

Computational Biology
(BIOSC 1540)
Mar 25, 2025
Lecture 11A
Protein structure prediction
Foundations
Announcements
Assignments
Quizzes
Final exam
After today, you should have a better understanding of
Identify what makes structure prediction challenging
Protein structure is essential for understanding biological function
Proteins are molecular machines; their 3D shape determines how they interact with substrates, DNA, other proteins, etc.
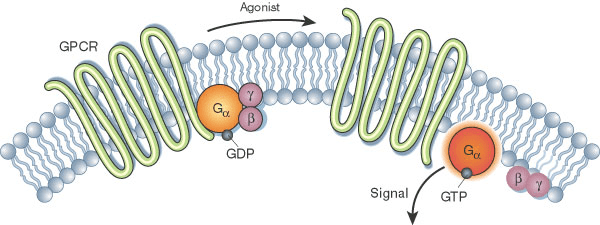
Experimental methods for structure determination are powerful but limited
As of 2021, >200 million protein sequences exist, but <200,000 structures are known

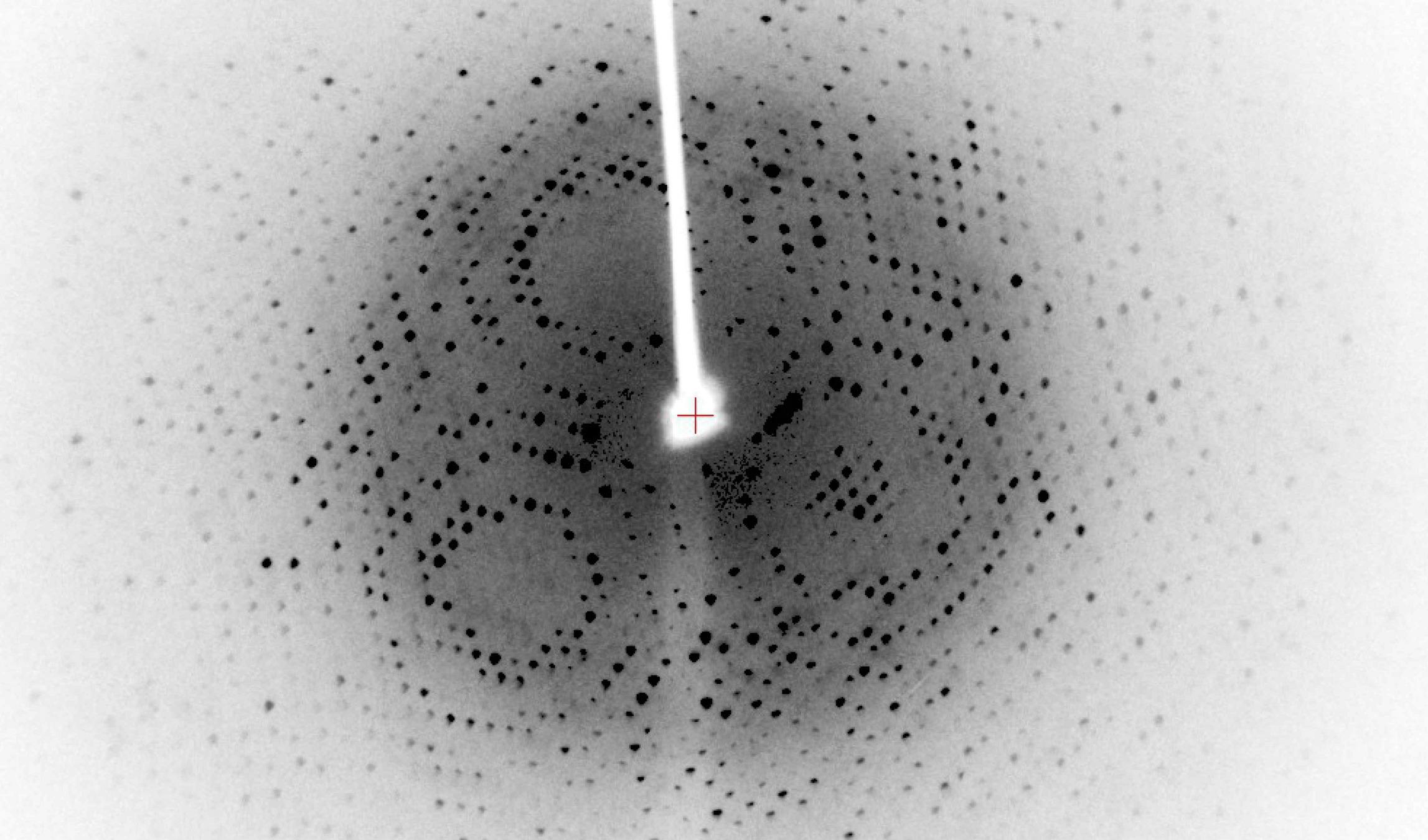
X-ray crystallography, NMR, and cryo-EM provide high-resolution data.
However, these methods are time-consuming, expensive, and often fail for specific proteins.
Protein structure prediction fills a crucial gap in biological discovery
Enables structural understanding of sequences with no experimental structure. This accelerates many research fields and democratizes access to atomistic insights
Example: Our collaborators (Dr. Cahoon) crystallized Lm PrsA1 in 2016, but we need a structural model of PrsA2. Instead of potentially years, AlphaFold 3 gives us a decent prediction within minutes
PrsA1 (X-ray)
PrsA2 (AF3)
Protein folding is computationally hard due to the vast conformational space
Proteins fold in milliseconds—implying nature doesn't sample all conformations.
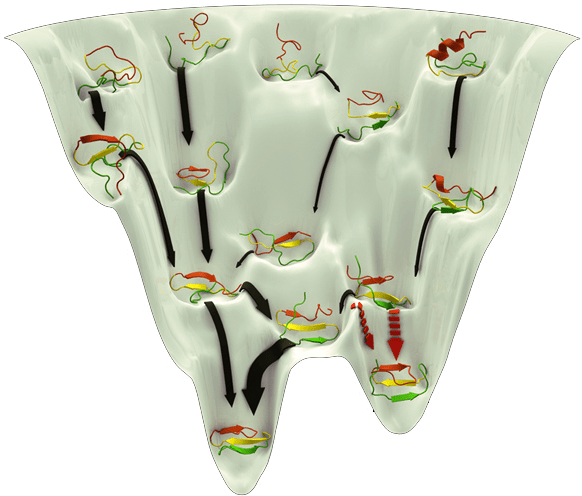
Levinthal’s Paradox: A protein can’t sample all conformations in a biologically reasonable time, yet it folds quickly
Example: A protein with 100 amino acids, each capable of adopting about 3 torsion angles, results in ~ possible conformations3100
Proteins fold into their native structure by minimizing free energy
Scoring functions attempt to model this with statistical or physics-based potentials
Proteins adopt conformations that minimize thermodynamic free energy
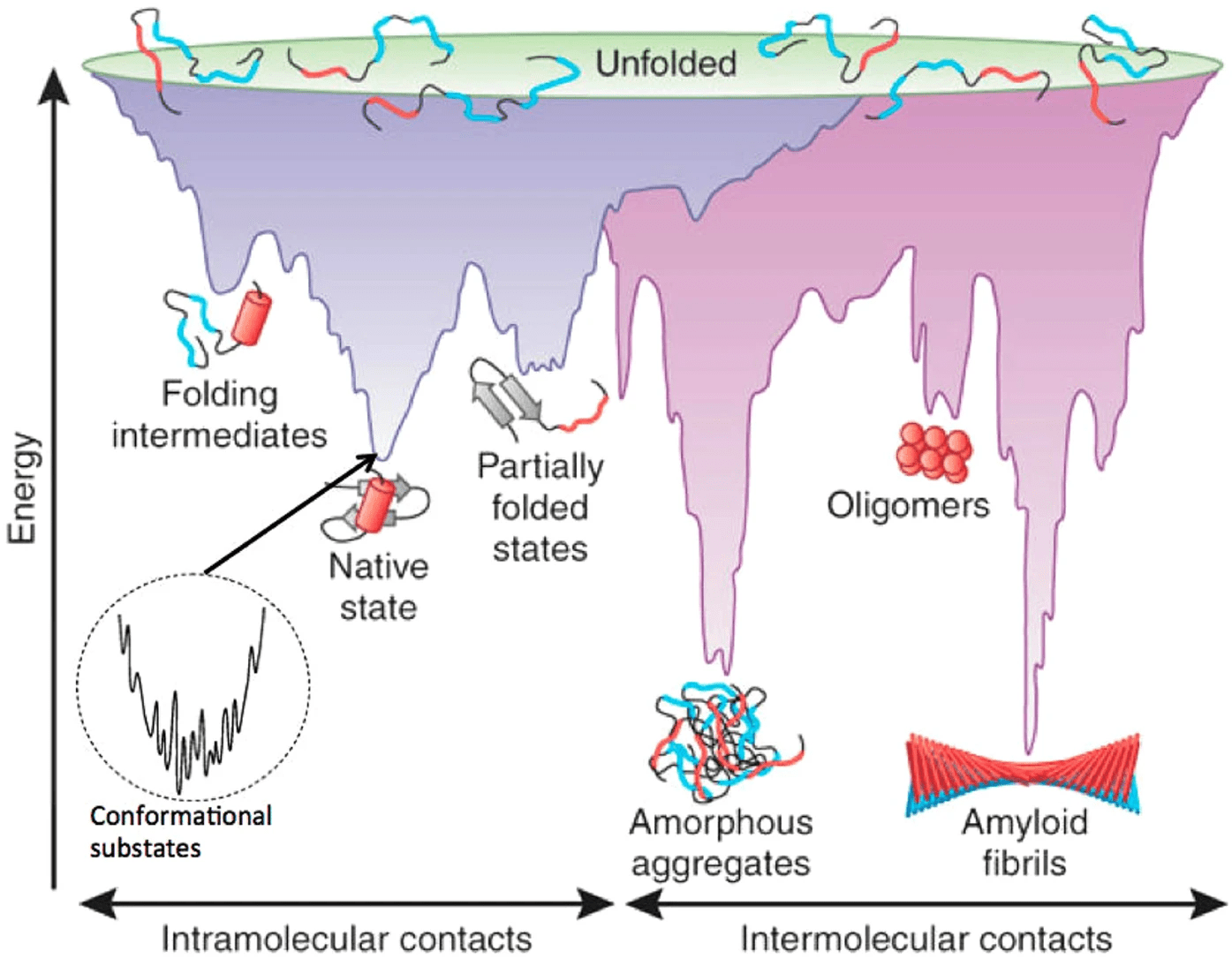
A potential energy surface (PES) represents the energy of a system as a function of the positions of its atoms
Allows us to understand how the system's energy changes upon reactions or movements
Flexible and disordered regions add complexity to structure prediction
Many proteins exist in ensembles of structures or are natively disordered.
