Lab Meeting 11/05/2023
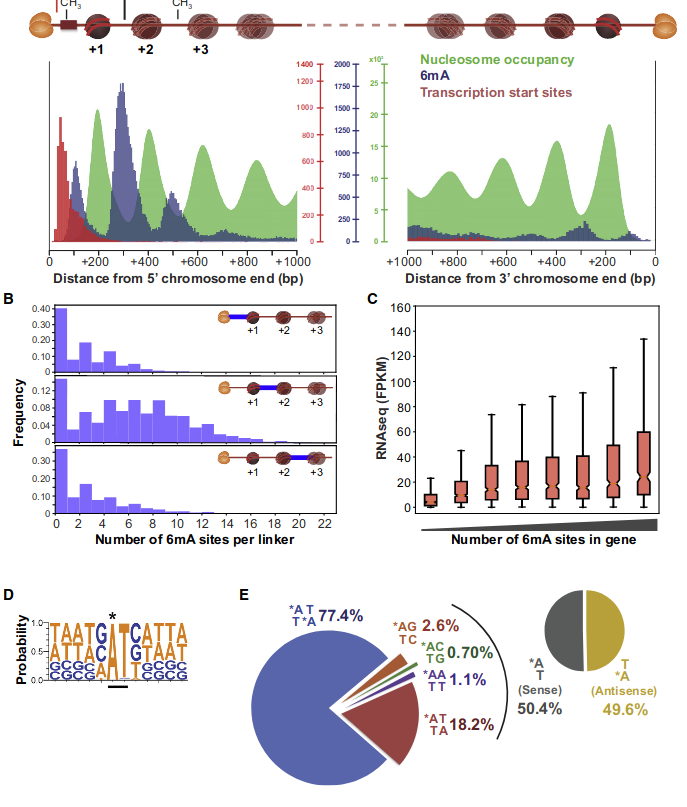
Role of DNA methylation in the programmed rearrangements in P. tetraurelia
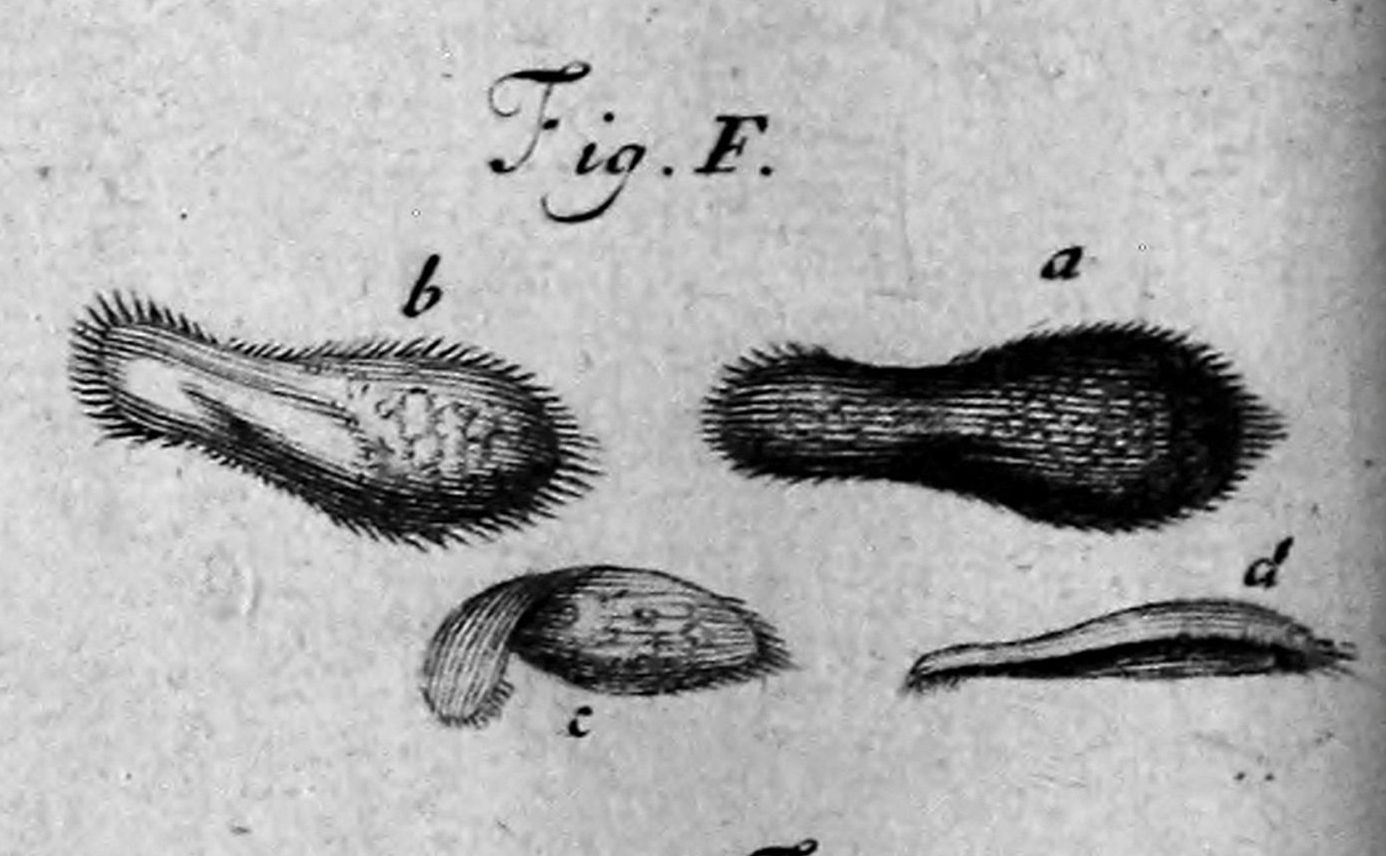
Philosophical Transactions, vol. 23, issue 288. 1703.
CNRS, Leica Microsystems, 2022
My background

- 2011-2018 Pharmacy (Lille)


-
2022: PhD degree at ENS Paris (bioinformatics)




-
2018: MSc Bioinformatics - Paris Diderot


My work history
Code
Stats
Hematology (HSCT)
ML
Ciliates
NGS data analysis
Structural biology



MCMC
DNA methylation
Introducing Paramecium
"The white rats of ciliophora"
Studying ciliates
A 350 years old story



A. van Leeuwonhoek (1668)
"Animalcules"
Pasteur (1862)
Spontaneous generation
HS Jennings (~ 1900)
Paramecium as a model
T. Sonneborn (1937)
Non-mendelian inheritance
of sexual type
in Paramecium

