LarvaMap
RPS ASA
LarvaMap is...
LarvaMap is a parallelized Lagrangian transport model that uses remote distributed environmental data to perform simulations.
The model is implemented in Amazon's Cloud for biologists to study the settlement patterns of larvae.
It includes the combined effects of environmental forcing and larvae behavior in numerous life stages.
Lagrangian vs. Eulerian
Lagrangian fluid dynamics follow a specific fluid element as it moves in space and time - sitting in a boat floating down a river
Eulerian fluid dynamics use a reference frame that is fixed in space in which fluid elements move through it in space and time - sitting on a dock watching a boat float by
Applications
- Volcanic plumes
- Oil/chem transport
- Sediment transport
- Larval fish
Motivation
Support
Modeling System
- Behavior library to create, catalog, and share larva behavior descriptions
- Lagrangian fate and transport model written as a Python library (paegan)
- Job queue web services implementing the transport model in a cloud architecture
- Web client for interacting with web services through REST API
paegan library
The core of LarvaMap is a Python library called paegan
- Transport model
- Data access
- Geospatial and astronomical utilities
- Transport model
- Data access
- Geospatial and astronomical utilities
Python lacks a broad common data model (CDM) library for array based met/ocean data stored in netCDF or distributed over OpenDAP.
Paegan attempts to fill this need.
-
Grids and meshes
- Coordinate identification
- Performance
>> from paegan.cdm.dataset import CommonDataset>> url = "http://thredds.axiomalaska.com/thredds/dodsC/PWS_DAS.nc">> pd = CommonDataset.open(url)>> test = pd.restrict_vars("u").restrict_bbox((-74, 40, -70, 42)).restrict_depth((3, 50)).nearest_time(datetime(2011, 5, 1, 0, 0, tzinfo=pytz.utc))
Paegan attempts to fill this need.
-
Grids and meshes
- Coordinate identification
- Performance