Their function may depend on transient interactions or induced folding.
Environmental context dramatically impacts protein folding
Proteins fold differently in different environments
Predictions need to capture interactions with solvent molecules, ions, and cofactors
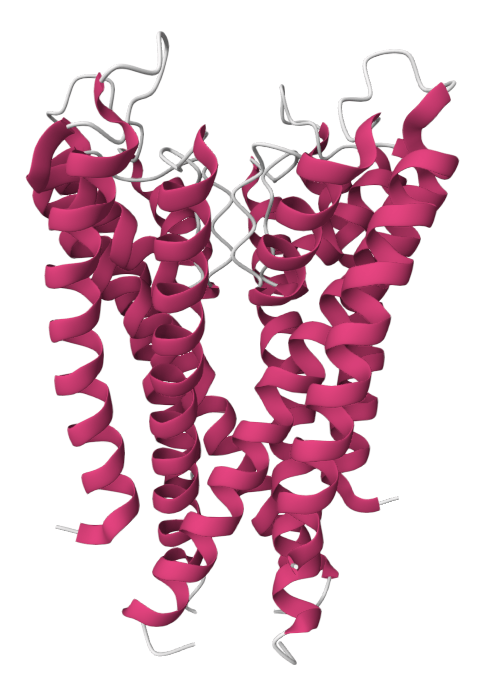
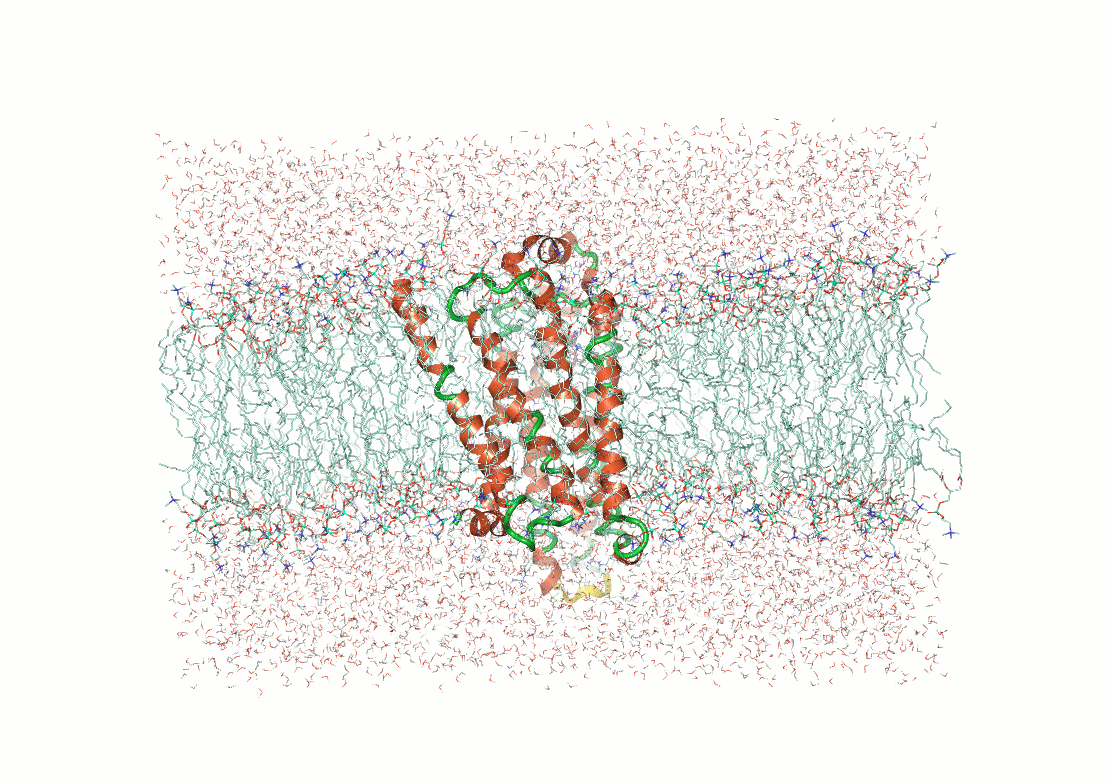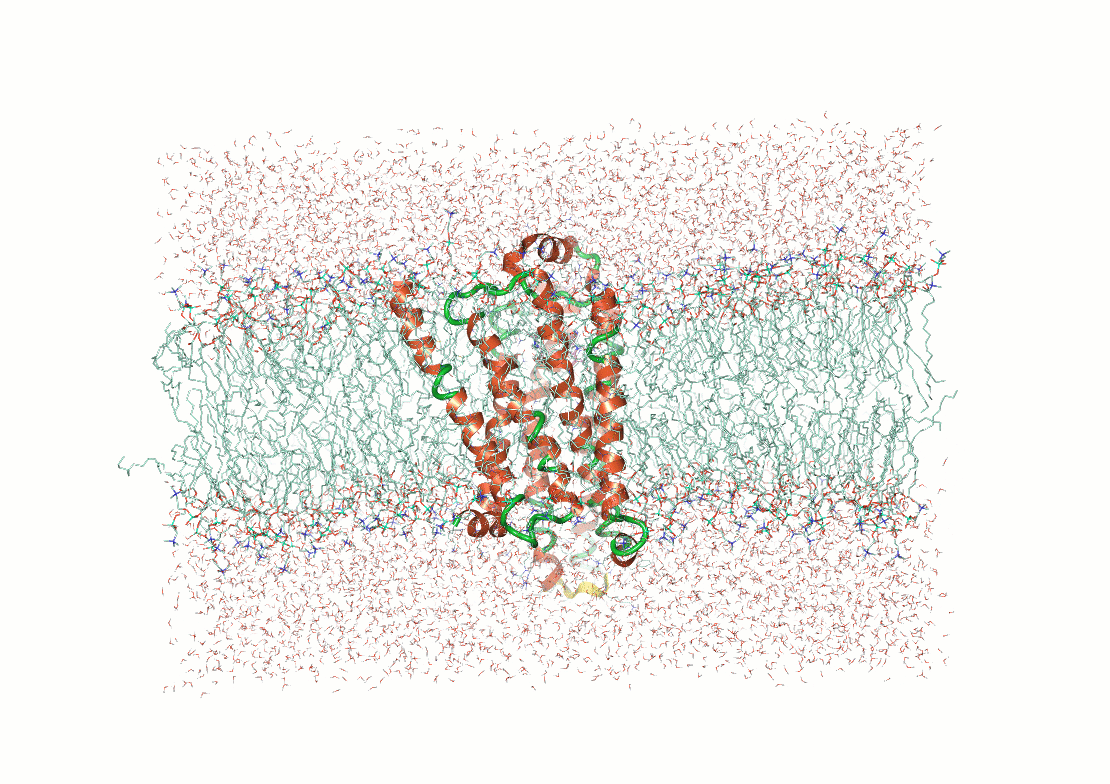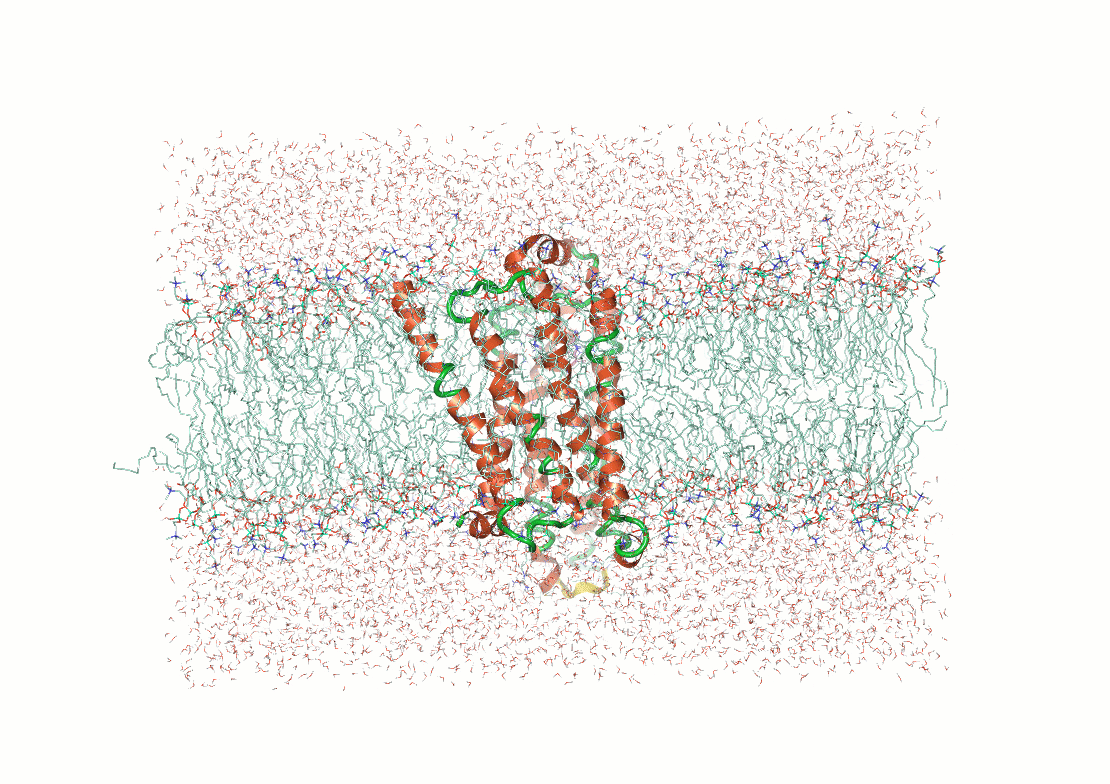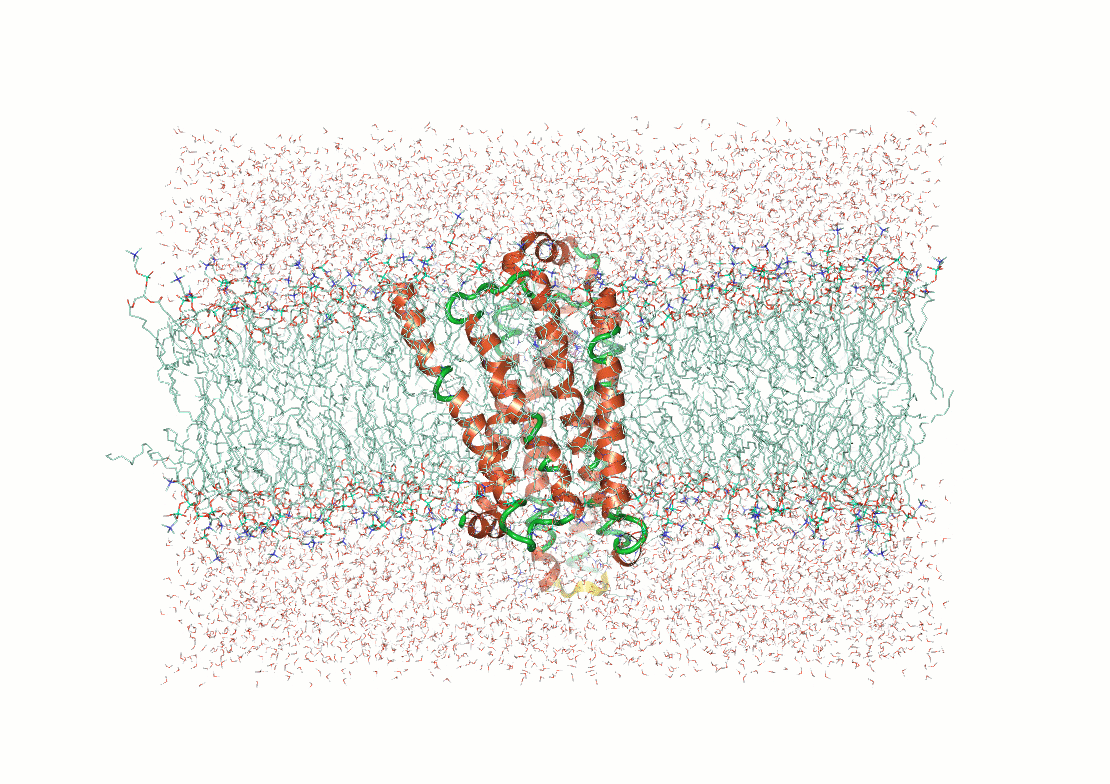
Example: Predicting transmembrane protein structures, where the lipid bilayer plays a key role in folding, is particularly complex.
AlphaFold 3
pH-gated K+ channel
Environmental context dramatically impacts protein folding
PTMs such as phosphorylation, glycosylation, and methylation can alter protein folding and function
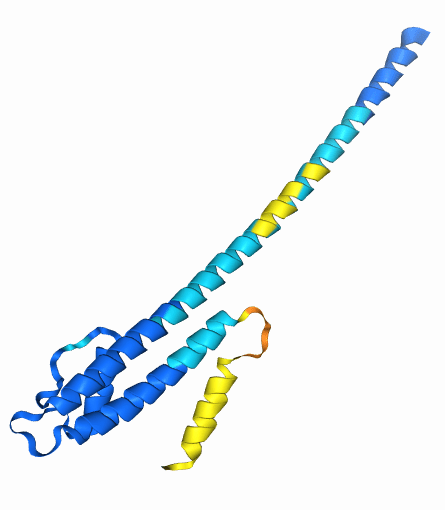
Example: eIF4E is a eukaryotic translation initiation factor involved in directing ribosomes to the cap structure of mRNAs
Ser209 is phosphorylated by MNK1
AlphaFold 3 accurately predicts these changes when they are already known
After today, you should have a better understanding of
Homology modeling
Hidden Markov Model alignments
Homology modeling predicts structure using evolutionary relationships
Requires a template—a known structure with detectable sequence similarity.
Often the first modeling strategy attempted due to simplicity and reliability.
Based on the principle that proteins with similar sequences tend to adopt similar structures.
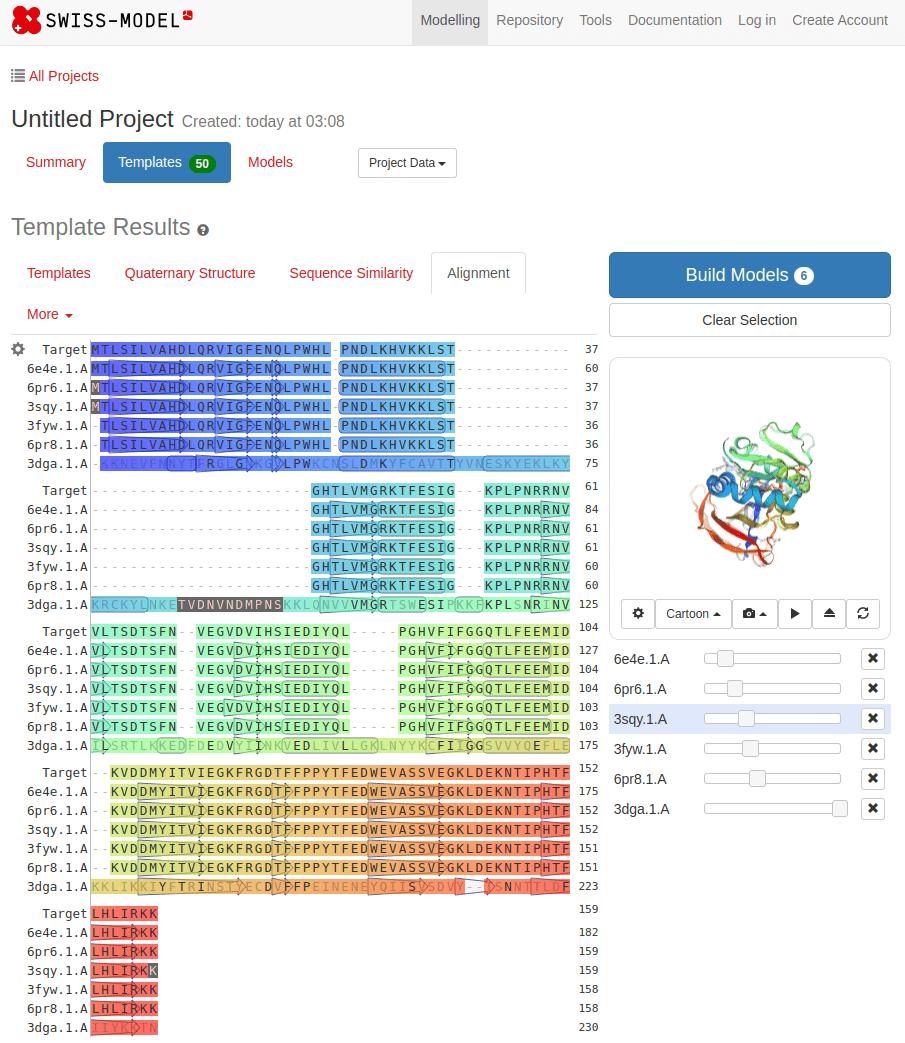
The first step in homology modeling is to search for similar sequences
You begin with a query sequence
(the protein you want to model)
> PrsA2
CGGGGDVVKTDSGDVTKDELYDAMKDKYGSEFVQQLTFEKILGDKYKVSDE
DVDKKFNEYKSQYGDQFSAVLTQSGLTEKSFKSQLKYNLLVQKATEANTDT
SDKTLKKYYETWQPDITVSHILVADENKAKEVEQKLKDGEKFADLAKEYST
DTATKDNGGQLAPFGPGKMDPAFEKAAYALKNKGDISAPVKTQYGYHIIQM
DKPATKTTFEKDKKAVKASYLESQLTTENMQKTLKKEYKDANVKVEDKDLK
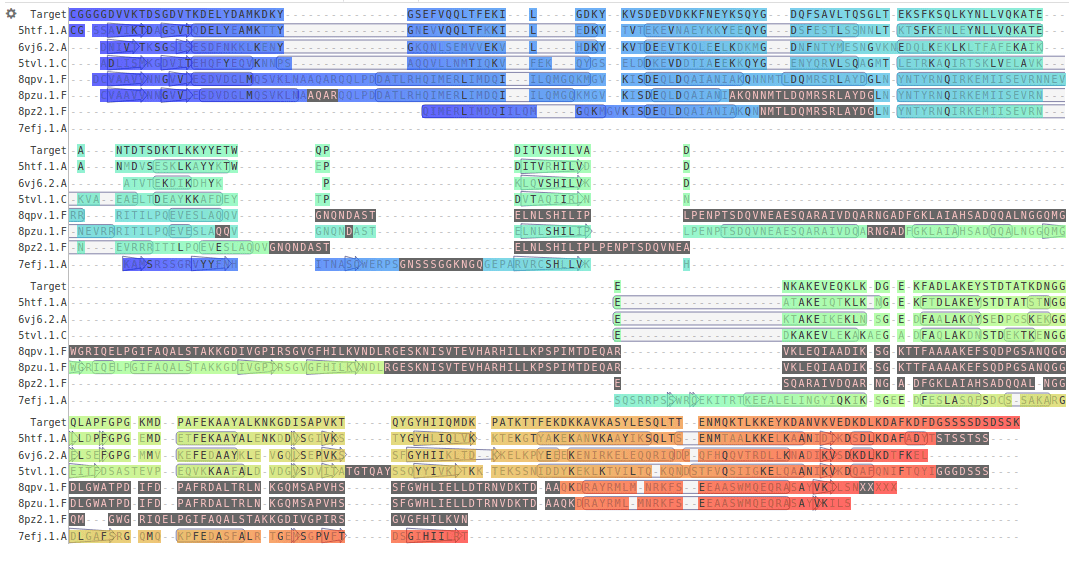
DAFKDFDGSSSSDSDSSKSequence alignment maps residues from the target onto the template structure
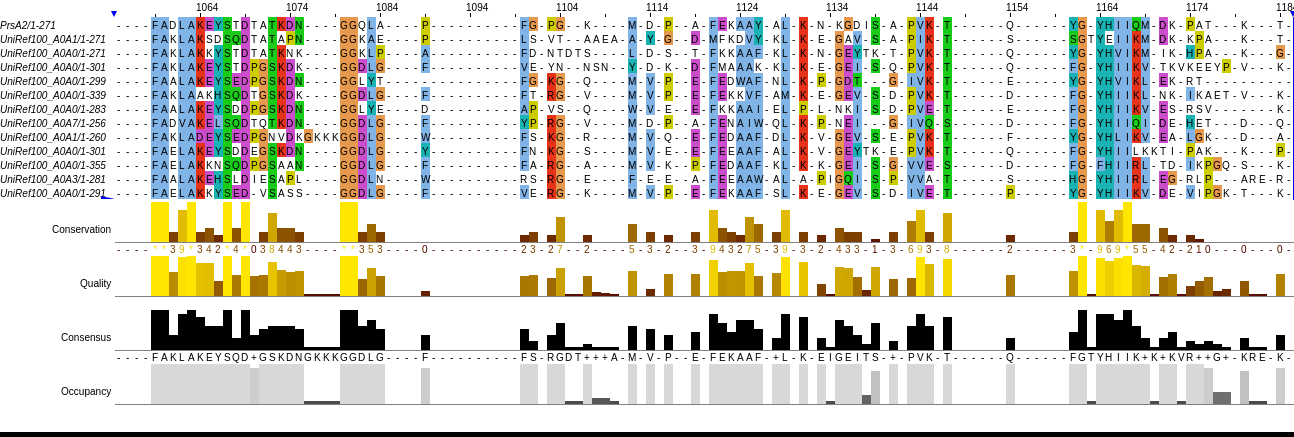
This is an MSA of Listeria monocytogenes PrsA2 to related proteins
Basic alignment algorithms are too simplistic for distant homolog detection
Methods like Smith-Waterman use direct pairwise alignment based on similarity scores (e.g., BLOSUM62).