Carol Greider &
Elizabeth Blackburn
Telomeres (1985) - Nobel prize

Meyer and Duharcourt (2014)
Sexual type is inherited via maternal RNAs, in Paramecium
There is more...
-
First known organisms that do not use the "universal" genetic code
- Paramecium (Caron and Meyer 1985)
- Tetrahymena (Preer et al. 1985)
-
Histone Acetlyases (HAT)
- Tetrahymena (Brownell et al. 1996)
-
Self-splicing introns (ribozymes)
- Tetrahymena
- Tubulin post-translational modifications
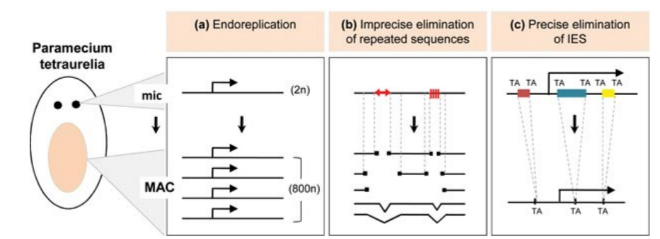
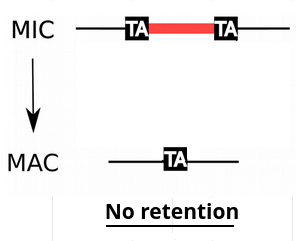
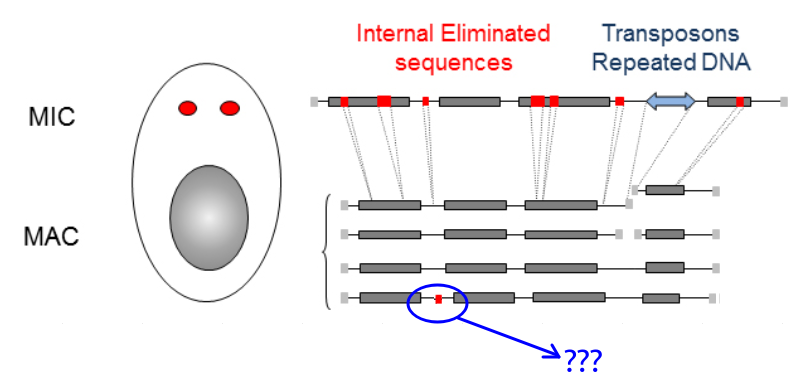
> The genome-wide programmed rearrangements <
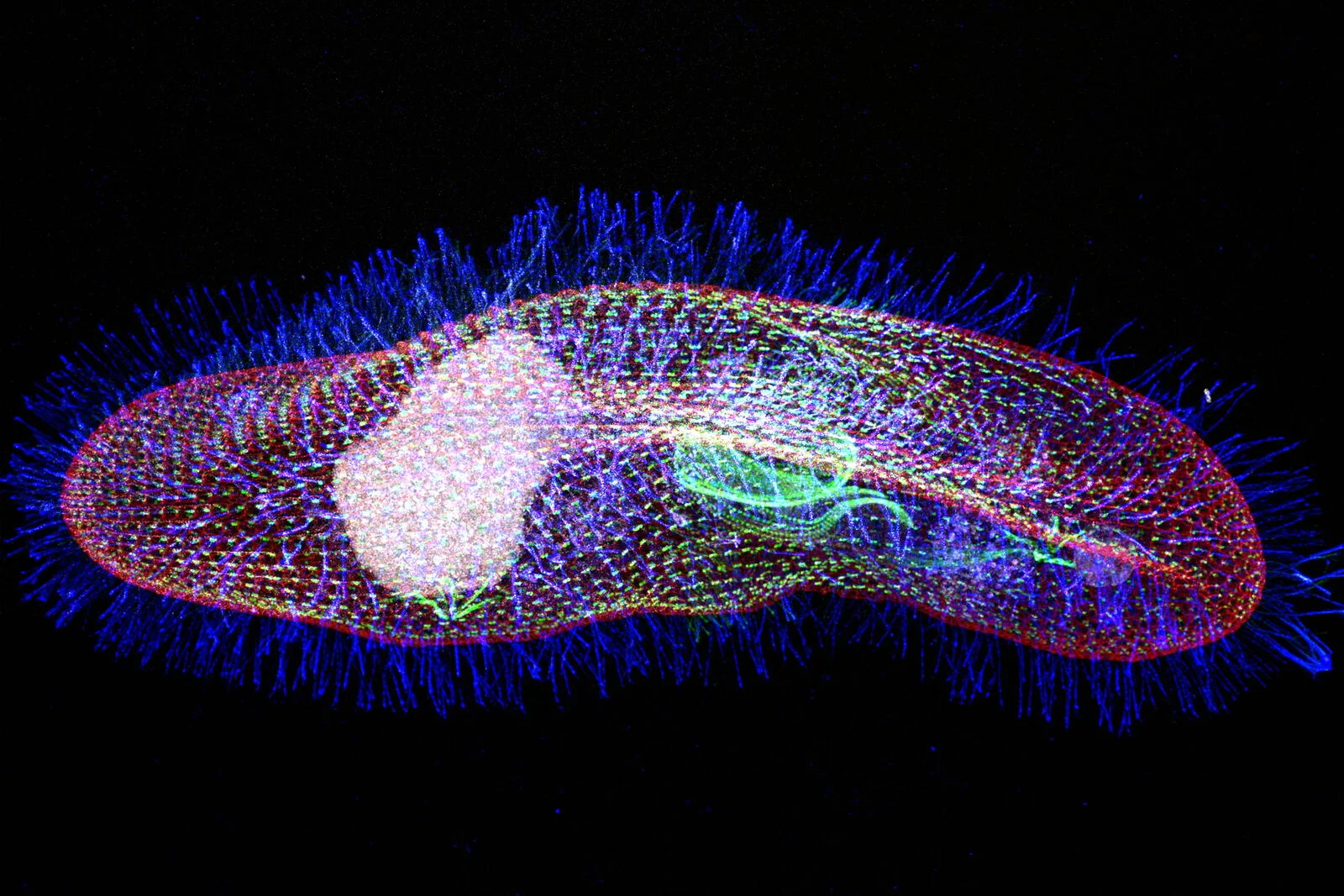
Paramecium tetraurelia

Unicellular eucaryote with 3 nuclei:
- 1xMAC nucleus (up to 800n)
- 2xMIC nuclei (2n)
DNA ratio: 1 MIC for 200 MAC
The difference ?
MAC = Somatic genome
- Transcriptionnally active
- Amplified
- Freed from TEs
- Freed from >49.260 unique "IES"

MIC = Full germline genome
- Transcriptionnally inactive
... I love great stories

Very rare picture of me, explaining my PhD during the lab meeting Pere Castor et Al. (Flammarion FR - Circa 2023 - Colourized)
Surviving the apocalypse of transposons
Lessons learned from 3.5 Bn years of evolution

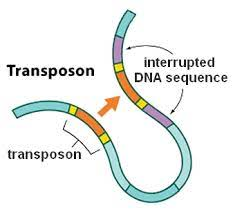
1948
Barbara McClintock : The jumping genes


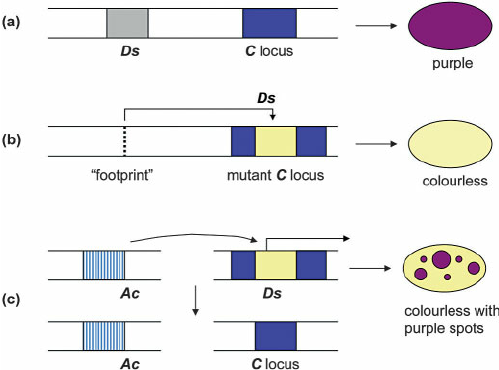
Historical classification
(Finnegan)
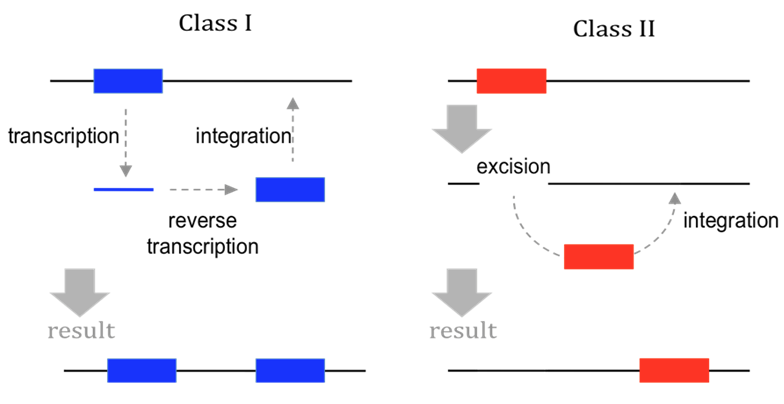
Copy/Paste
Cut/Paste
Both can be autonomous or dependant
Horizontal transfer of TE
- TE are considered as "sharing an ancestor with viruses"
- They could transfer horizontally by their own or through pathogens, pollinators, symbiosis, plasmids...

