Crystallography
- Flexible or disordered regions do not pack into crystals well, often leading to failure in obtaining high-quality crystals.
- Even in cases where crystallization is successful, flexible or disordered regions often do not show up clearly in the electron density map.
- Crystals capture a single conformation of the molecule, often ignoring the flexibility or dynamic range.
Cryo-EM and Conformational Flexibility
One strength of Cryo-EM is its ability to capture multiple conformational states of a molecule, providing insights into flexibility and structural heterogeneity.
Challenge: A major issue in Cryo-EM is that highly flexible or disordered molecules may appear as fuzzy or low-resolution regions in the final structure
Advanced computational techniques are required to sort out different conformations present in the Cryo-EM data
Intrinsically Disordered Proteins (IDPs)
Intrinsically disordered proteins (IDPs) or regions lack a stable 3D structure under physiological conditions but are still functional, often gaining structure upon binding to partners
Conformational Heterogeneity and Biological Function
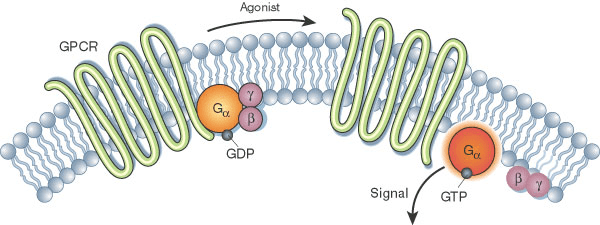
Many proteins function by switching between different conformations, which is essential for their activity (e.g., enzymes, transporters, and receptors).
- Example: G-protein coupled receptors (GPCRs) adopt different conformations when bound to different ligands, triggering different cellular responses.
After today, you should be able to
Know why protein structure prediction is helpful
Challenges in Experimental Structural Biology
Technical Limitations
- Difficulty in capturing dynamic and flexible regions.
- Incomplete structures due to unresolved disordered regions.
Biological Complexity
- Dynamic conformational ensembles not represented in static snapshots.
Resource Constraints
- Time-consuming and costly experiments.
Before the next class, you should
- Review today's lecture
Lecture 11:
Structural biology
Today
Thursday
Lecture 12:
Protein structure prediction