They do not consider evolutionary variation, insertions, or residue-level probabilities.
We need methods that detect evolutionarily distant but structurally conserved relationships.

Profile-based aligners improve sensitivity by modeling residue variability at each position
A profile captures how conserved each position is across an MSA
Instead of a single residue, each position becomes a probability distribution over all 20 amino acids.
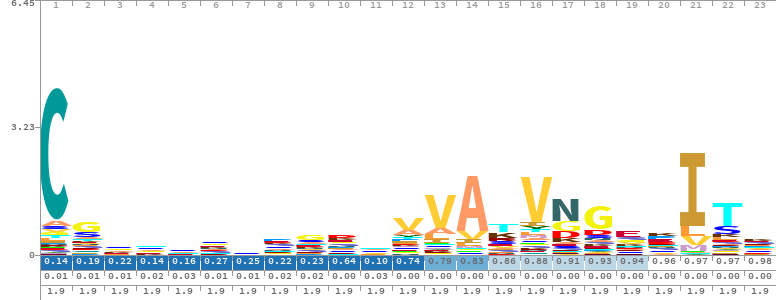
Made with skylign
HHblits starts by converting the query sequence or MSA into a profile HMM
A profile HMM models the amino acid probabilities at each position, plus insertion and deletion likelihoods.
The result is a probabilistic model that captures both conservation and structural variability.
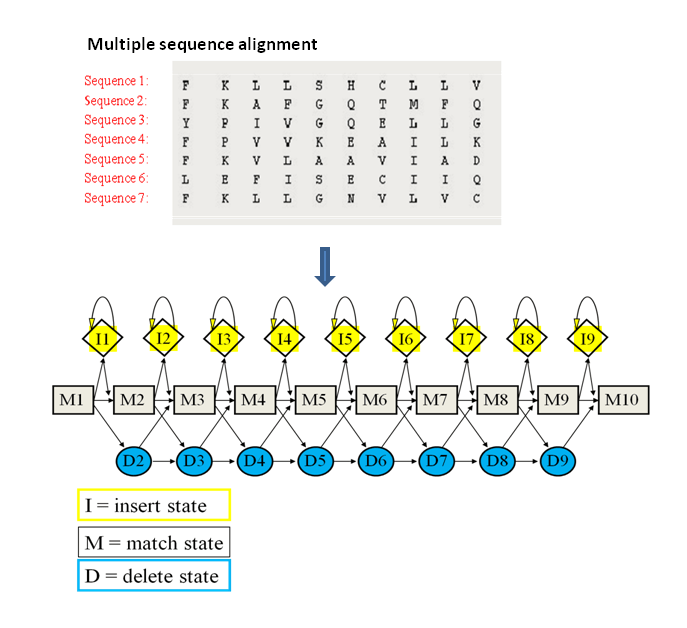
Sequence likelihood can be computed by walking along the profile HMM
HHblits performs accurate HMM–HMM alignments on the best candidates
Full alignments are done using the Viterbi algorithm to find the best path through the HMM state space.
A maximum accuracy (MAC) alignment is also computed to optimize for correct residue–residue matches.
These alignments return E-values to estimate match confidence.
Only statistically significant hits (e.g., E < 1e-3) are retained for the next iteration.
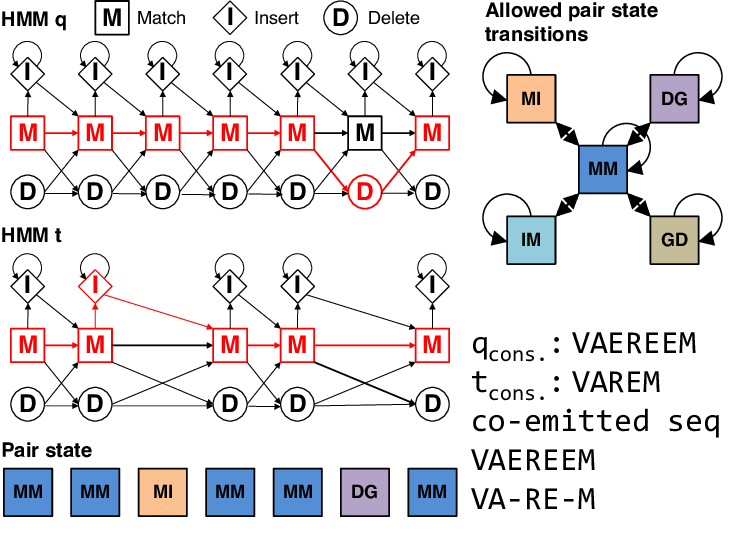
The alignment is represented as red path through both HMMs
Homology modeling is most accurate when sequence identity is >30%
>50% identity: high-accuracy models (~1 Å RMSD) are achievable
Between 30–50%: moderate accuracy; errors appear in loops, side chains
<30% identity: The "twilight zone" where structural similarity is uncertain
HHblits compares HMMs using a fast, two-step prefilter before full alignment
Step 1: Converts HMM columns into a discretized profile alphabet of 219 letters to speed up comparison.
Step 2: Performs a fast local alignment of the query HMM to these compressed database representations.
These steps eliminate ~99.9% of irrelevant comparisons—reducing millions of alignments to thousands
HHblits iteratively refines the profile HMM by adding new homologs from each search round
Sequences from matched HMMs are added to the query MSA.
A new query HMM is built from this updated MSA.
Each iteration improves sensitivity by capturing more distant, diverse sequences.
This process continues for 1–4 rounds, depending on the desired depth and quality.
After today, you should have a better understanding of
Homology modeling
Template building
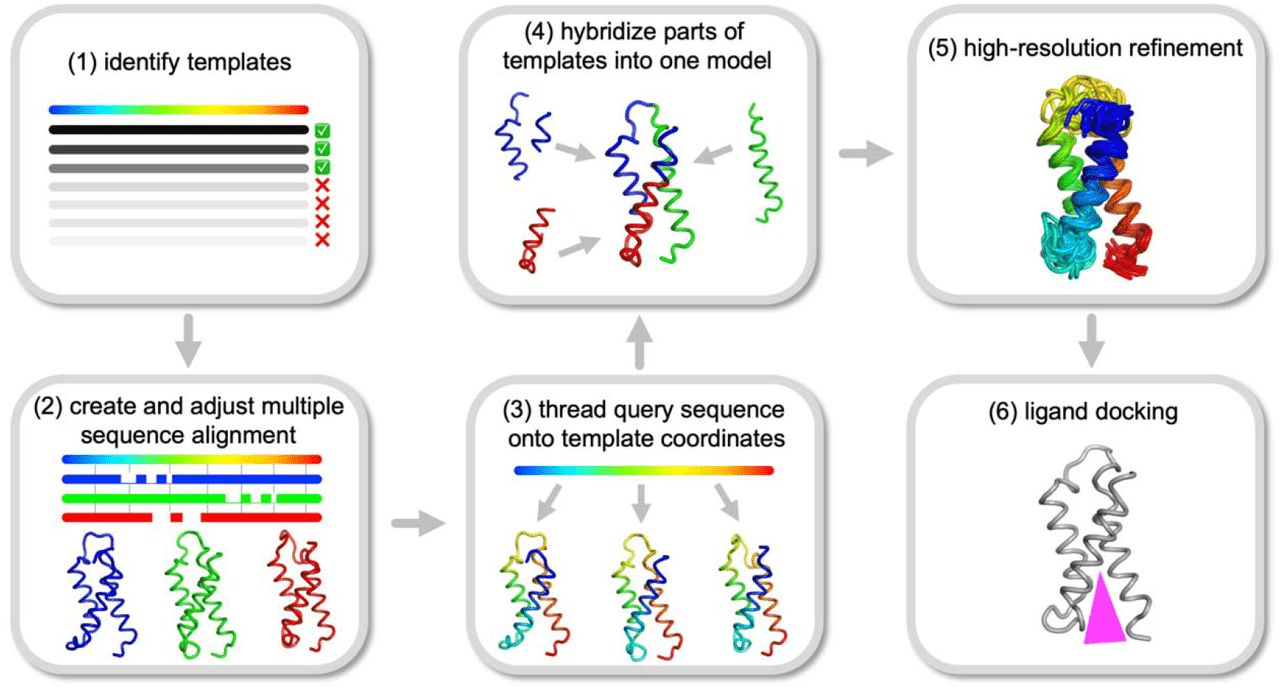
The model is built by copying the template structure and modeling variable regions
Conserved regions: Backbone atoms are copied from template directly.
Variable regions (loops, inserts): Built using fragment libraries or loop modeling algorithms.
Side chains are adjusted using rotamer libraries to fit target sequence.
Model refinement improves geometry and resolves steric clashes
After model construction, the structure often contains bad bond angles, clashes, or unrealistic torsions.
Refinement includes energy minimization using force fields or statistical potentials.
Some tools use molecular dynamics or Monte Carlo sampling.
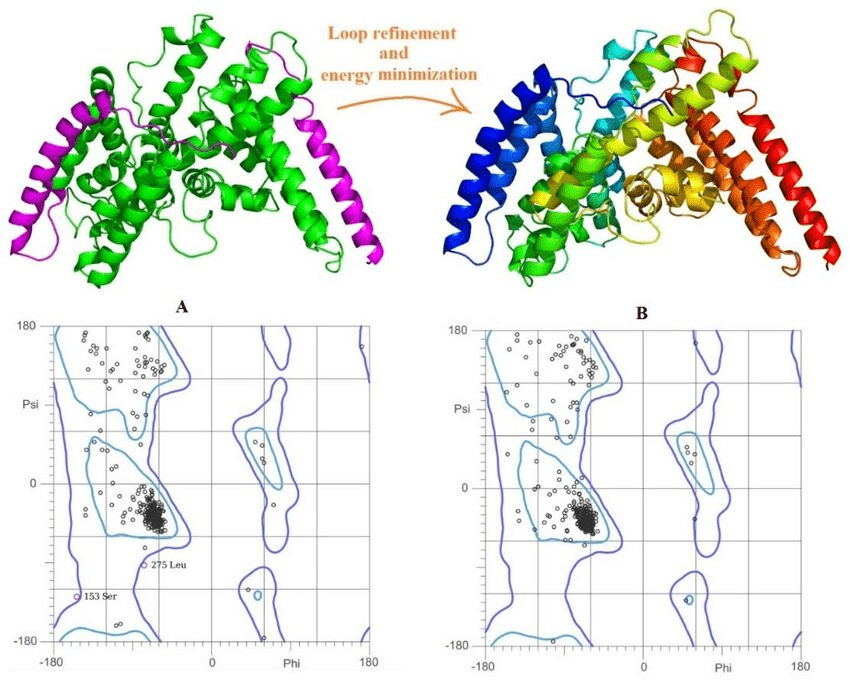
Model validation helps assess confidence and detect errors
Ramachandran plots visualize backbone torsion angles.
Statistical scores (e.g., DOPE, QMEAN, GA341) evaluate nativeness.
Residue-by-residue assessment helps identify weak regions (e.g., VERIFY3D, ERRAT).
Good models have:
-
Most residues in favored Ramachandran regions
-
Low-energy scores
-
No large clashes
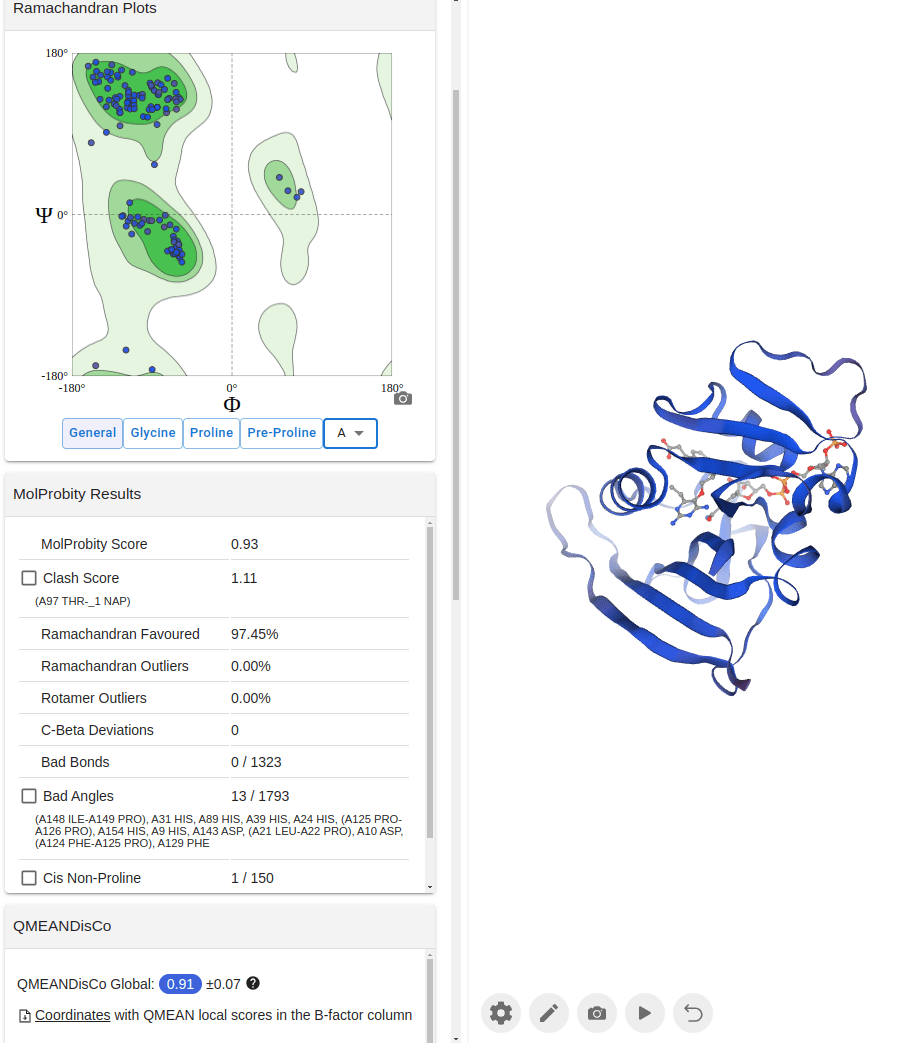
Homology modeling works best when you iterate and re-evaluate
If a model fails validation, revisit earlier steps:
-
Try a different template
-
Refine the alignment
-
Adjust loop modeling parameters
Multiple models are often built and ranked—choose the one with the best validation metrics.