>> from paegan.cdm.dataset import CommonDataset>> url = "http://thredds.axiomalaska.com/thredds/dodsC/PWS_DAS.nc">> pd = CommonDataset.open(url)>> test = pd.restrict_vars("u").restrict_bbox((-74, 40, -70, 42)).restrict_depth((3, 50)).nearest_time(datetime(2011, 5, 1, 0, 0, tzinfo=pytz.utc))Workflow
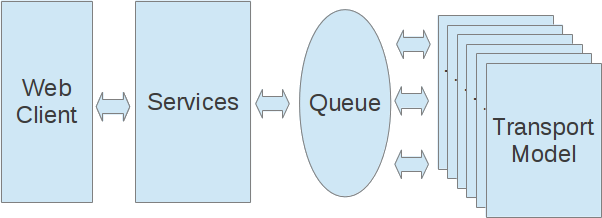
- Configure larva behavior and model scenario
- Model job sent to service and validated
- Modeling job placed in queue
- Amazon EC2 instance picks up run from queue and executes
Scaling
Each transport model is an unaware self-contained "server"
--------------------------------------------------------->
| Scale to modeling demand
| by adding more instances to
| Amazon pool
|
|
|
| Scale individual run speed
| by adding more cores to
V individual instances
The Transport Model
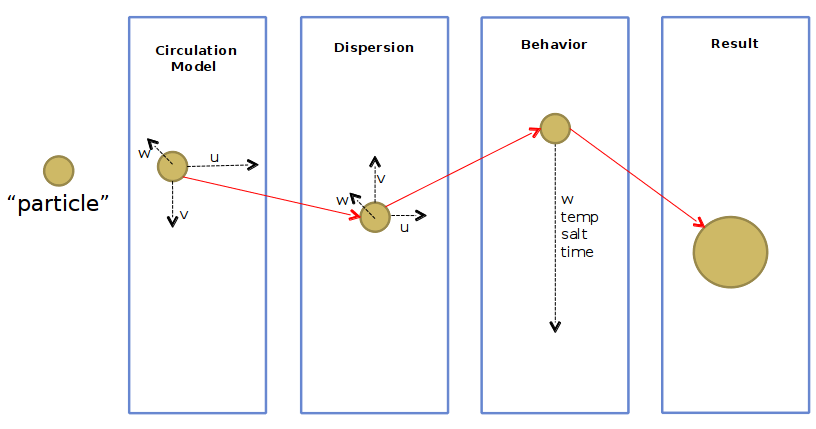
"Grid-less"
(or native grid)
-
shoreline
- bathymetry
- spawning region
Distributed Data Access
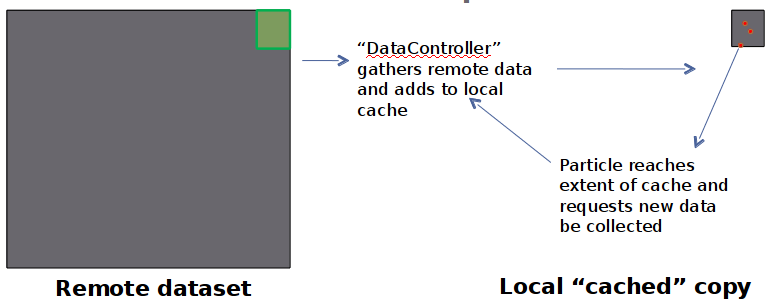
- Data requested "as needed" from remote OpenDAP server