... And many others
TEs = Chaotic invaders ?
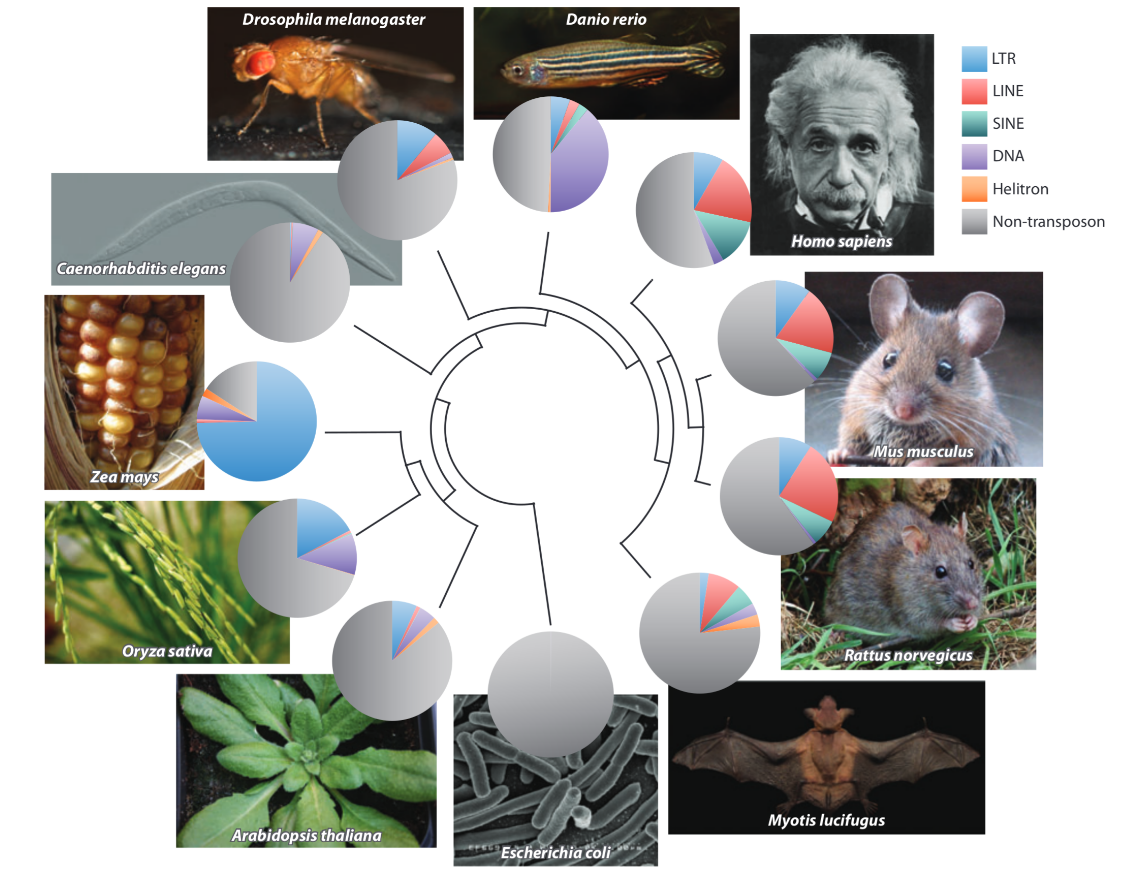
Extreme case 1)
Maize

Up to 86%
He dead ?
Extreme case 2)
- Prokaryote
- One of the smallest genome ever sequenced
- TE ? Transposases ?

2 hypothesis (none proved):
1 - Very high multiplication rate + Very high vulerability to TE
2 - Defense mechanism so great it's not affected
Prochlorococcus marinus SS120
Historical understanding
Junk/Selfish DNA
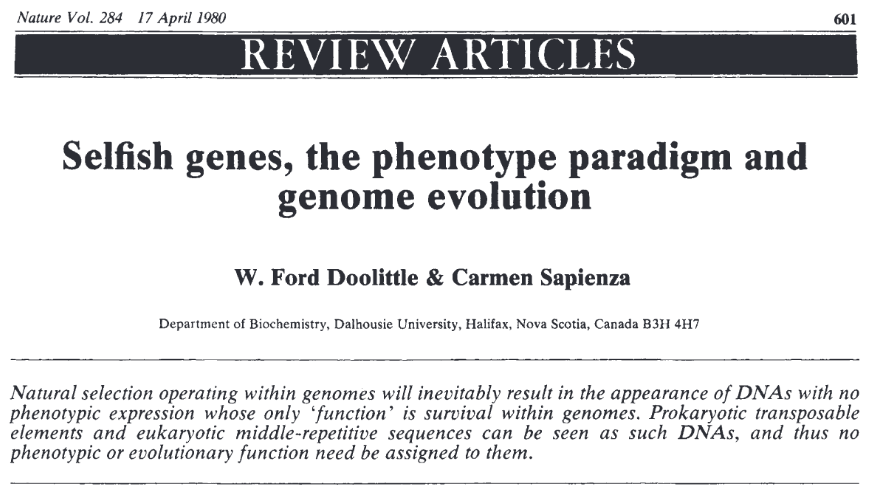

TEs were first considered as :
-
"Selfish DNA"
- They do not perform any "function" for their host
-
Neutral or mildly deleterious
- That is, evolutionary burdens
TEs are conserved not because they provide an additional fitness to their host, but despite the fact that they don't. This is the non-phenotypic selection.
Doolittle, Orgel, Crick and Sapienza (1980)
The same day in Nature journal
The selfish gene - 1976