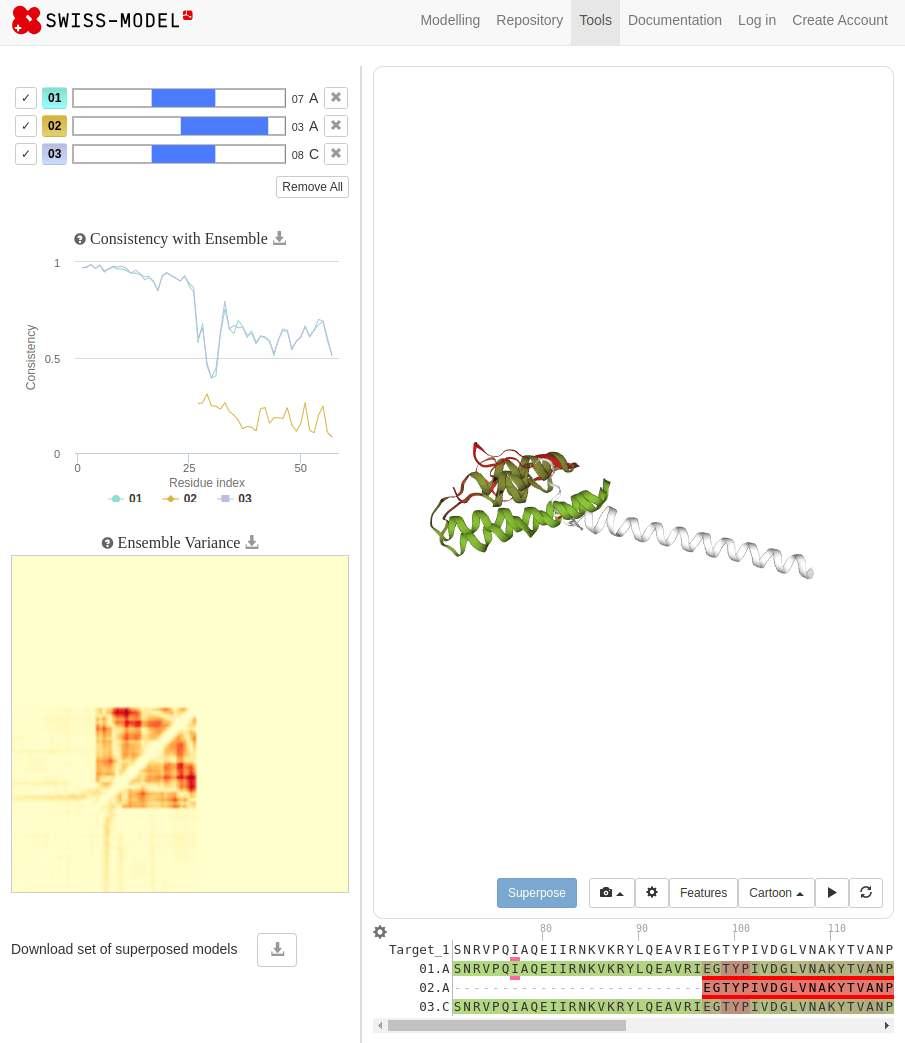
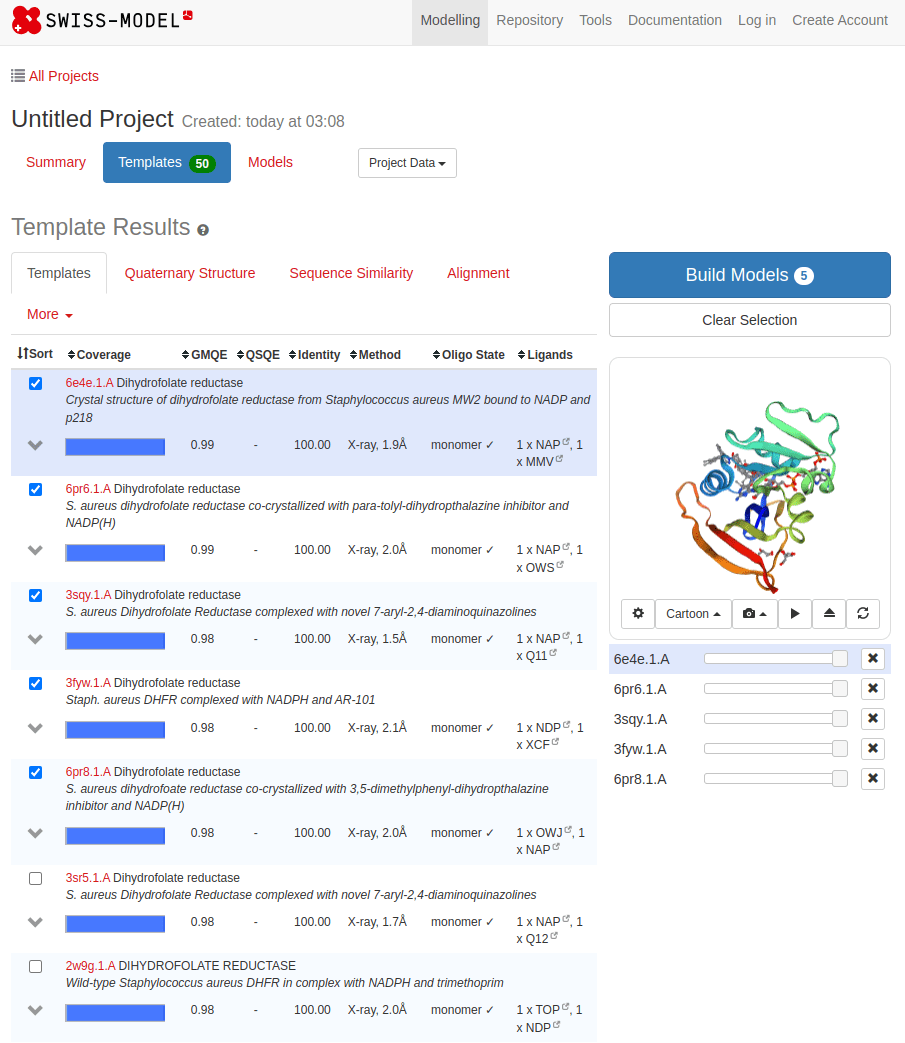
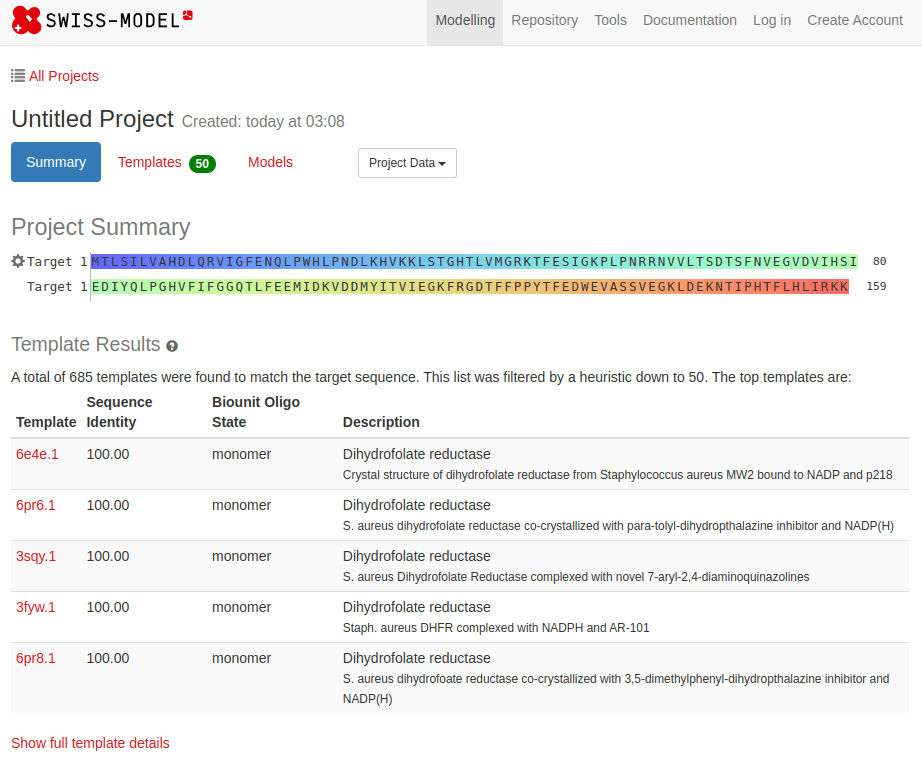
SWISS-MODEL
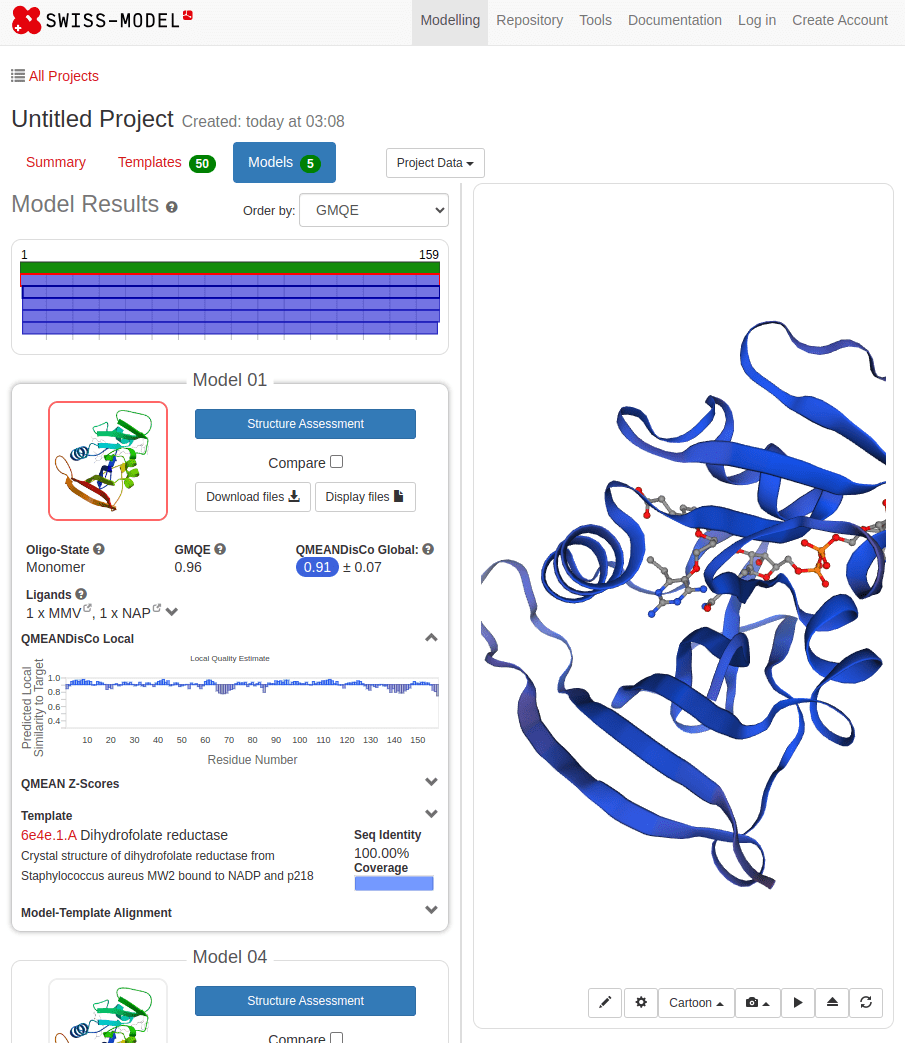
MTLSILVAHDLQRVIGFENQLPWHLPNDLKHVKKLSTGHTLVMGRKTFESIGKPLPNRRNVVLTSDTSFNVEGVDVIHSIEDIYQLPGHVFIFGGQTLFEEMIDKVDDMYITVIEGKFRGDTFFPPYTFEDWEVASSVEGKLDEKNTIPHTFLHLIRKKDHFR (UniProt)
SWISS-MODEL

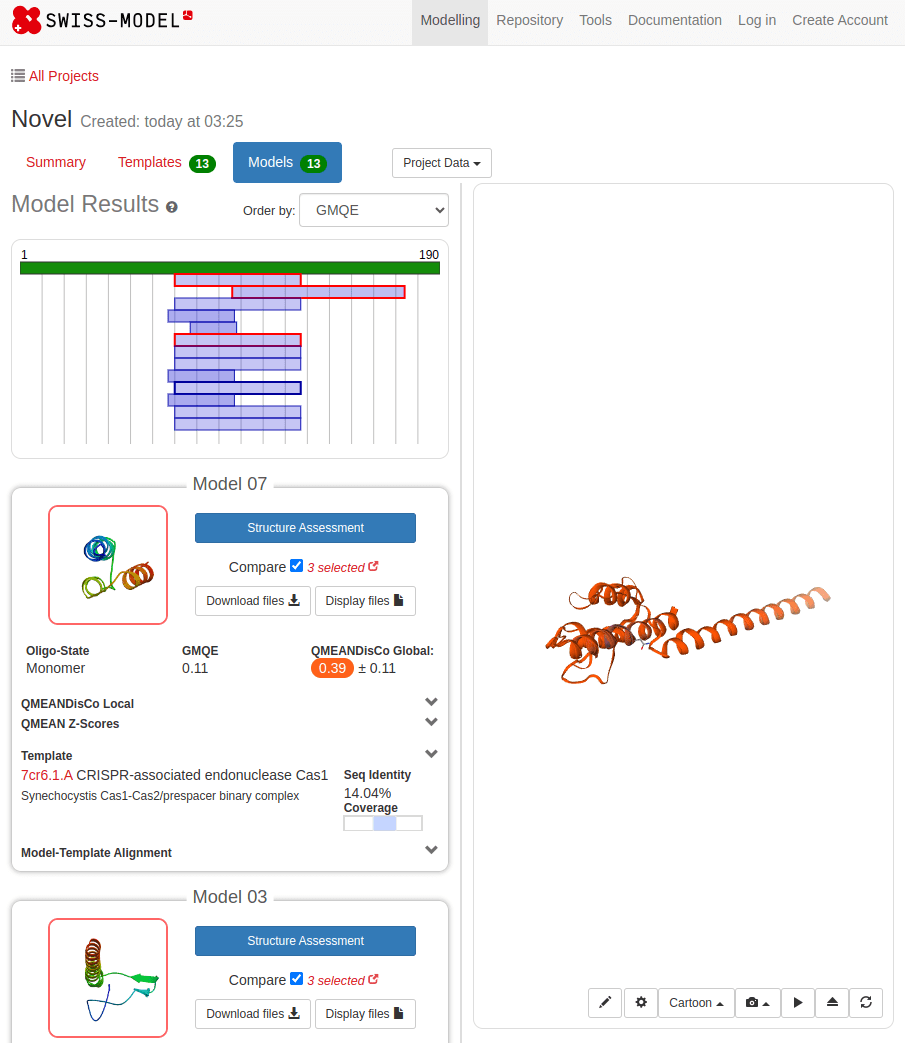
Novel proteins are too challenging
After today, you should have a better understanding of
Know when to use threading instead of homology modeling
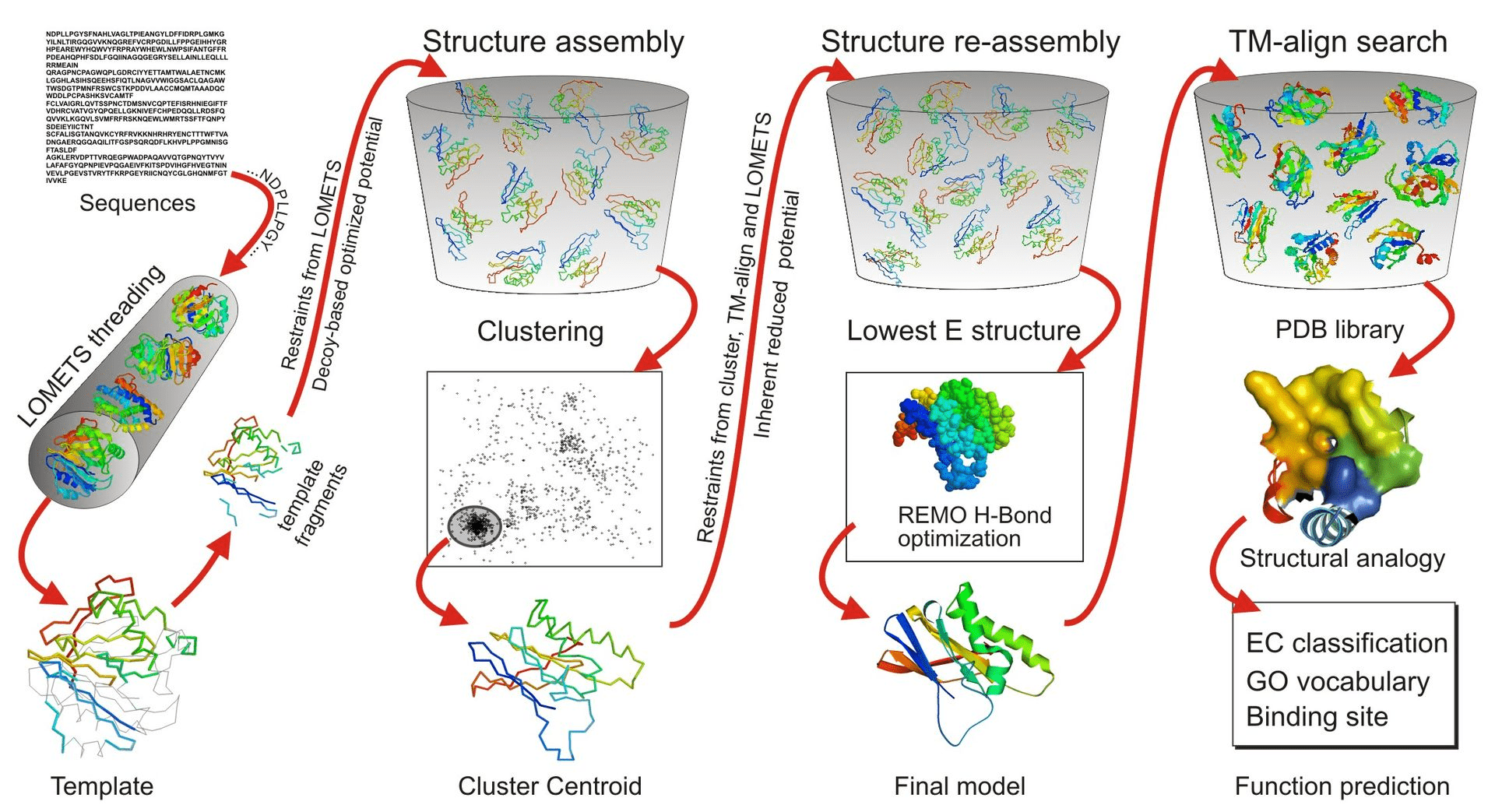
Why Use Threading?
