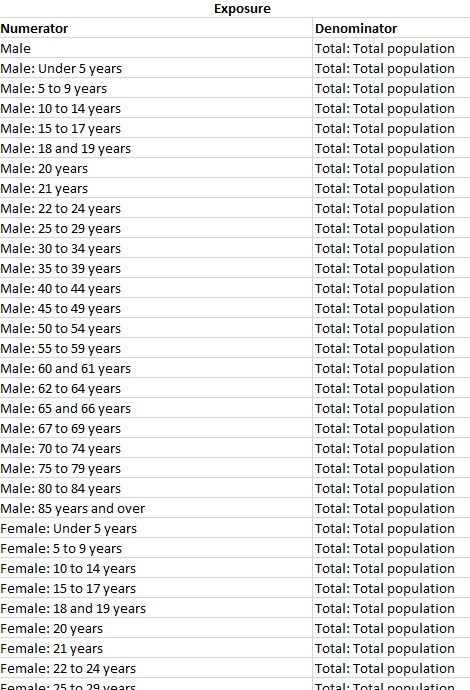
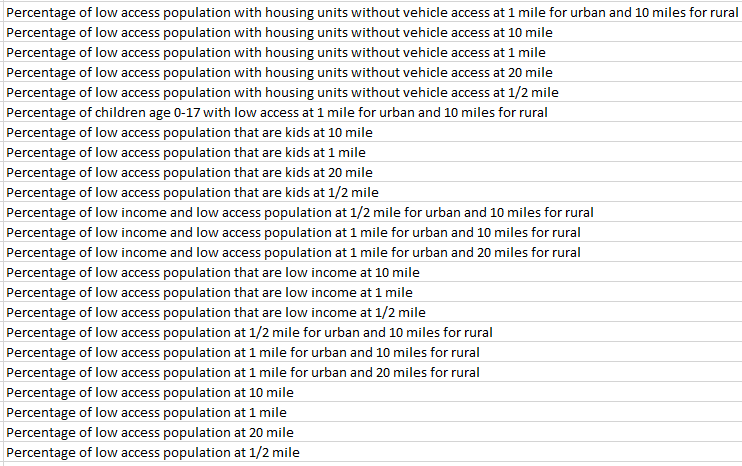
Methodological Challenges in
Spatial and Contextual Exposome-Health Studies
Hui Hu Ph.D.
Assistant Professor of Medicine
Associate Epidemiologist
Channing Division of Network Medicine
Brigham and Women's Hospital and Harvard Medical School
July 27, 2022
The Exposome

To draw attention to the critical need for more complete environmental exposure assessment
"encompasses all life-course environmental exposures from the prenatal period onwards, complementing the genome"
"in a broader sense of all lifestyle, infections, radiation, natural and man-made chemicals and occupational exposures"
Environmental Exposures
Two domains:
- The internal exposome
- The external exposome

Source: Hu et al. 2022

Examples of publicly available spatial and contextual exposome data sources
Source: Hu et al. 2022

Challenge 1:
Engineering of the spatial and contextual exposome data

- Multiple data sources available to assess the same spatial and contextual measure
- PM2.5:
- Air Quality System (AQS) - EPA
- Fused Air Quality Surface Using Downscaling (FAQSD) - EPA
- LUR modelled estimates - CACES
- GWR modelled estimates - ACAG
... - Walkability:
- Walk Score - Redfin
- National Walkability Index - EPA
...
Data Source Identification
Source: Hu et al. 2022
- How to choose?
- Based on spatiotemporal coverage and scale
- What if there are still multiple options available with similar spatiotemporal coverage and scale?
- Traditional studies: domain knowledge + sensitivity analyses
-
This approach is increasingly challenging in spatial and contextual exposome studies:
- Lack of expertise
- Infeasible to conduct sensitivity analyses
-
Potential solution: establish reference spatial and contextual exposome databases with gold-standard measures
- on different spatial and contextual exposome constructs
- across different geographic areas and time periods
Variable Selection
- Large variabilities in the number of variables included in existing exposome-health research
- 8 ~ 14,663 based on our systematic review in 2020
- A data source may include multiple variables measuring similar spatial and contextual exposome constructs
- Example: ACS includes thousands of variables characterizing contextual-level social environment
Two approaches with different assumptions/hypotheses:
- Include all individual variables:
- Assumes represent different constructs
- Seeks to understand the impact of each variable separately
- Perform dimension reductions and use indices:
- Assumes these variables matter in aggregate
- Quantifying their individual contributions is difficult or not of interest
vs
- Neighborhood deprivation index
- Index of concentration at the extremes
...
Source: Hu et al. 2020
Variable Selection
- For many spatial and contextual exposome factors, it is possible to generate multiple variables with the exposures aggregated at different spatiotemporal windows
- Example: ACS, FARA
- Large impacts on downstream studies:
- different p-value cut points used to account for multiple testing
| Total number of variables | p-value of var1 | p-value cut point | Statistically significant? |
|---|---|---|---|
| 100 | 0.0001 | 0.0005 | Yes |
| 1,000 | 0.0001 | 0.00005 | No |
- Potential solution:
- develop ontology-based approaches to standardize variable selection and the approaches of making these choices
Source: Hu et al. 2020

Challenge 2:
Spatiotemporal linkages of spatial and contextual exposome data to indivdiuals
Spatiotemporal Linkages
| id | long | lat | startDate | endDate |
|---|---|---|---|---|
| id | exp1 | exp2 | ... |
|---|---|---|---|
- Approach 1: Preserve the original spatial scale (treat the exposures as area-level factors)
- Approach 2: Use buffers to generate individual-level exposure estimates