- Finalist/Anthropomorphist analogy: "The DNA is selfish and only 'wants' to reproduce itself" --> Parasitic DNA
- With limited resources, the best DNA replicators "win"
- The evolution and natural selection could be better described by "DNA replicators" rather than species
Modern taxonomy
Wicker et al. 2007
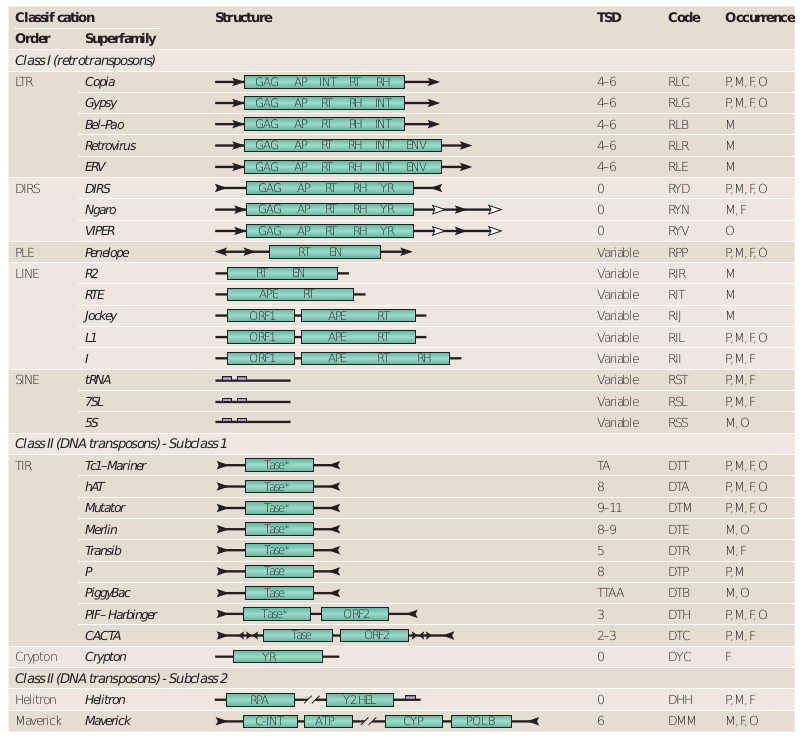
- 3 classes
- 9 orders
- 29 superfamilies
Based on:
- Mechanistic / Enzymatic criteria
- Structural data
The cut-paste versus copy-paste comparison turned out to less relevant over time
TEs are ubiquitous

Modern understanding
- TEs Generate selection :
- Purifying: growth rate = Genome-wide invasion
- Adaptative: Exon shuffling, transcription regulation ++,
exaptation (e.g Rag1 & Rag2 in mammals), etc.
- Are transferred vertically (++) and horizontally (HTT)
- Several thousands known HTT
- P-element invaded Drosophilia worldwide in less than 100 years !
- Are present in all cellular organisms
- Probably exist since the ~ begining of the cellular life
The paradigm is currently shifting from "Junk DNA" to "Major actors of evolution".
$$\sim2^N$$

Regulation mechanisms

> Any particulary vulnerable organisms has been wiped out in the past
> Logical conclusion : All remaining life forms have some kind of resilience towards TEs.
> All virulent TEs have wiped out their host (and disappeared with them)
Many epigenetic regulations exist :
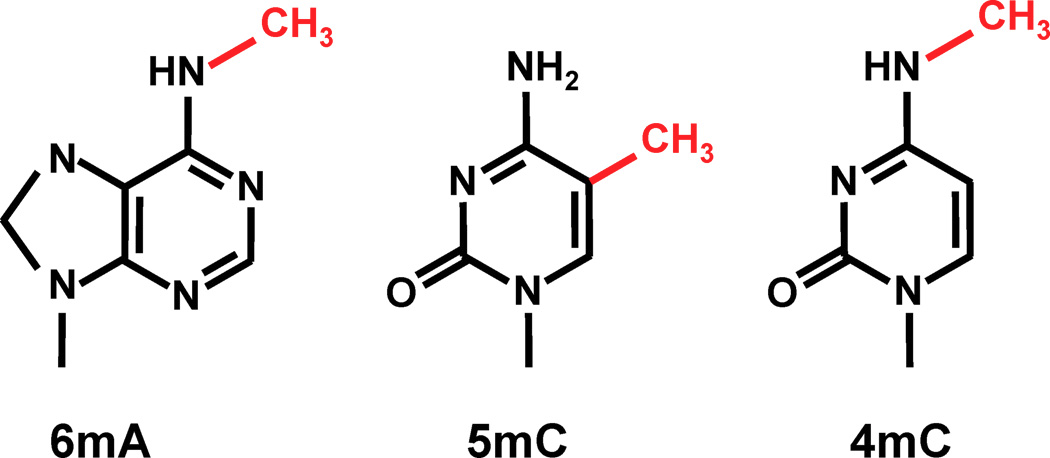
- 5-methylcytosine (5mC) silences TEs in H. Sapiens
- 6mA in Drosophila
- piRNA in animals
- 5mC and H1 Histone methylation in Arabidopsis
- ...
P. tetraurelia has an original way of dealing with TEs
Reconstructing the story of transposable elements

Hints of a hidden past
Reconstruct TE's story is something feasible
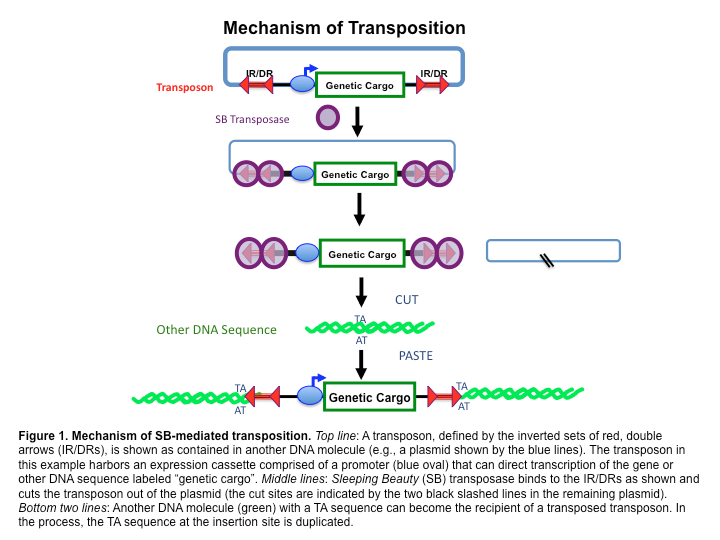