In cases where sequence similarity to known structures is low (< 30%), homology modeling becomes unreliable
Phyre2, RaptorX, MUSTER, and I-TASSER are commonly used for threading and takes much longer than homology modeling
Threading matches sequences to known structural folds based on structural rather than sequence similarity
Identifying the Right Fold
After today, you should have a better understanding of
Interpret a contact map for protein structures
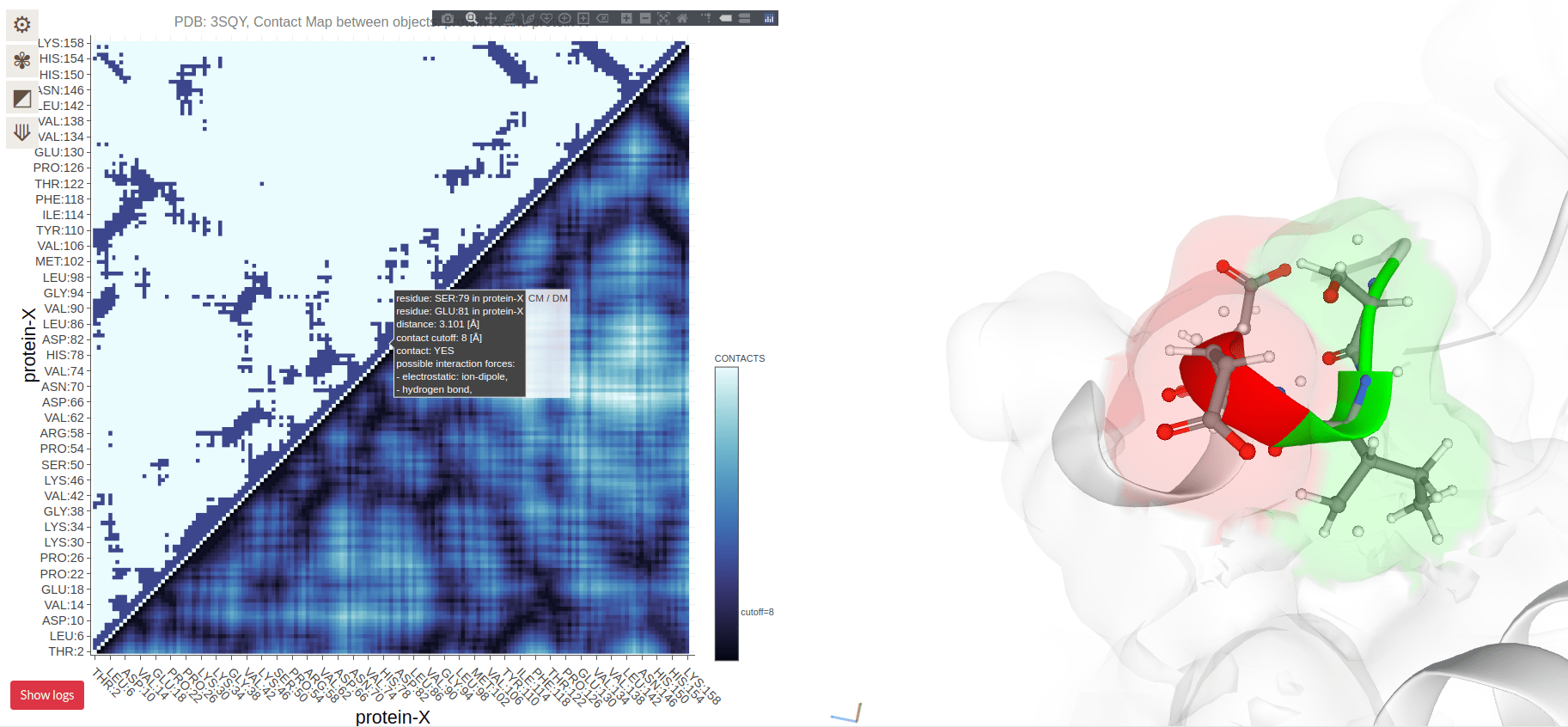
Contact Maps Visualize Residue Interactions in Proteins
A contact map is a 2D representation of which residues are in close proximity
Each point on the map corresponds to two residues that are close in 3D space
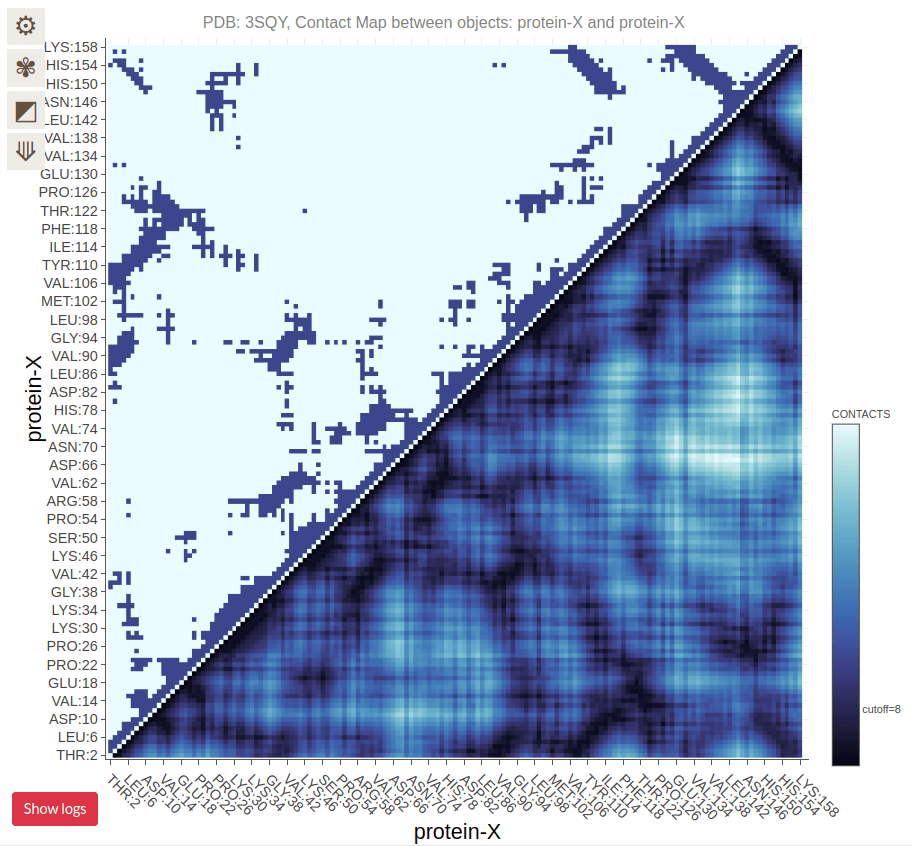
Contact Maps Represent Spatial Proximity, Not Sequence Order
Contacts are determined by spatial proximity, typically within a certain distance threshold
Residues far apart in the sequence can still be close in the 3D structure, reflected in the contact map
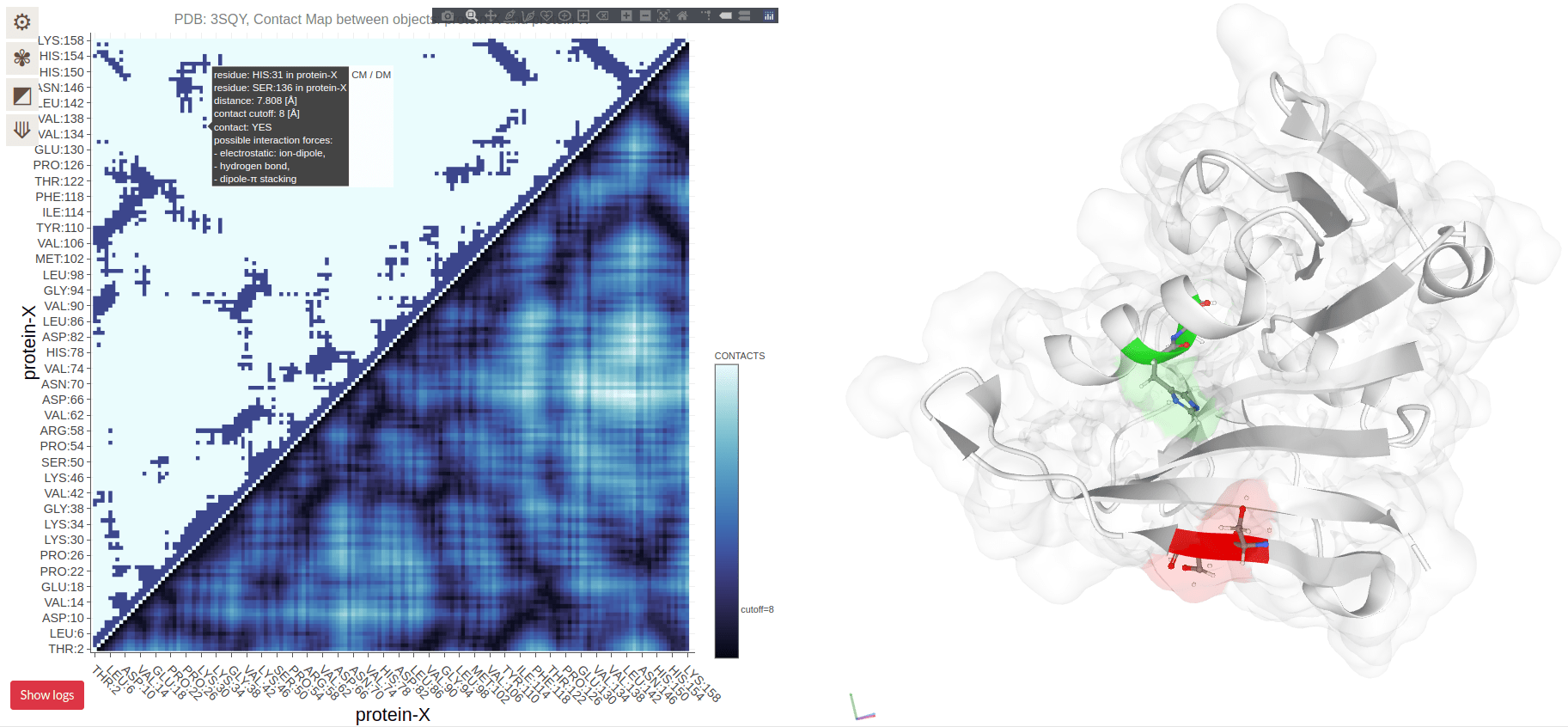
Residues on the diagonal are adjacent in sequence (and spatially)
After today, you should have a better understanding of
Comprehend how coevolution provides structural insights
The Rise of Machine Learning in Structural Biology
Traditional methods like homology modeling and threading rely on templates and known structures
AlphaFold (DeepMind) and RosettaFold (Baker Lab) lead the charge in this area
ML predicts 3D structures only from sequence data
What is AlphaFold?