- Different buffer sizes
- Area-/population-weighted averages
- Not necessarily circular buffers, can also use other shapes

Source: Lovasi et al. 2011
- Challenges in scalability using buffer-based approach:
- large number of exposures
- large number of sample size
- most existing packages implements in-memory processing
- Potential solutions:
- Google Earth Engine
- PostGIS with PG-Strom

Challenge 3:
Statistical methods for spatial and contextual exposome-health studies

Many statistical methods have been developed/applied in exposome-health studies
Predominantly developed to handle exposures measured at the individual-level
Source: Hu et al. 2022
Differences between toxicant/chemical mixtures and the spatial and contextual exposome
| Toxicant/Chemical Mixtures | The Spatial and Contextual Exposome | |
|---|---|---|
| Number of variables | 10-10 | 10 -10 |
| Common sample size | 10 -10 | ≥10 |
| Spatial structure | No | Yes |
| Temporal structure | Minimal | Yes |
2
4
3
2
4
5
Scalability
- Some methods have been applied to studies with thousands of exposures
ExWAS and elastic-net
p=5,784
N=819,399



ExWAS
p=337
N=3,108

Scalability
- Most existing methods have only been applied to studies with relatively small number of exposures and sample size
- existing simulation studies often consider small scale scenarios

p=237
N=1,200
p<20
N<250


Lack of consensus to handle heterogeneous spatiotemporal scales
- Different spatiotemporal aggregations can lead to subsequently different associations
- the modifiable areal unit problem
- the modifiable temporal unit problem
- Two approaches widely used in the field to address data heterogeneity:
- area-/population- and time-weighted averages based on pre-selected spatiotemporal exposure windows
- preserve the original spatiotemporal scales and account for them in analyses using appropriate statistical methods
- Largely unknown:
- performance of these two strategies
- whether and how the modifiable areal/temporal unit problems impact results
- whether the performance and impacts are different by exposures

Challenge 4:
Using spatial and contextual exposome data for disease prediction
- Spatial and contextual exposome data are appealing to be used in disease prediction:
- wide availability of geolocation data in both clinical and research settings
- low cost to obtain and append these data to large number of individuals
- Most existing efforts so far only used single or very few spatial and contextual factors



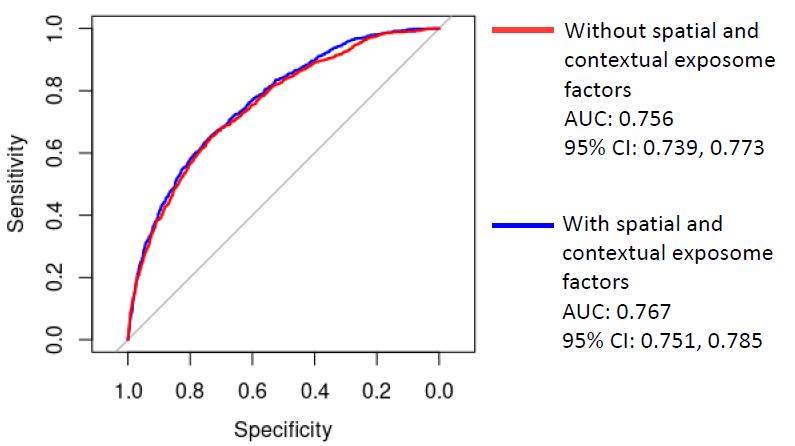
50,368 patients with COVID-19 between March 2020 and October 2021
Predictors:
- Sociodemographic factors: age, gender, race/ethnicity, health insurance
- Comorbidities
- County-level COVID-19 related factors (#days since first case, vaccination rates, hospital bed capacity)
- With or without the spatial and contextual exposome







Linked data from:
- Florida Vital Statistics Birth Records (VSBR)
-
Florida Pregnancy Risk Assessment Monitoring System (PRAMS)
- We developed predictive models using information from:
- VSBR only
- VSBR + PRAMS
- VSBR + PRAMS + Exposome
VSBR only: 0.63 (0.59, 0.67)
VSBR+PRAMS: 0.68 (0.65, 0.72)
VSBR+PRAMS+
Exposome: 0.74 (0.70, 0.77)



Testing AUC
Gradient boosting decision trees:
- CatBoost
- A total of 6,989 women randomly divided into 80% training and 20% testing sets
- Hyper-parameters tuned by a grid search based on 5-fold cross validations
- Traditional machine learning models have been predominantly used
- manual spatiotemporal aggregations -> loss of spatiotemporal structures -> loss of model performance
- Deep learning approaches are appealing
- perform automatic feature selection and engineering
- especially useful for data with spatiotemporal structures

- Spatial and contextual data include spatiotemporal structures, which cannot be fully leveraged by traditional machine learning models
- Deep learning has been shown to outperform traditional machine learning to preserve spatiotemporal structures in image and time series data
- Differences between spatial/contextual data and image/time-series data
- thousands of variables vs. few variables
- different spatiotemporal resolutions vs. common resolution
- for certain exposures (e.g., air pollution, green space), only a few 'pixels' matters (based on time-activity pattern)
- Existing deep learning model architectures cannot fully leverage the predictive power of spatial/contextual data
Summary
- Engineering of the spatial and contextual exposome data
- Spatiotemporal linkages of spatial and contextual exposome data to individuals
- Statistical methods for spatial and contextual exposome-health studies
- Using spatial and contextual exposome data for disease prediction
Ackowledgements
- NIH/NHLBI K01HL153797
- NIH/NIEHS R21ES032762
- NIH/NHLBI OTAHL161847
Thank you!