The case of the "Sleeping beauty" transposon
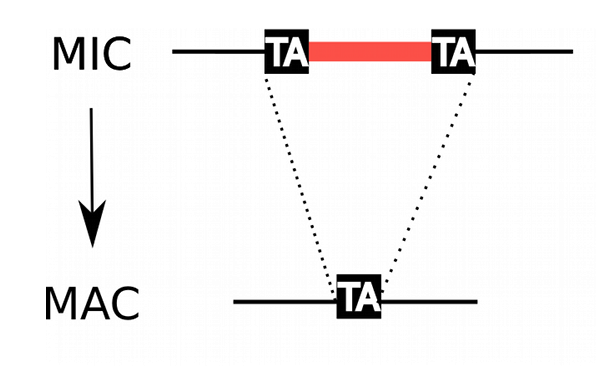
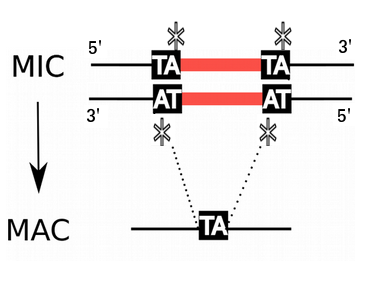
Programmed rearrangements
After sexual processes, a new MAC is formed, with important genome re-arrangements
Results in a MAC DNA almost purely made of coding sequences
Coyne et al. 2012
Profiling IESs (1/2)
- Non-coding
- Remnants of Tc1/Mariner ?
-
All excised by Pgm
- Excised with a single-nucleotide precision
- Life or death issue : Genes interrupted
-
IES excision was exapted many times
- e.g The mating-type !
- Size shrinks with age, most IESs are very short (26-150bp)
49.260 unique sequences
O. Arnaiz et al 2012
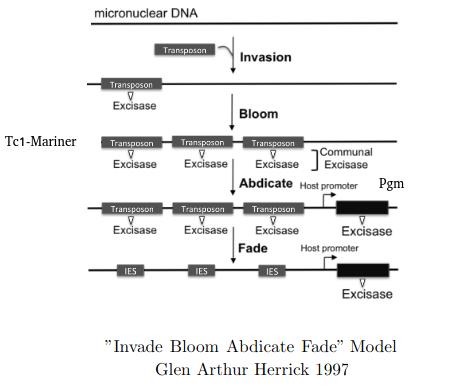
"Invade Bloom Abdicate Fade" model
Adapted from Glen Arthur Herrick 1997
Excision = 100% PiggyMac (Pgm)
Profiling IESs (2/2)
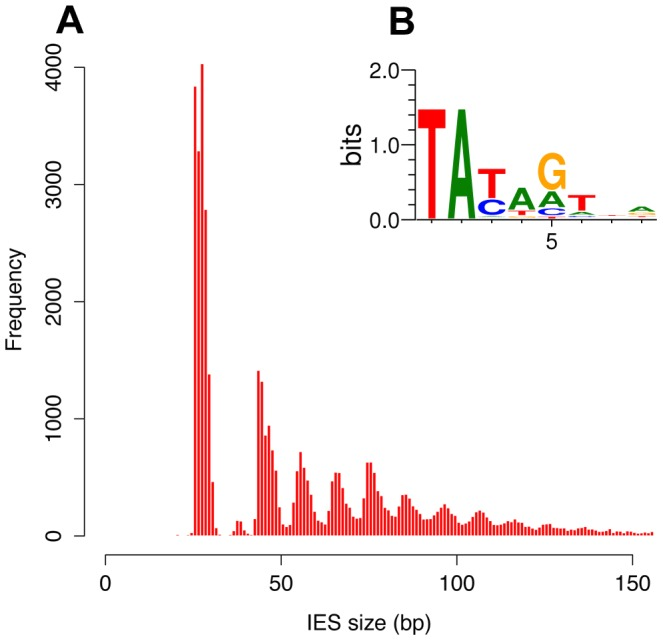
- 100% TA-Bounded
-
Weak consensus TAYAG
-
~ Tc1/Mariner
-
~ Tc1/Mariner
- Periodic size distribution ~10bp
O. Arnaiz et al. 2012
Not sufficient to distinguish IESs from the rest of the genome
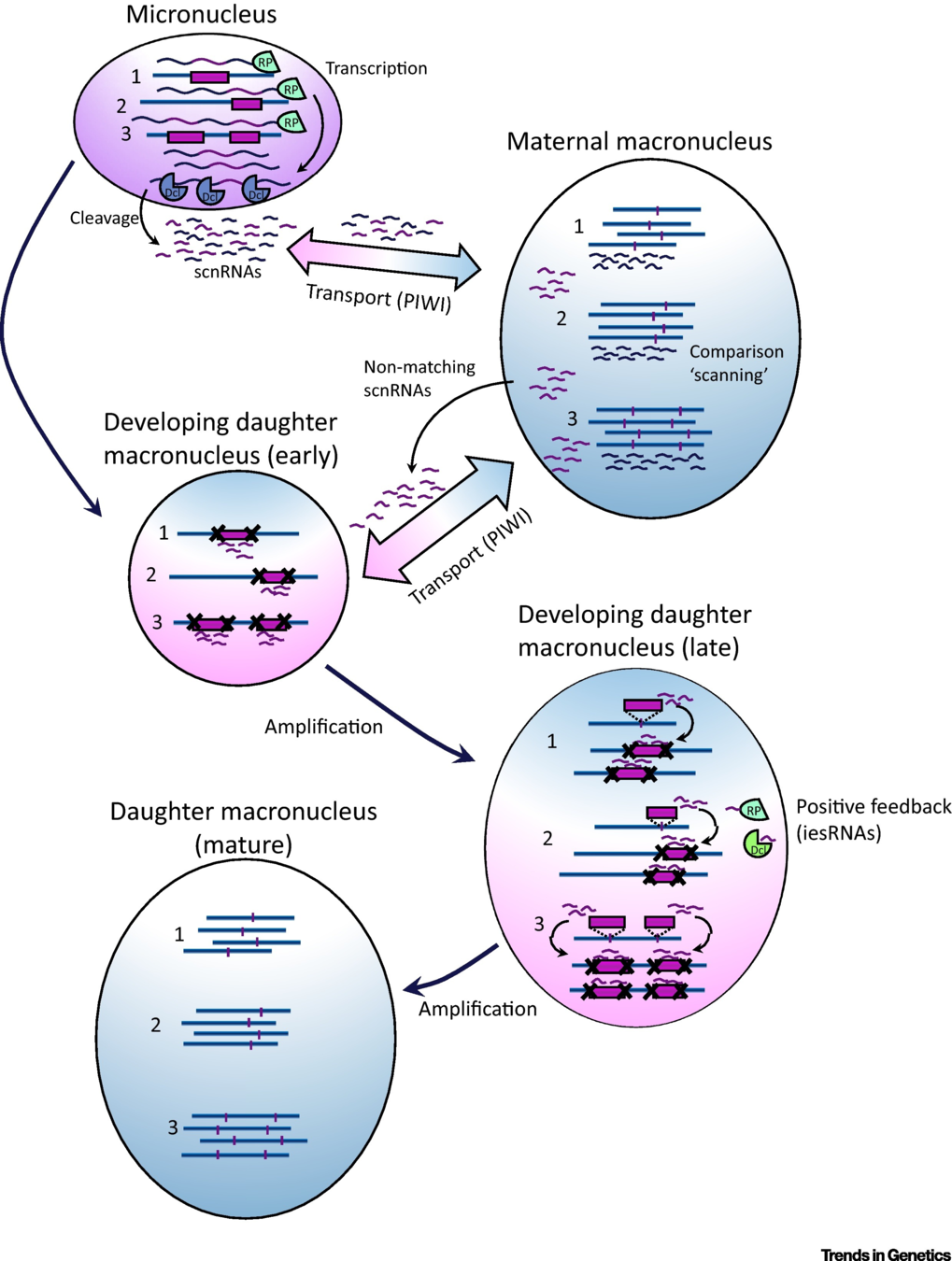
IES recognition: scnRNA pathway
S. E. Allen and M. Nowacki - 2017
If not in the maternal MAC : Recognized and excised
How are IESs recognized ?
We understand the recognition of ~30% of IESs (small ncRNA)
...How does the cell recognizes the other 70% ?
But, almost nothing was known about DNA Methylation in both the MIC, and the MAC

- 100% TA-Bounded
-
Weak consensus TAYAG
-
~ Tc1/Mariner
-
Transposable element
-
Transposable element
-
~ Tc1/Mariner
- Periodic size distribution ~10bp
- Small non-coding RNAs++

O. Arnaiz et al 2012
The IBAF model
- Non-coding
- Remnants of Tc1/Mariner ?
-
All excised by Pgm
- Excised with a single-nucleotide precision
- Life or death issue : Genes interrupted
-
IES excision was exapted many times
- e.g The mating-type !
- Size shrinks with age, most IESs are very short (26-150bp)
O. Arnaiz et al 2012