Developed by DeepMind, AlphaFold predicts protein structures with atomic accuracy by using deep learning models trained on large structural datasets
Breakthroughs
- AlphaFold 2 achieved near-experimental level accuracy in the 2020 CASP14 competition (Critical Assessment of protein Structure Prediction)
- AlphaFold 3 (2024) predicts proteins, DNA, RNA, ligands, and post-translational modifications
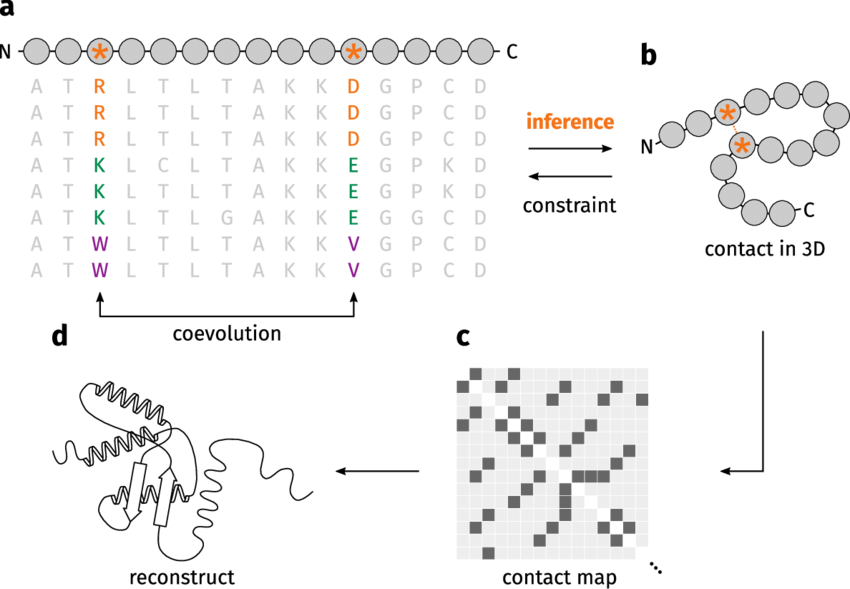
Coevolving residues mutate in a correlated manner
Mutations in one residue often result in compensatory mutations in its interacting partner
This is observed across species through analysis of homologous protein sequences
Correlated mutations indicate functionally significant residue pairs
Arg (positive)
Asp (negative)
Lys (positive)
Glu (negative)
Trp (hydrophobic)
Val (hydrophobic)


Evolution
Evolutionary Analysis Reveals Structural Insights
Coevolution analysis helps predict which residues are close in the 3D structure
Residues showing correlated mutations are likely to be spatially close in the folded protein
This is particularly useful when no experimental structure is available

Multiple Sequence Alignments Enable Coevolution Detection
Coevolution is detected using large MSAs from homologous proteins
The more diverse the sequences in the MSA, the better the resolution of coevolving residues
Evolutionary information from MSAs guides predictions for residue-residue contacts
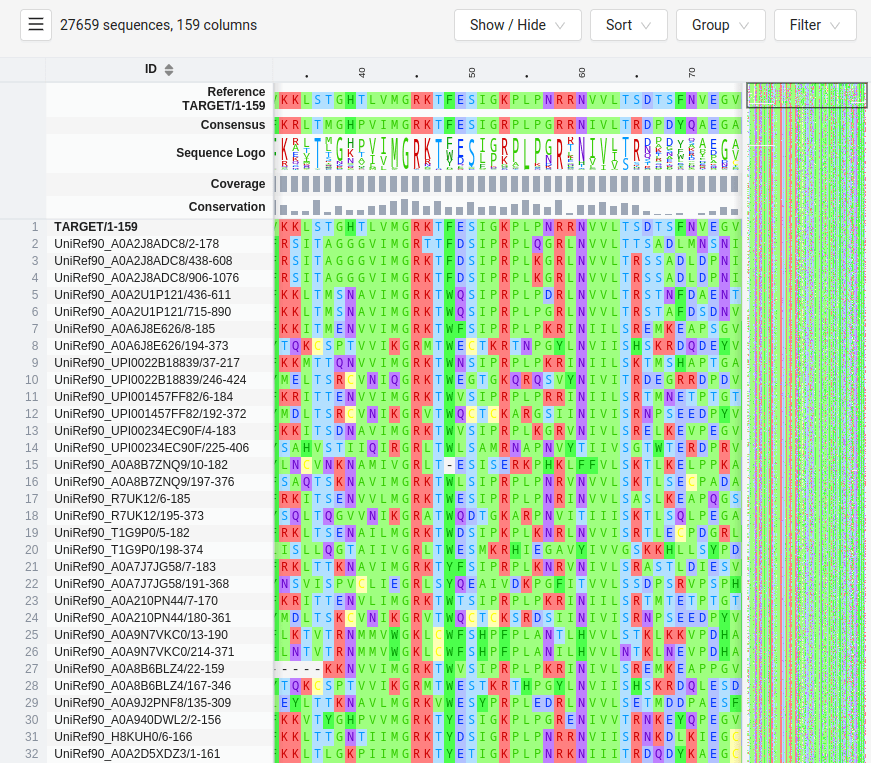
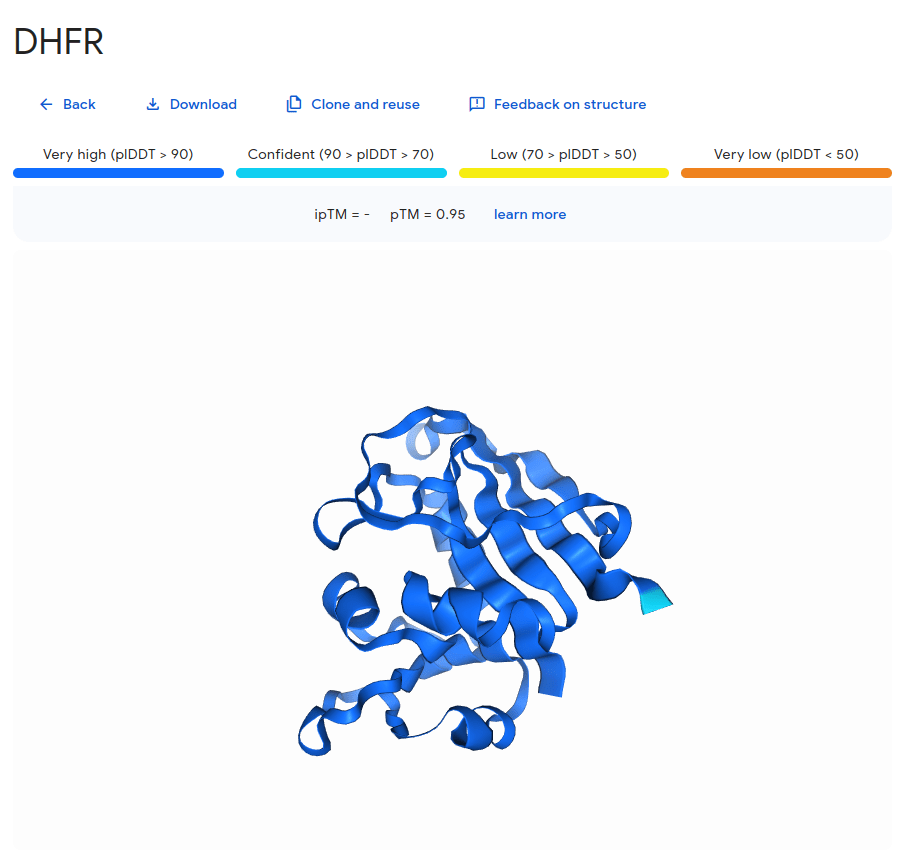
Coevolution example: DHFR
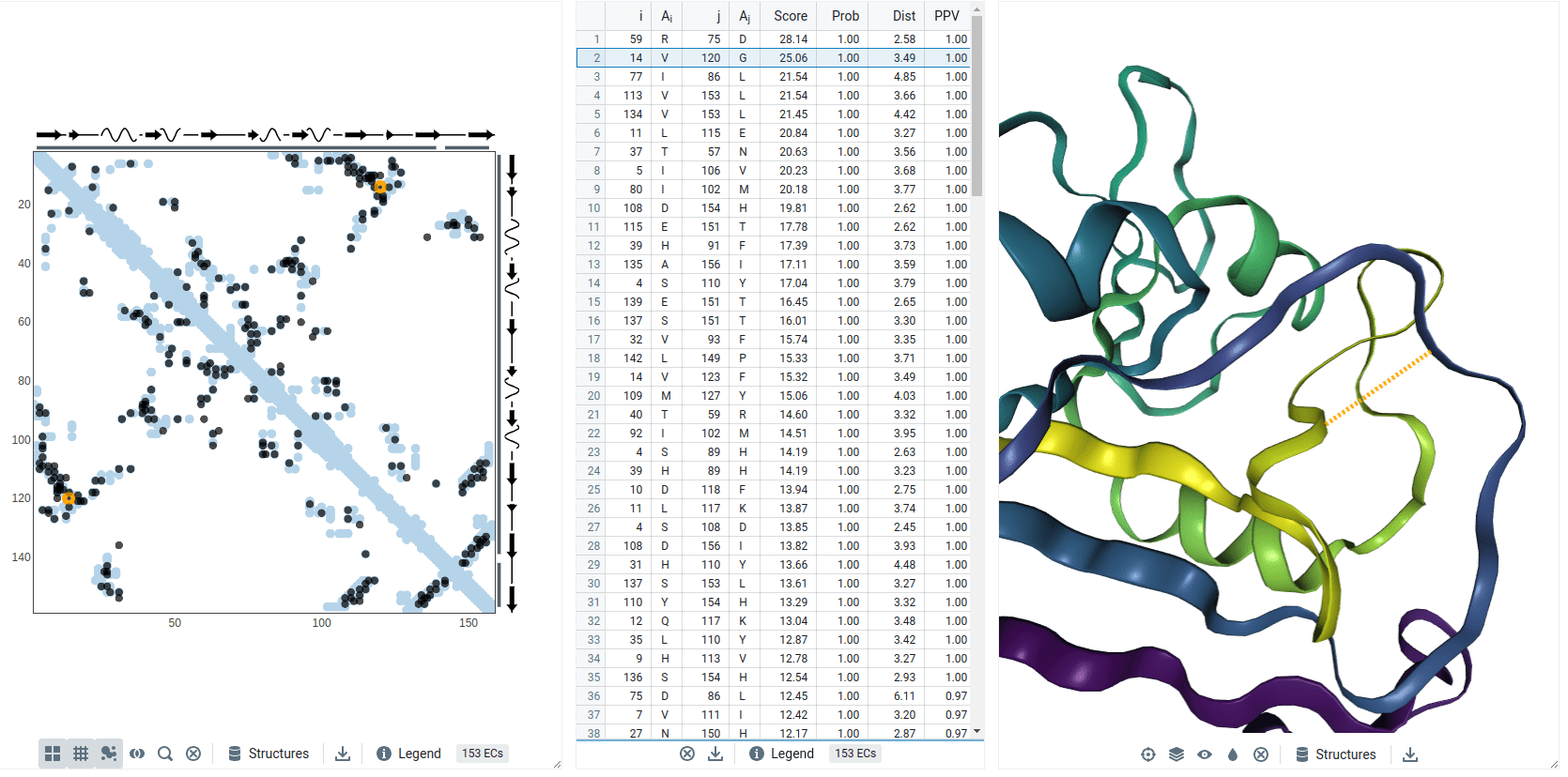
Residues with a high Score (i.e., coevolve) are near each other in the protein's structure (i.e., small distance)
Val14 and Gly120 coevolved
Models predict these residues are spatially close
Coevolutionary signals can be noisy
Not all correlated mutations are due to direct physical interactions; some may be indirect
Noise in the data can come from random mutations or insufficient evolutionary diversity.
Large and diverse sequence data sets are needed for reliable coevolution predictions.
Machine learning leverages coevolution for high-accuracy predictions
AlphaFold and RosettaFold utilize coevolutionary data from MSAs to predict residue interactions
These models incorporate evolutionary information along with structural features, leading to highly accurate predictions
After today, you should have a better understanding of
Explain why ML models are dominate protein structure prediction
AlphaFold pipeline, simplified
Given the following data
MTLSILVAHDLQRVIGFENQLPWHLPNDLKHVKKLSTGHTLVMGRKTFESIGKPLPNRRNVVLTSDTSFNVEGVDVIHSIEDIYQLPGHVFIFGGQTLFEEMIDKVDDMYITVIEGKFRGDTFFPPYTFEDWEVASSVEGKLDEKNTIPHTFLHLIRKKInput sequence
Multiple Sequence Alignment
Predict
Atomistic structure
ML models

AlphaFold 2 pipeline: Evoformer
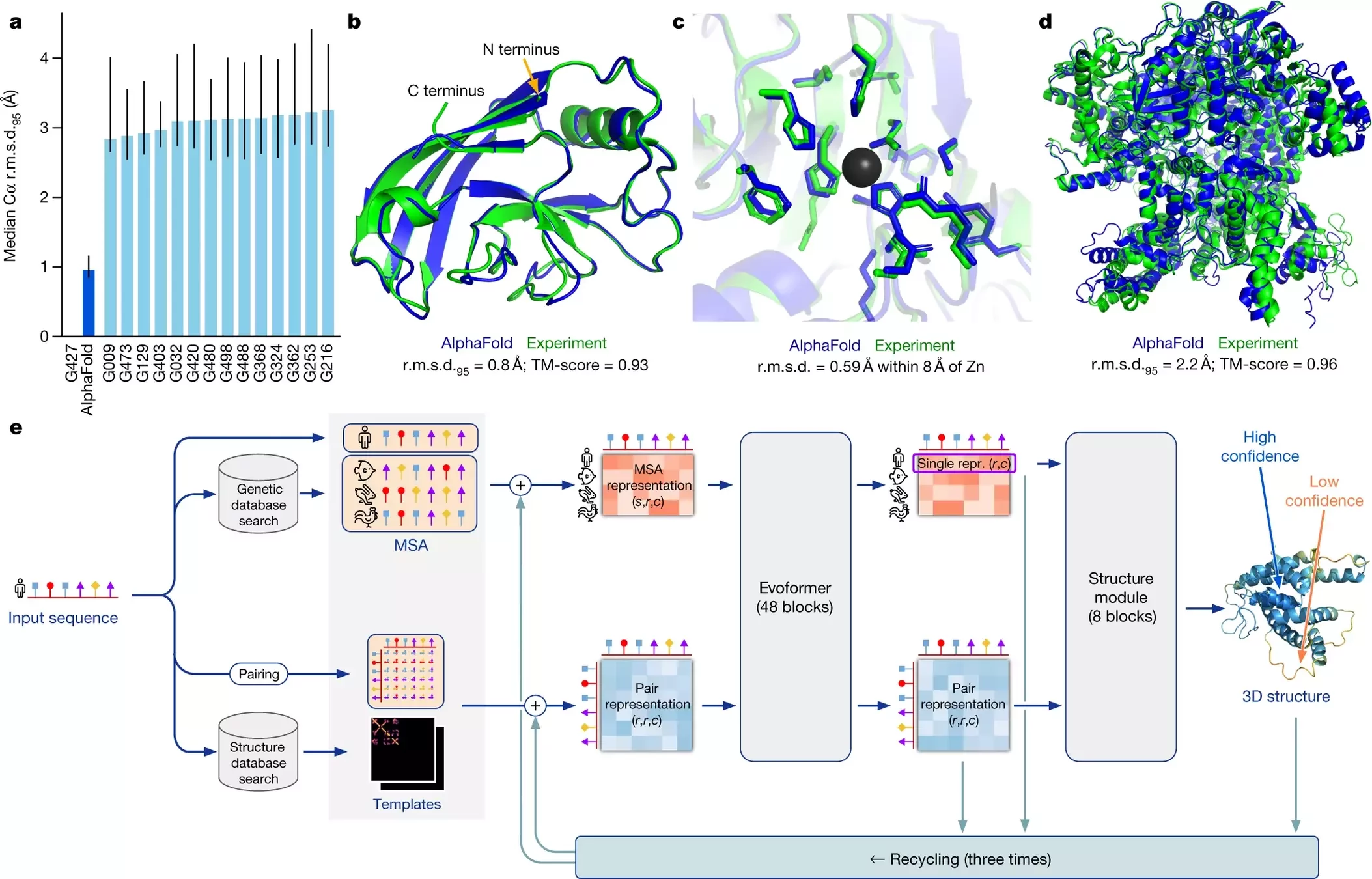
Using MSAs and contact maps, DeepMind trained a model to predict protein structures
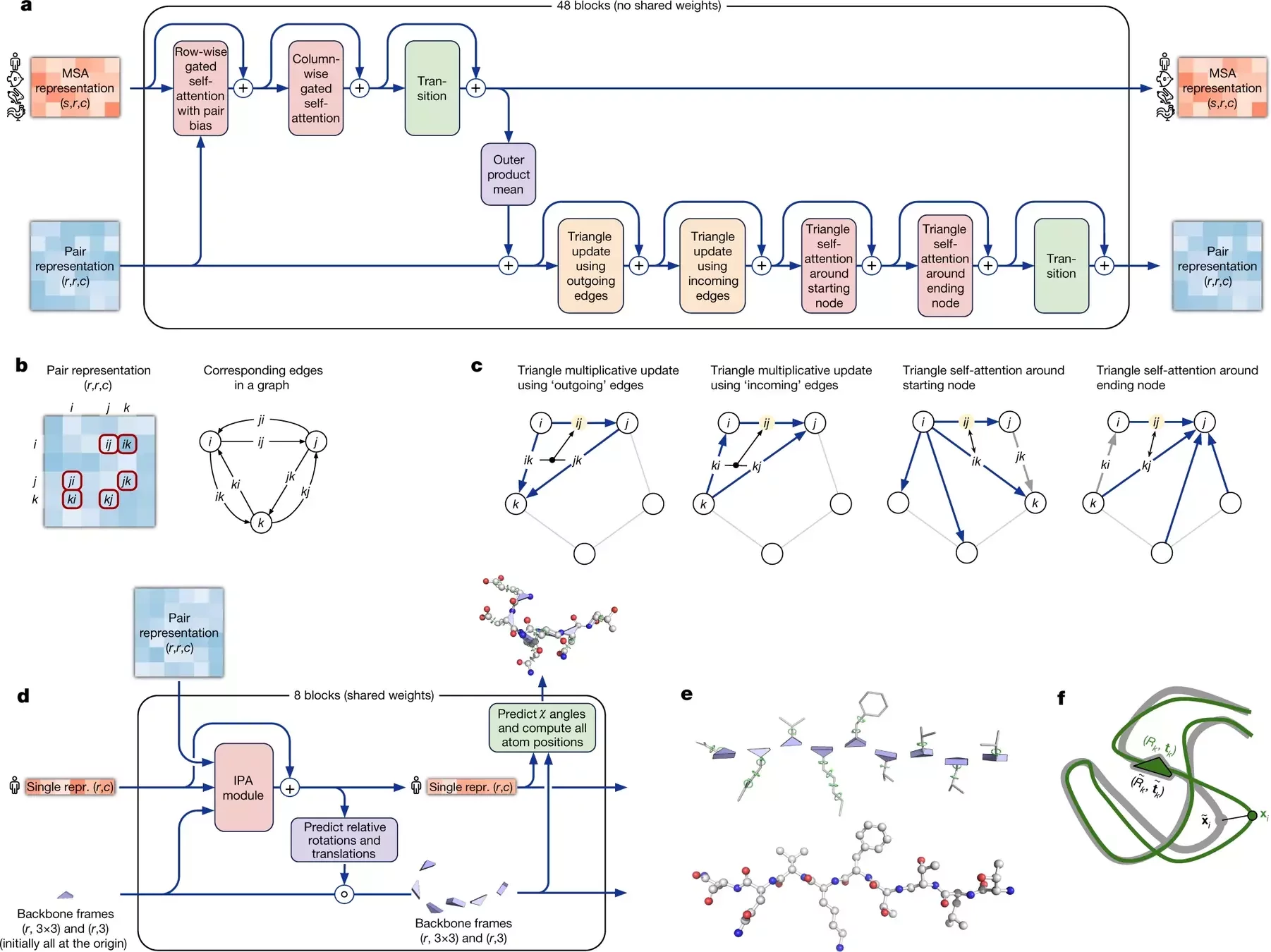
Contact maps are converted into dihedral angles
AF2 iterations
What is new in AlphaFold 3?