- Particles running in parallel all use local cached copy in forcing algorithms
Distributed Data Access
Distributed Data Access
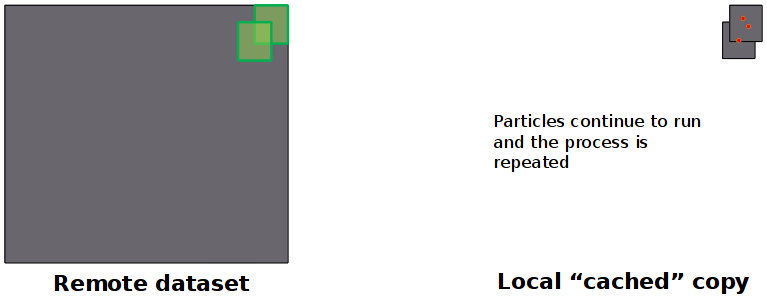
Distributed Data Access
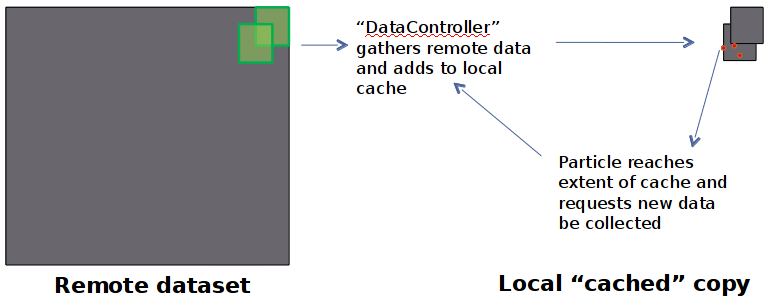
Distributed Data Access
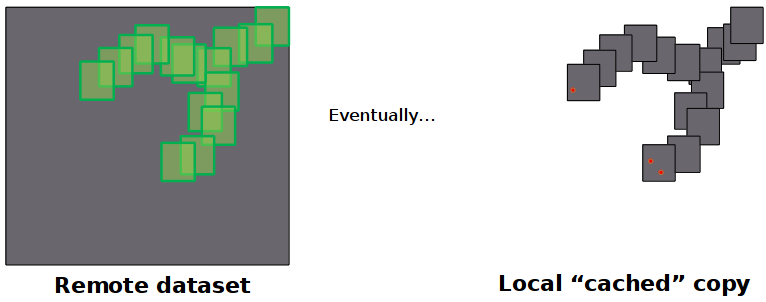
Model Output
The model outputs the trajectories of each of the Lagrangian "particles"
- netCDF format using CF-1.6
- ESRI Shapefile (Point features)
Case Study
Prince William Sound
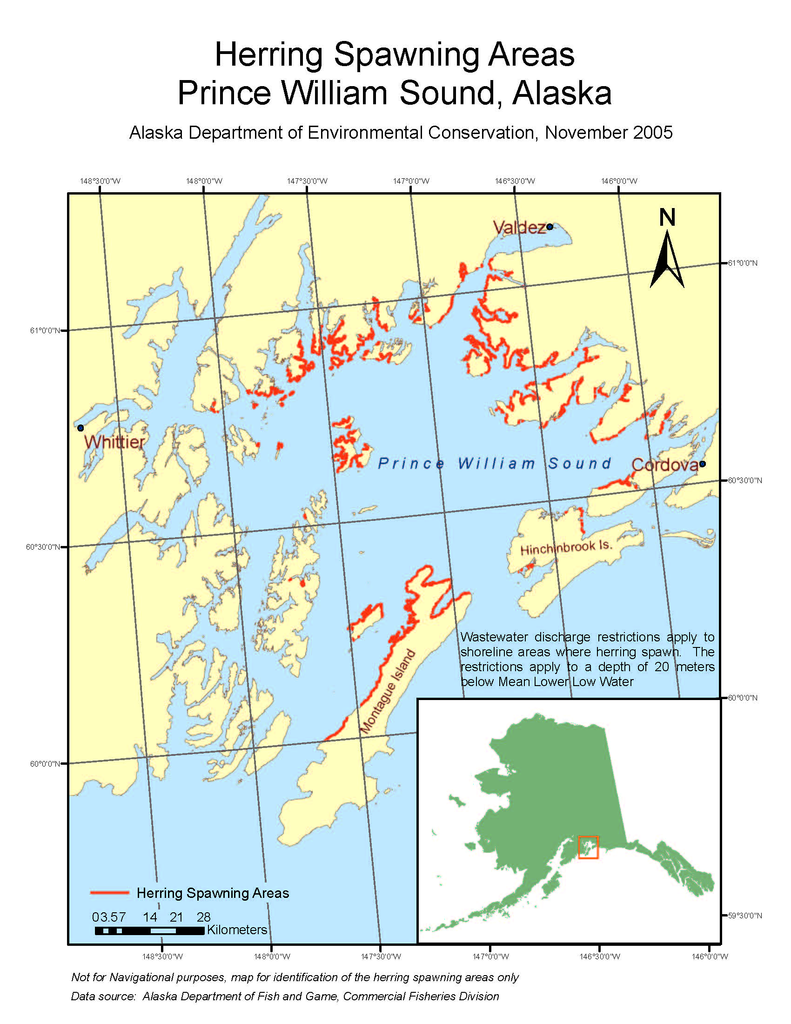
Trajectory
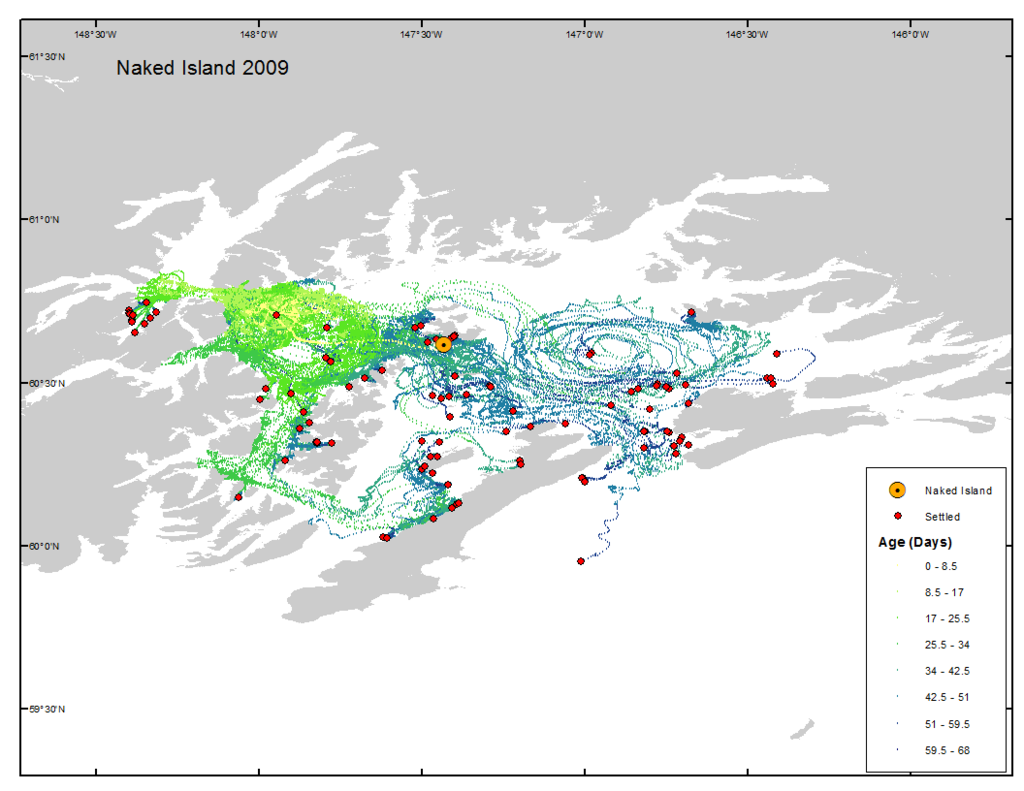
Probability
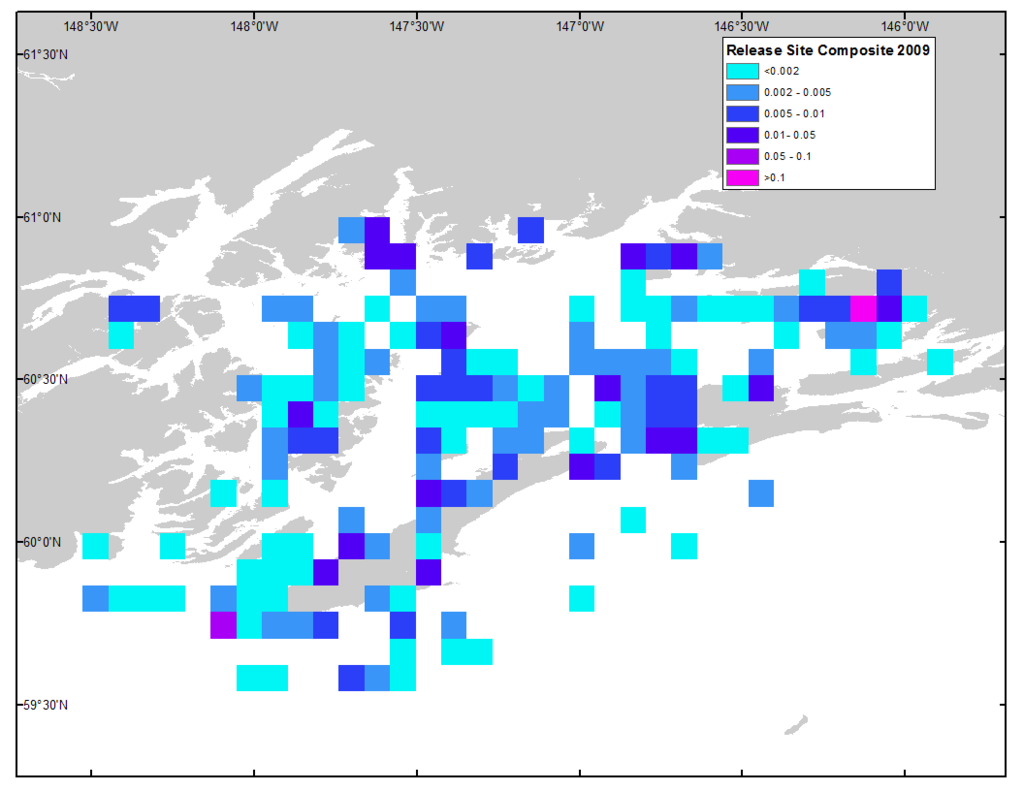
Lessons
- Stability of remote services and data
- Complicates code (defensive coding)
- Slows down model runs
- Parallel arch + distributed data
- Easy to add new model instances to cluster
- Very hard on remote data servers
Next Steps
- Better support in data model
- More analysis tools
- More visualizations
- New types of Lagrangian elements
- Move some code to C and Cython
- Leverage GPU computing