"Invade Bloom Abdicate Fade" model
Adapted from Glen Arthur Herrick 1997
Excision = 100% PiggyMac (Pgm)
Invade, Bloom, Abdicate, Fade

IES recognition: scnRNA pathway
S. E. Allen and M. Nowacki - 2017
If not in the maternal MAC : Recognized and excised
Problematic
Inactivation of scnRNA and iesRNA pathways:
- ~30% of IESs are retained
- Their retention is not even complete
- Oldest = More independent to small RNAs
- IES features = insufficient to explain the recognition
All IESs may be recognized through the small RNAs
... but is there a redundant system for the oldest/shortest ones ?
Problematic ~ Self VS non-self recognition
Hypothesis : Role of DNA modifications

Two possible roles of DNA methylation
DNA modifications could play a role :
- In the recognition of IESs +++
- i.e : It is permanently present in the MIC
- In their excision
-
i.e : It is transiently present in the new forming MAC, right when the IESs are excised.
- ~ Actor of the scnRNA pathway
-
i.e : It is transiently present in the new forming MAC, right when the IESs are excised.

Hypothesis


The fifth element
-
2.1-2.5% in the MAC and MIC of P. aurelia (Cummings et al. 1975)
-
Detection by SMRT in the MAC (A. Hardy et al. 2020)
- 0.8% and 1.6% of adenines
- 81.5% are located in AT sites
- Enriched downstream the Transcription Start Sites (TSS)
- In other ciliates :
- AT sites ++ and TSS ++ :
- Oxytricha by L. Landweber et al. (2019)
- Tetrahymena (No 6mA in MIC)
- AT sites ++ and TSS ++ :
... Could be N6-methyladenine (6mA)
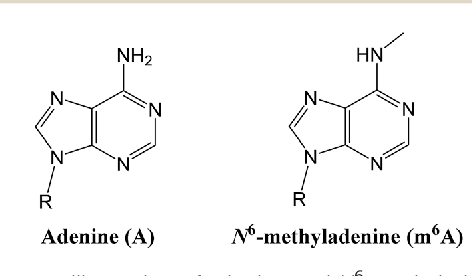
?
Other:
- 4mC ?
- No 5mC in the MAC

Police-sketch of the methylation pattern
If the pattern allows the recognition of IESs, it must allow :
- Single-nucleotide precision
- Distinction between a TA of an IES, from another TA elsewhere
- Conservation through replication
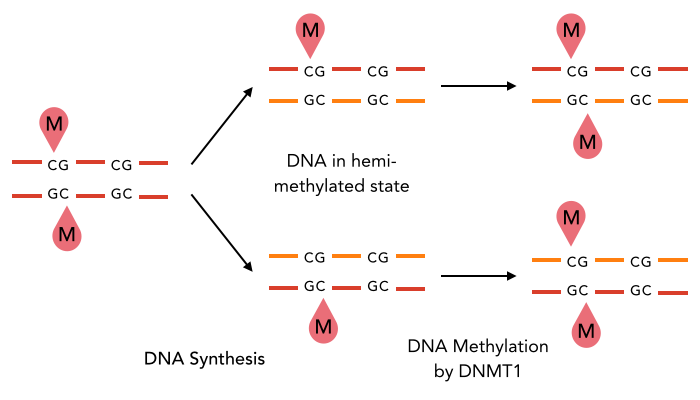
A typical example : DNMT1-Like system
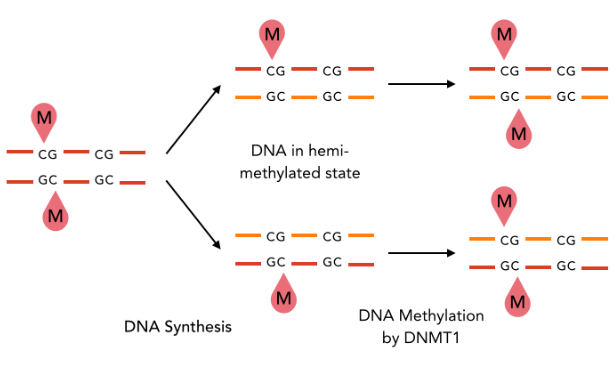
The DNA methylation hypothesis
Examples of possible patterns
And many other possibilities...

Maintenance
through replication ?

Single-nucleotide precision OK
Maintenance through replication : OK if DNMT1-Like


Single-nucleotide precision ?
One hypothesis : DNA methylation (6mA)
Constant 6mA pattern in the MIC (only) ?

Other:
- 4mC ?
- No 5mC in the MAC
2.1-2.5% in the MAC and MIC of P. aurelia (Cummings et al. 1975)
Experimental approach
Long before I arrived in the lab !
Methylase candidates
-
DAMT-1 in C. elegans (6mA) Greer et al. 2015
- MTA-70 domain of DAMT-1 identified in P. tetraurelia too

Expr. Sexual events (collaborators)
RNA methyltransferases ?
Our 6 proteins
- NM family
- NM4
- NM9
- NM10
-
MT family
- MT1A
- MT1B
- MT2
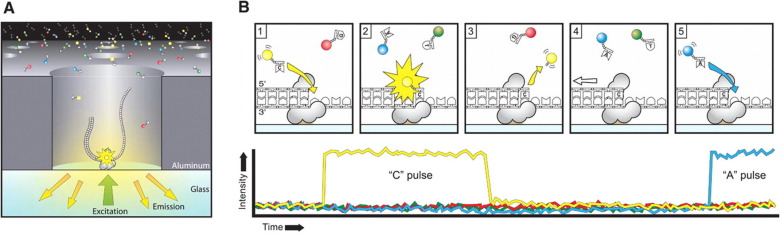
Sequencing strategy
Protocol :
- Silence the methylase candidates by RNA interference
- Sequence with PacBio SMRT sequencing