Biggest change is the use of a diffusion model
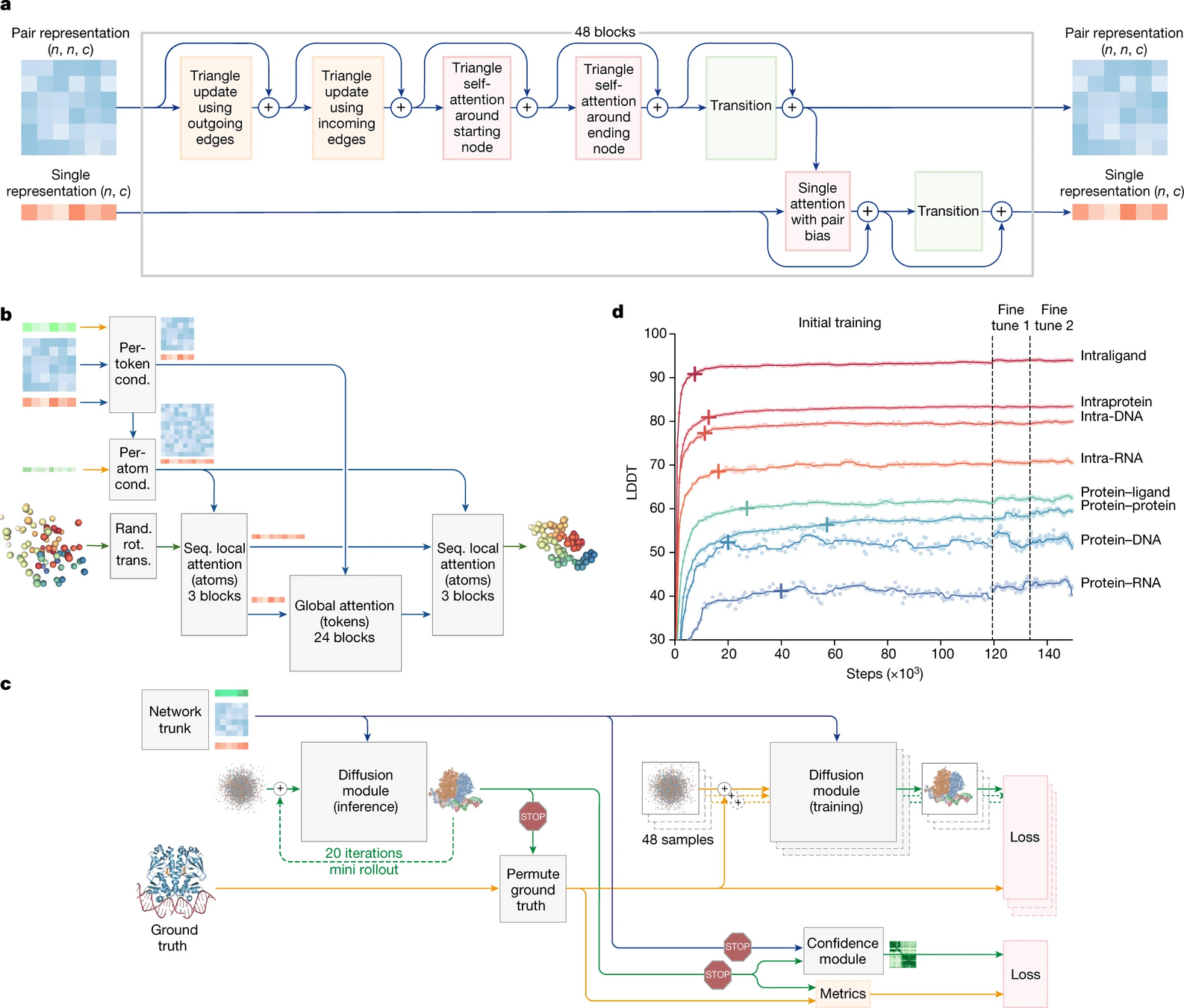
Diffusion models essentially learn to unscramble atoms into a structure
AlphaFold 3 is supercharged for any biomolecule
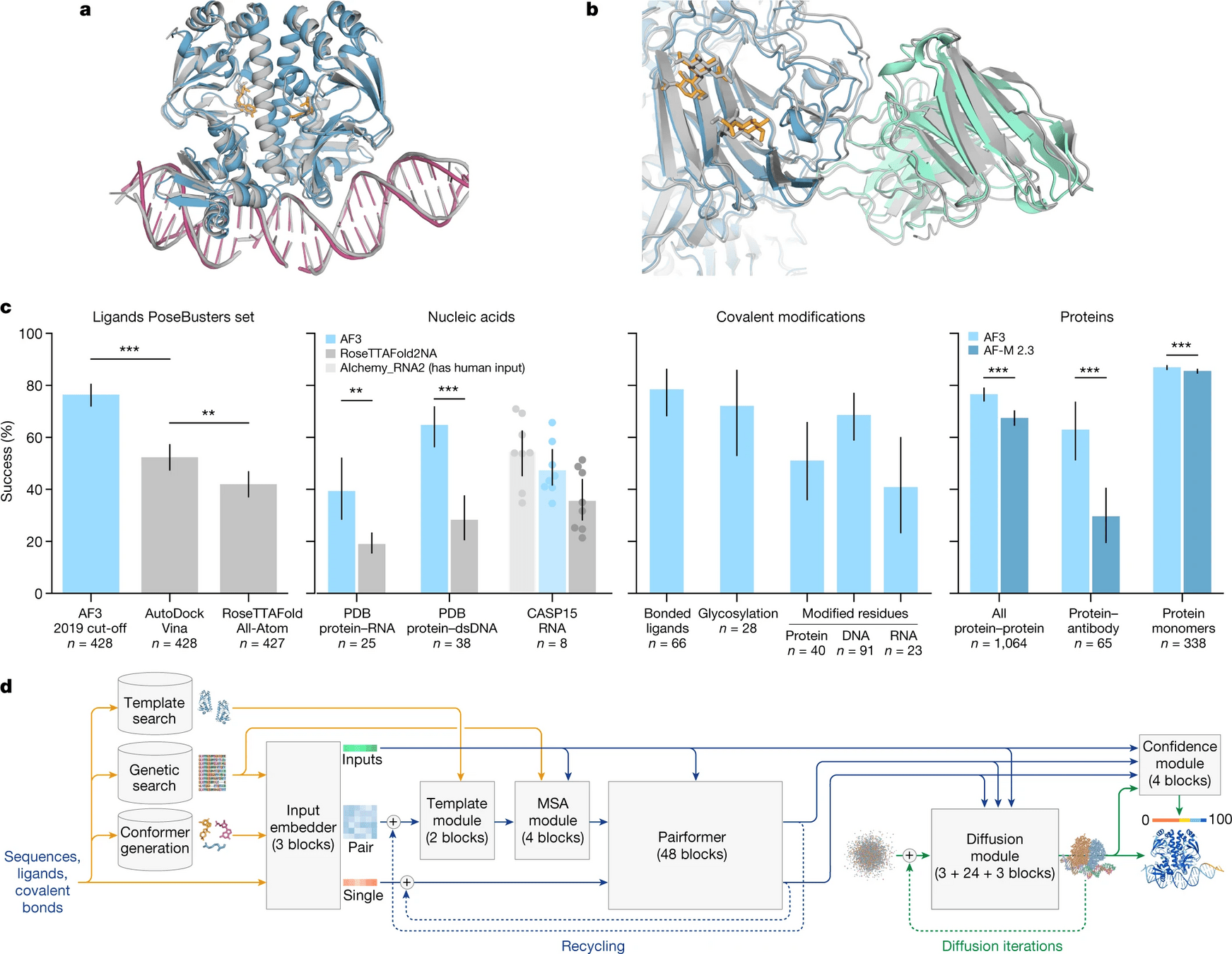
Proteins, DNA, RNA, ligands, PTMs, protein-proteins, etc.

AlphaFold 3
MTLSILVAHDLQRVIGFENQLPWHLPNDLKHVKKLSTGHTLVMGRKTFESIGKPLPNRRNVVLTSDTSFNVEGVDVIHSIEDIYQLPGHVFIFGGQTLFEEMIDKVDDMYITVIEGKFRGDTFFPPYTFEDWEVASSVEGKLDEKNTIPHTFLHLIRKKDHFR (UniProt)
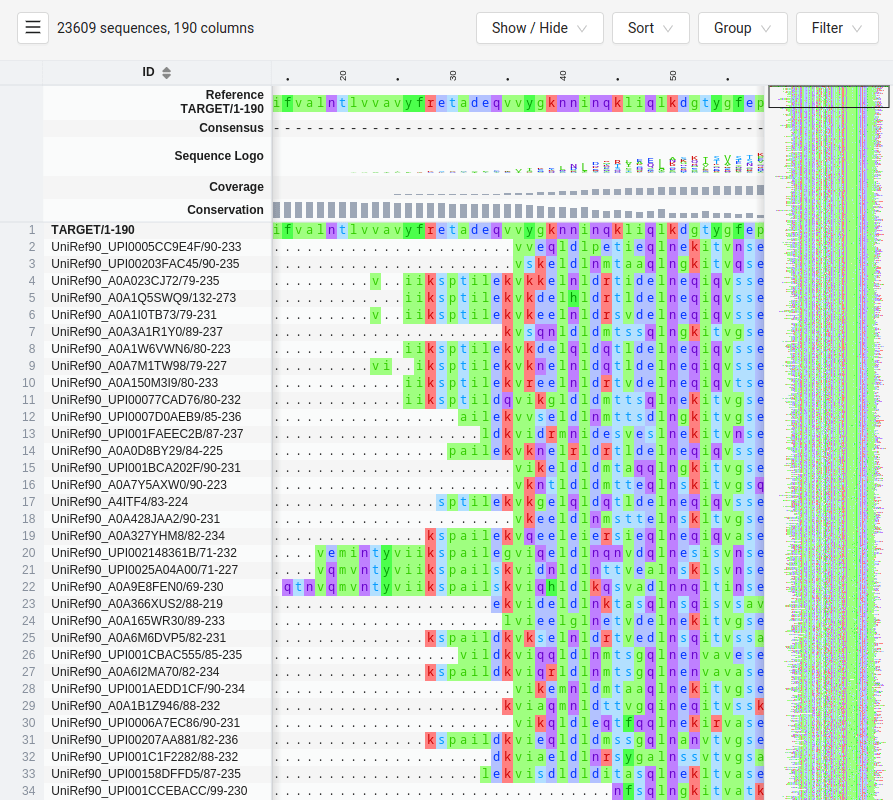
MGKKEVILLFLAVIFVALNTLVVAVYFRETADEQVVYGKNNINQKLIQLKDGTYGFEPALPHVGTFKVLDSNRVPQIAQEIIRNKVKRYLQEAVRIEGTYPIVDGLVNAKYTVANPNNLHGYEGFLFKDNVPLTYPQEFILSNLDGKVRSLQNYDYDLDVLFGEKEEVKSEILRGLYYNTYTRAFSPYKLNovel protein (ChatGPT)
AlphaFold 3 is a breakthrough, not the final solution

Caveat: Proteins are dynamic
What about intrinsically disordered proteins?
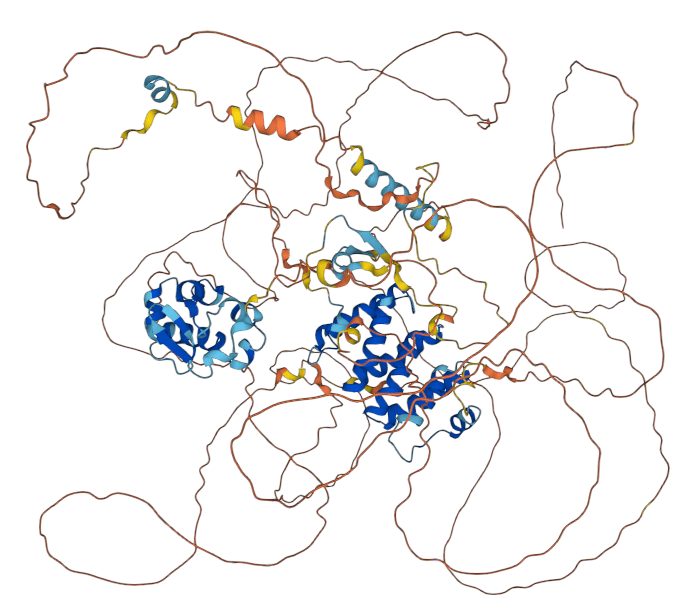
At least 40% of proteins have disordered regions
AlphaFold (and all other methods) struggle with disordered regions
Before the next class, you should
Lecture 11B:
Protein structure prediction -
Methodology
Lecture 11A:
Protein structure prediction -
Foundations
Today
Thursday
A Markov model predicts outcomes based on transitional probabilities
Suppose I collect weather data in Pittsburgh for the past 30 days: Sunny, Cloudy, or Rain
I want to figure out how to predict tomorrow's weather based on today's
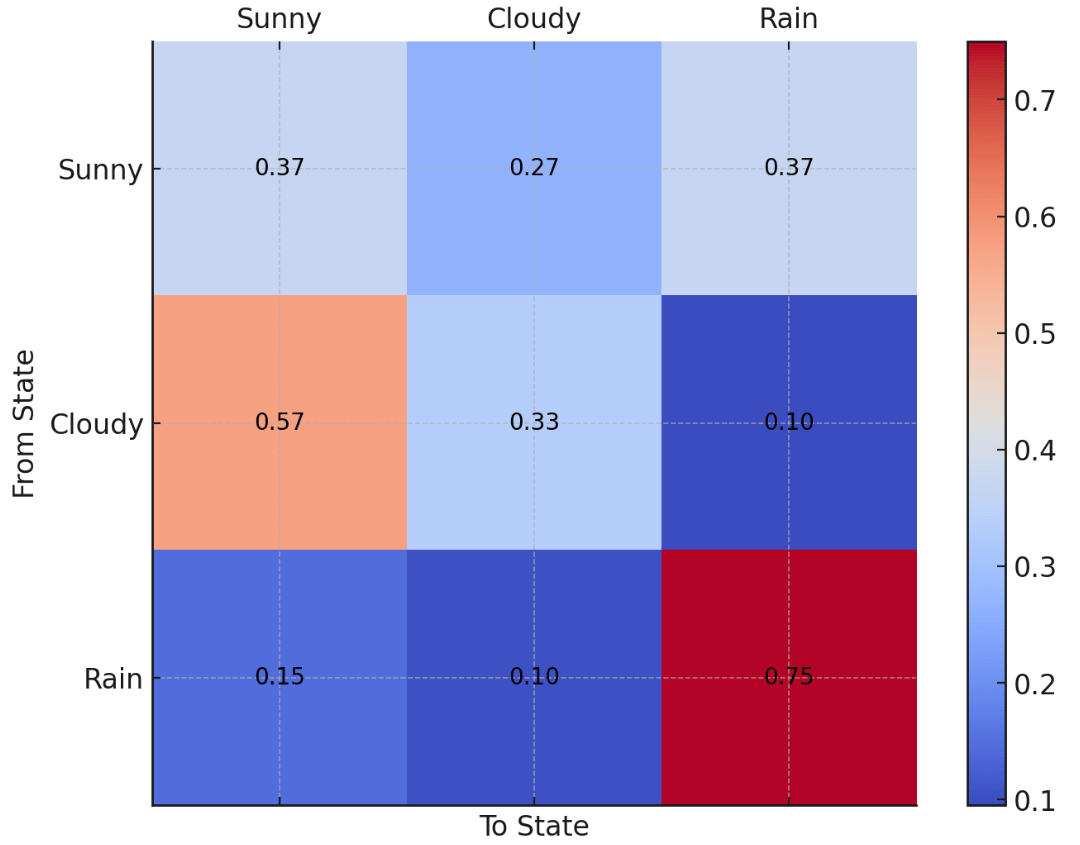
Today's weather
Tomorrow's weather
Transition probability
Example: If today is cloudy, there is a 57% chance it will be Sunny tomorrow
We can represent these states and probabilities as a (cursed?) graph

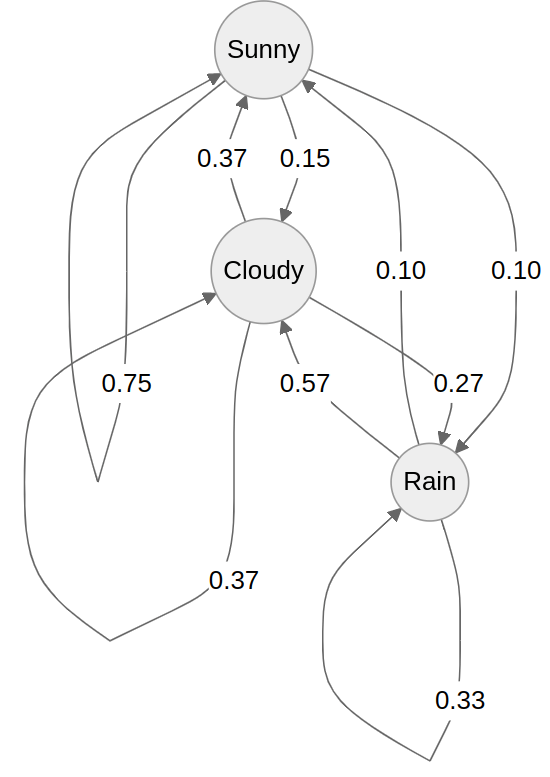
Each edge represents the probability of transitioning from one state to the next
Hidden Markov models also include additional information in "hidden states"
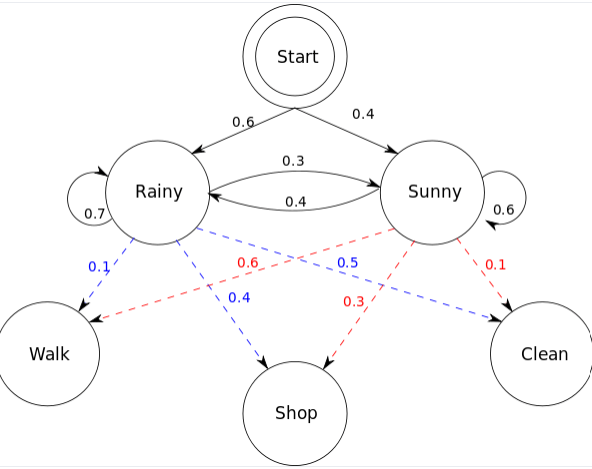
Suppose my friend lives in a remote location where it is either Rainy or Sunny
I cannot look up the weather but I have last year's weathers reports
- Walking
- Shopping
- Cleaning
We know how weather patterns transitions, but we don't have this information from our friend
Obervables
Hidden states
Note: If we had previous observable data, we could fit/learn transition probabilities of hidden states
My friend can only tell me
Hidden Markov Models (HMMs) Capture Evolutionary Patterns in Proteins
HMMs are statistical models representing sequences using probabilities for matches, insertions, and deletions
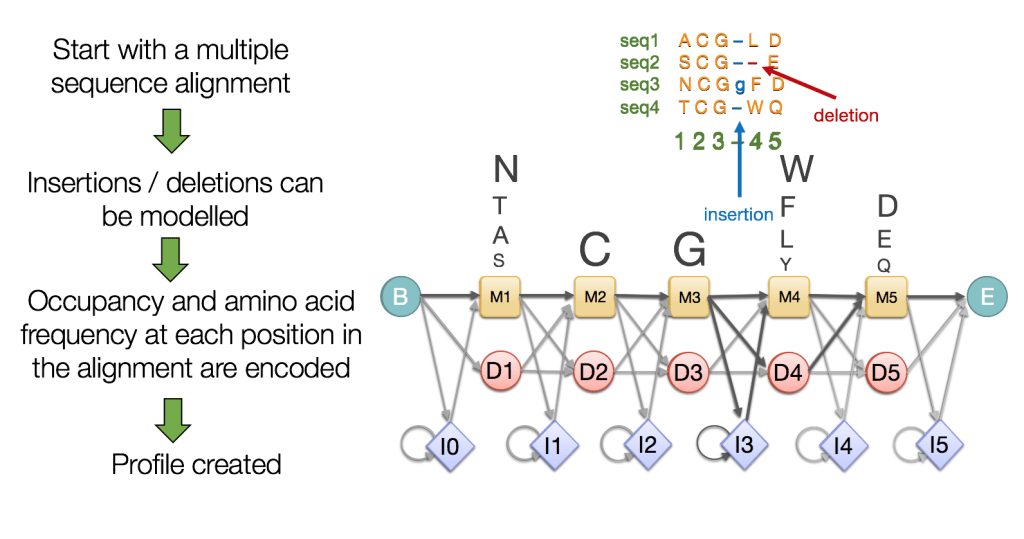
Essentially more robust alignments
HMMs Model Protein Sequences as a Series of Probabilistic States
Hidden states represent the underlying biological events that are not directly observable
Observables are the actual amino acids (residues) in the protein sequence that we can observe

- Match states: conserved positions in the sequence
- Insertion states: positions where extra residues are added
- Deletion states: positions where residues are missing
SWISS-MODEL
SWISS-MODEL
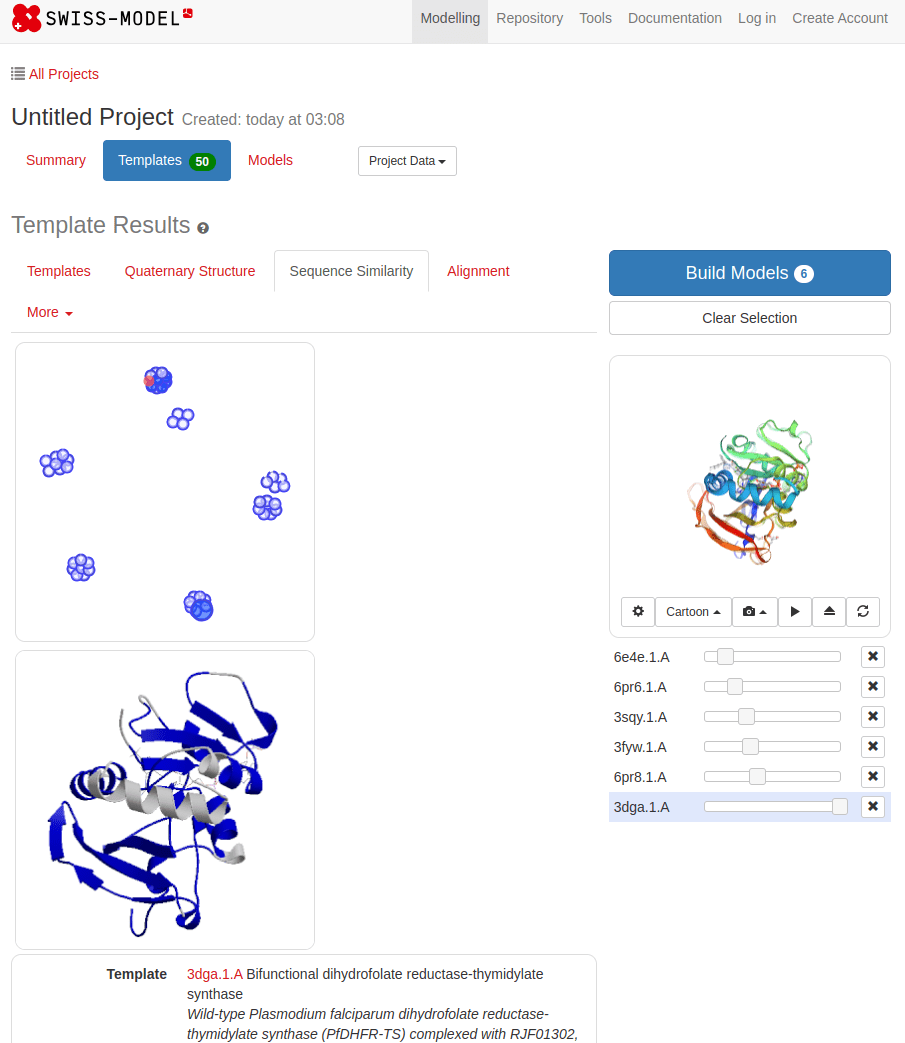

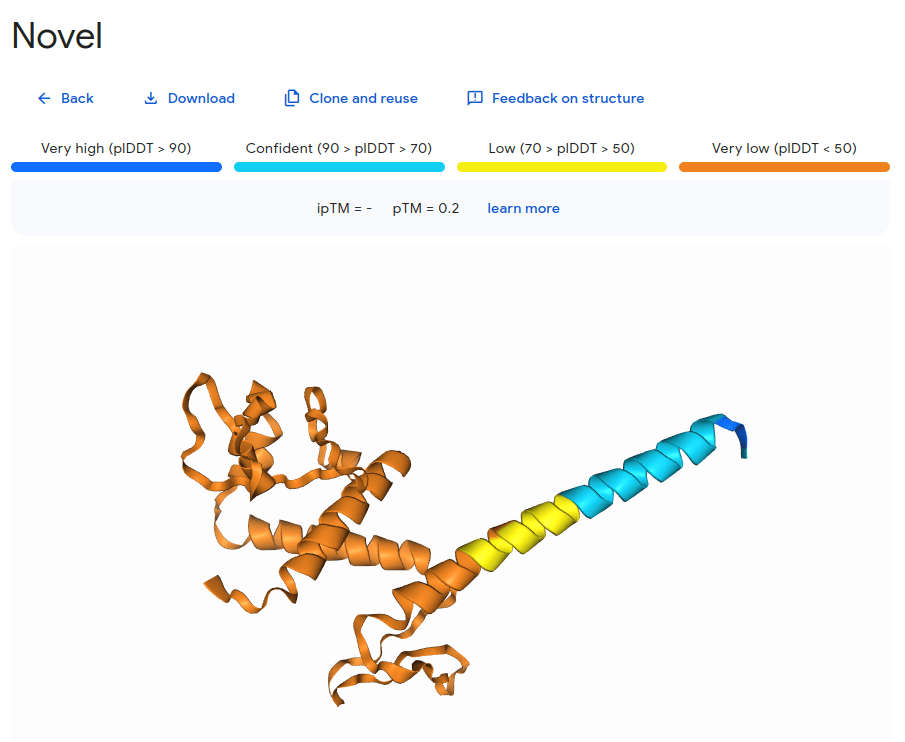
What happens with a novel protein?
MGKKEVILLFLAVIFVALNTLVVAVYFRETADEQVVYGKNNINQKLIQLKDGTYGFEPALPHVGTFKVLDSNRVPQIAQEIIRNKVKRYLQEAVRIEGTYPIVDGLVNAKYTVANPNNLHGYEGFLFKDNVPLTYPQEFILSNLDGKVRSLQNYDYDLDVLFGEKEEVKSEILRGLYYNTYTRAFSPYKLNovel protein (ChatGPT)
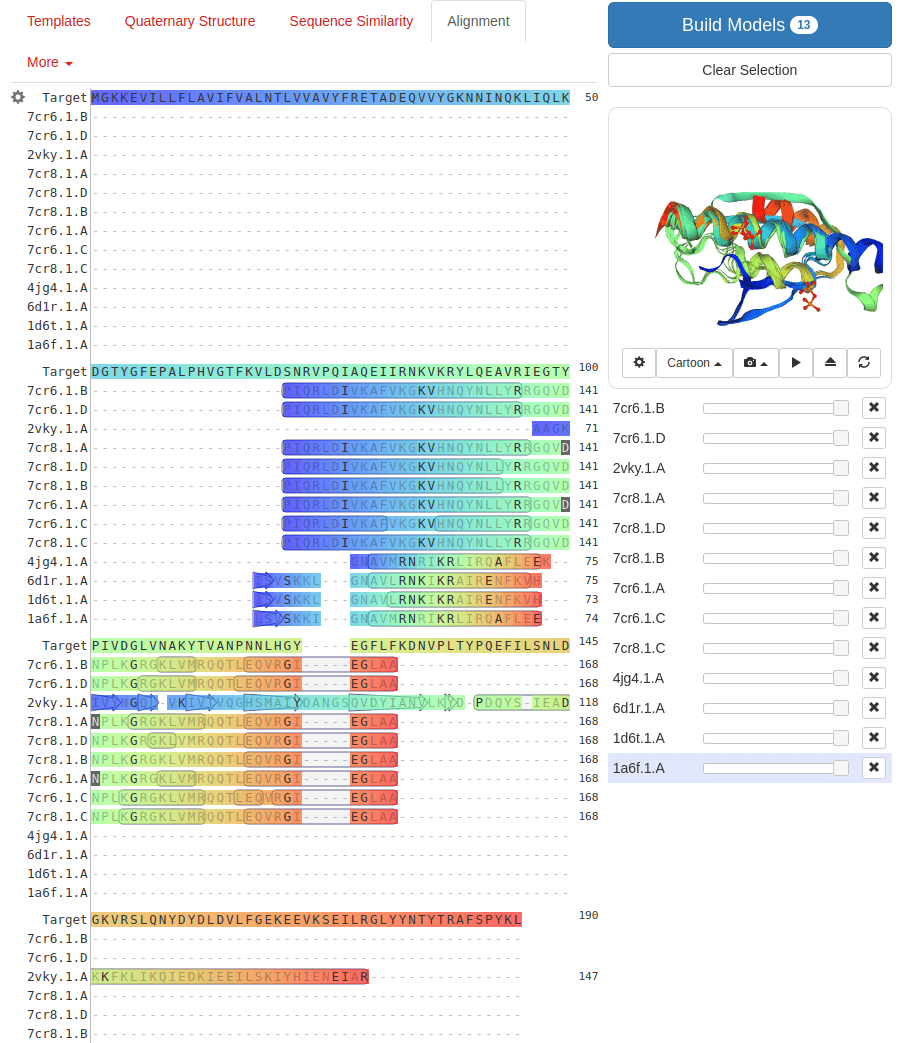
Prediction methods depend on known structures and evolutionary information
Example: AlphaFold has made strides, but predicting de novo structures remains challenging, especially for proteins with no templates
Our predictions rely on similarity to known structures, but novel sequences or folds (for which no homologous structures exist) are difficult to predict accurately