Vegetative
cells
Control
silencing


RNA interference






Candidate methylases
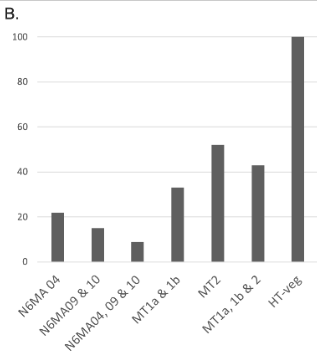
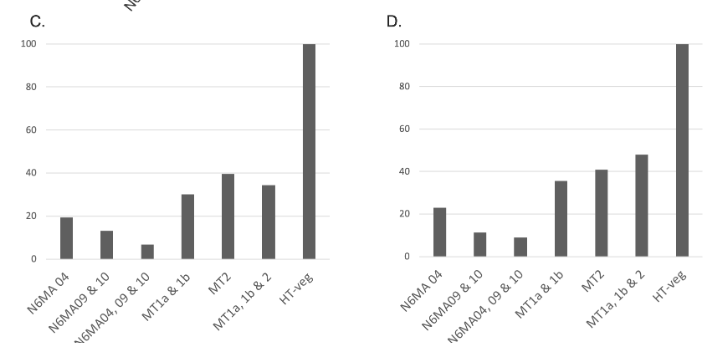
Preliminary results : Reduction 6mA
Southwesternblot
up to 90%
Total DNA
1:200 MIC !!!
Reduction of 6mA
P. tetraurelia
Mitochondrial
Total BET
Southwestern blot
PacBio SMRT sequencing
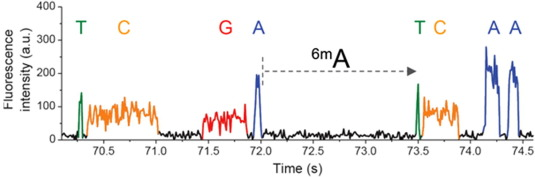
IPD
Nucleotide context (-3/+8nt)
Kinetic signatures
Depends on
DNA modifications
PacBio SMRT sequencing
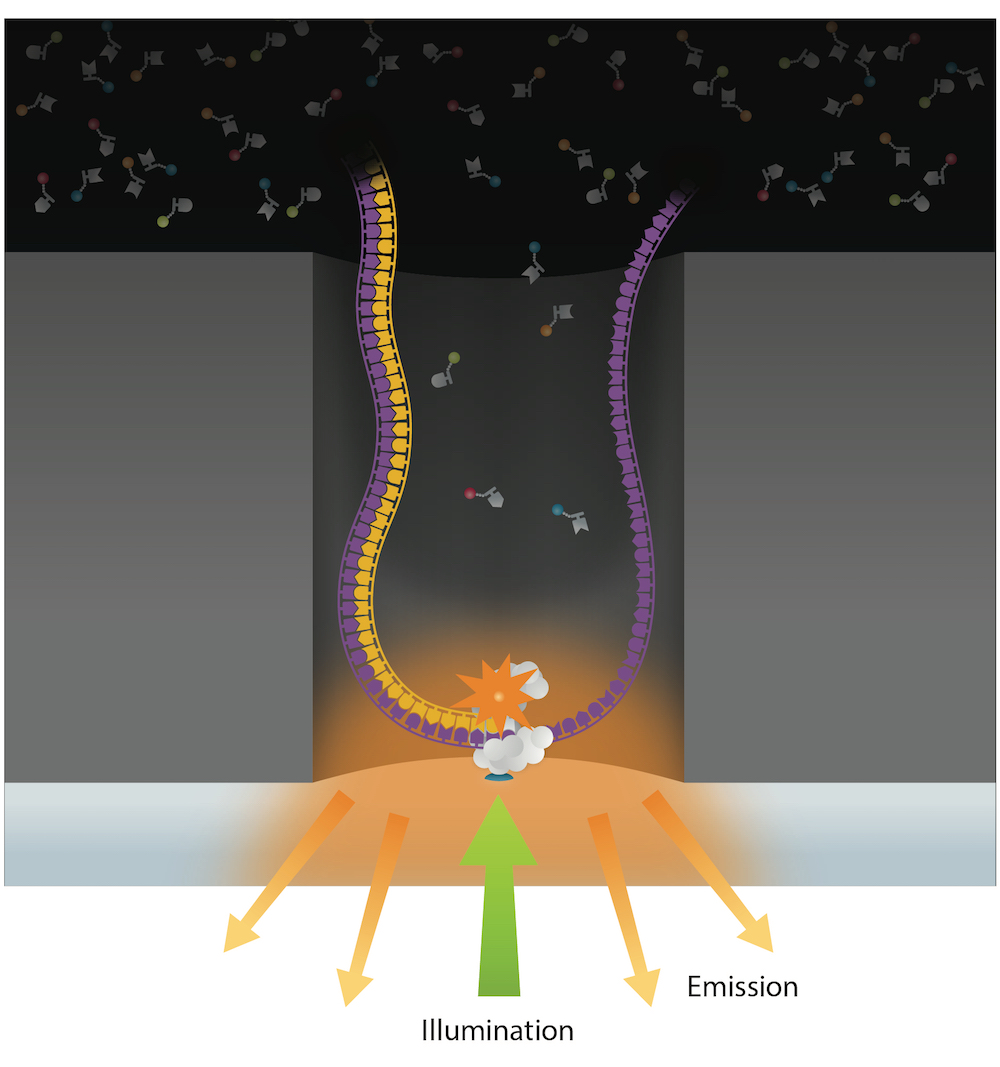
PacBio SMRT sequencing

~ 85% accuracy
~ 100% accuracy
if many passes
10 – 15 kb

IPD
Nucleotide context (-3/+8nt)
Kinetic signatures
Depends on
DNA modifications
Ideal approach
- Purify MIC DNA
-
Make libraries of long inserts from it
- Several kb
- Sequence it with PacBio SMRT
- Compare with PCR of full MIC genome
e.g

~O(20) to ~O(100) molecules
[...]
- Position n°7260 in the genome
- Strand +
- 52 molecules methylated out of 100
Problem = Purification of MIC DNA
(min. 25X)
- Fish IES+ molecules in a sea of MAC molecules
- Short inserts (~350bp)
- Same molecule sequenced multiple times
Random sampling strategy

each molecule
~O(20) to ~O(100) measures
(MIC)
Workaround : work on total DNA instead
- Molécule n°49256
- Nucléotide n°7, strand + (Adénine)
- ipdRatio : 20x
But purifying the MIC is problematic
(min. 25X)

Look at methylation (only) around (some) IESs
in-silico control required
Sorting (overview)

~ 85% accuracy
~ 100% accuracy
10 – 15 kb
~350bp
Alignment
MAC
MAC+IES
MIC
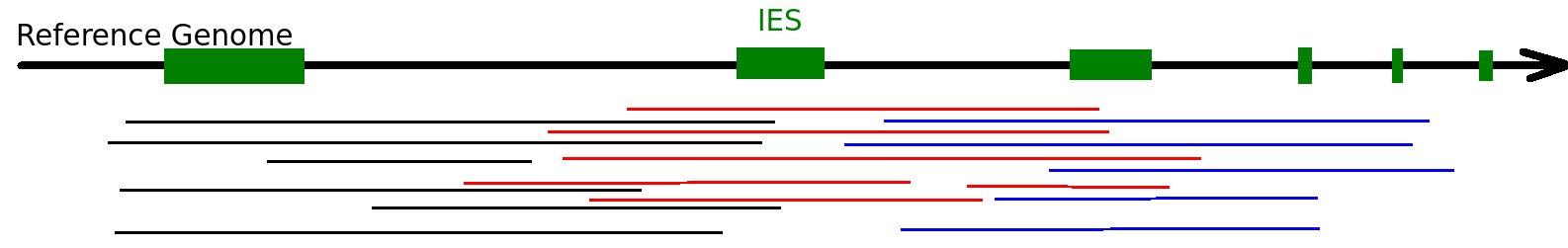
The random sampling strategy
IES
Other MIC
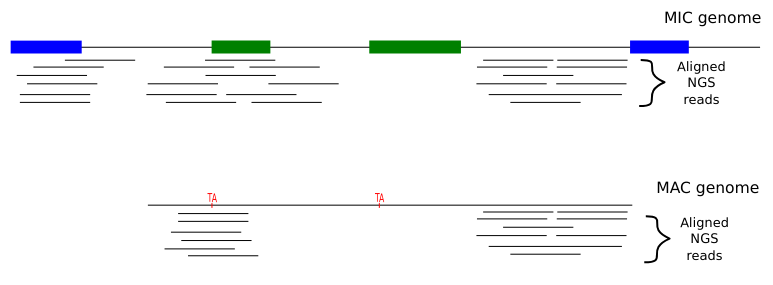
Other MIC
IES
Mac Destinated Sequences (MDS)
MAC
TA Junction
-
Most sequences are Mac Destinated Sequences (MDS) : We cannot guess their nuclear origin
- A vast majority of them (>99.5%) comes from the MAC
The MIC could not be purified
Total DNA was sequenced instead
IES
Other MIC

Other MIC
IES+
Mac Destinated Sequences (MDS)
IES-
Total DNA -> 99.5% of sequencing data = waste
Reminder : 1:200 comes from the MIC
Results
Results
- Very few IES+ reads !
- Most IES+ reads come from... The MAC !
- The MAC ploidy was ill-estimated: a 40 years old mistake
- A pipeline to analyze PacBio SMRT data
- DNA methylation in the MAC
- DNA methylation in the MIC
Few IES+ reads !
- Remove contaminants
- 1 molecule out of 200 comes from the MIC
- ~ 1/6 of MIC inserts will carry an IES
- 30% don't interest us
- # of PacBio reads per sample : 150.000 to 300.000
- ...
That is,
-
Expected ~100 to 300 IES+ molecules per experiment only
- Got 49 to 310
This is not much, but if had been right, 100% of them had to be methylated

Results
- Small amounts of IES+ reads
-
Most IES+ reads come from... The MAC !
- The MAC ploidy was ill-estimated: we fixed a 50 years old mistake !!!
- A pipeline to analyze PacBio SMRT data
- DNA methylation in the MAC
- DNA Methylation in the MIC
P(R)
1 - P(R)
> Not all IES+ molecules come from the MIC
Danger ! -> We are interested only in the *MIC*
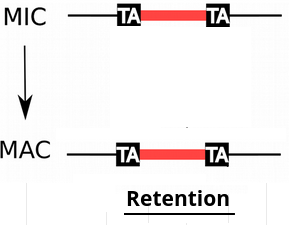
IESs are sometimes retained in the MAC
Even the slightest amount of retention is dramatic...

e.g : If retention = 1/200,
~4 IES+ reads come from the MAC
4 IES+ reads come from the MIC
P( MIC|IES+ ) = 50%
MIC IES+ reads
MAC IES+ reads
MAC IES- reads
MIC : 4n
MAC: 800n
-
Quantification: "IES Retention Score" (IRS)
- MIRET : Cyril Denby Wilkes, Olivier Arnaiz, Linda Sperling 2016, eg:
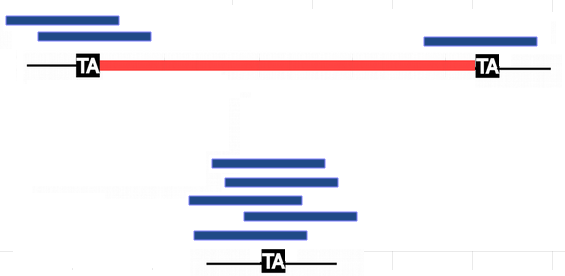
5 reads IES -
1 read IES +
2 reads IES +
$$IRS_L = \frac{2}{2+5} \approx 27\%$$
$$IRS_R = \frac{1}{1+5} \approx 16\%$$
The higher the IRS, the higher the retention.
Quantifying IES retention
If MAC ploidy = 800n than without retention :
$$E(IRS) = \frac{4}{800+4} \approx 0.005$$
If retention :
$$E(IRS) >> \frac{4}{800+4}$$
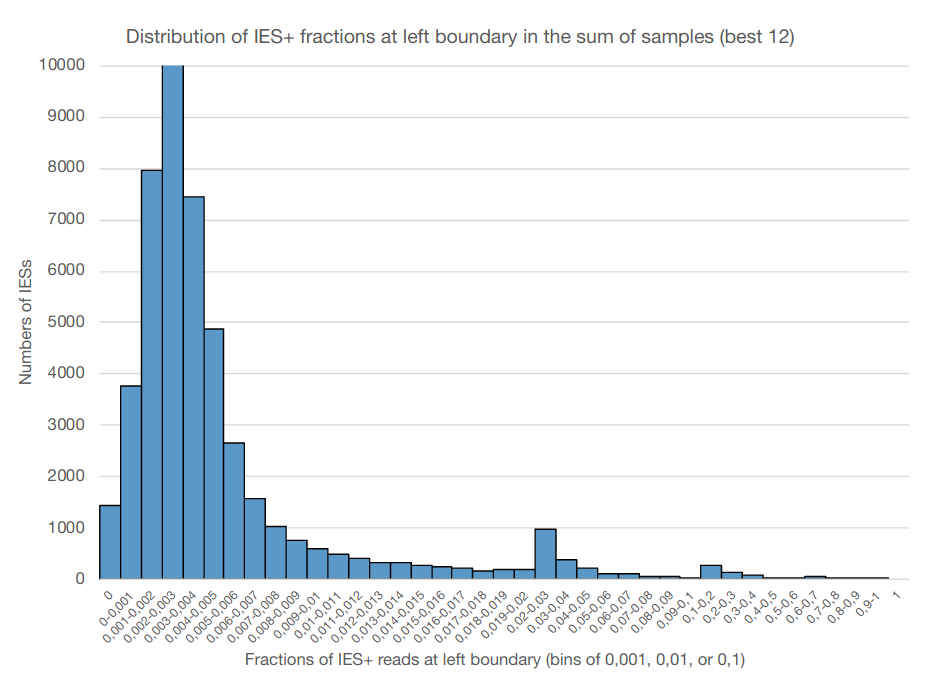
0.002-0.003
The distribution of IES retention scores indicates a 50 years old mistake in the MAC ploidy
The distribution of IES retention scores indicates a 50 years old mistake in the MAC ploidy
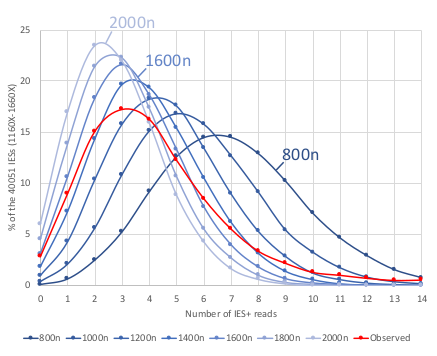
- The real ploidy is 1600N, not 800N !
- 50 years old mistake !!
Where do IES+ molecules come from ? (1/2)

- Bayesian modelling - "Urn problem"
- Hamiltonian Monte-Carlo with Stan
- + Old sequencing datasets
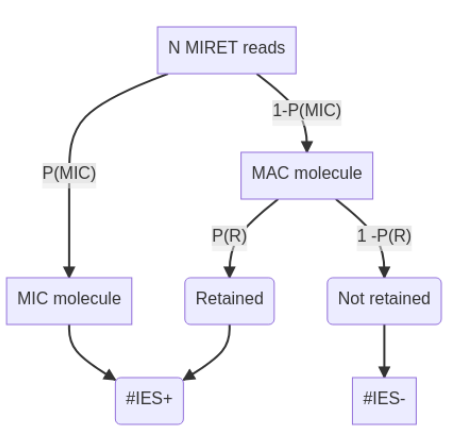




??
P(R) is the only
unknown variable
Numerical application
On a fake dataset (simulated retention)
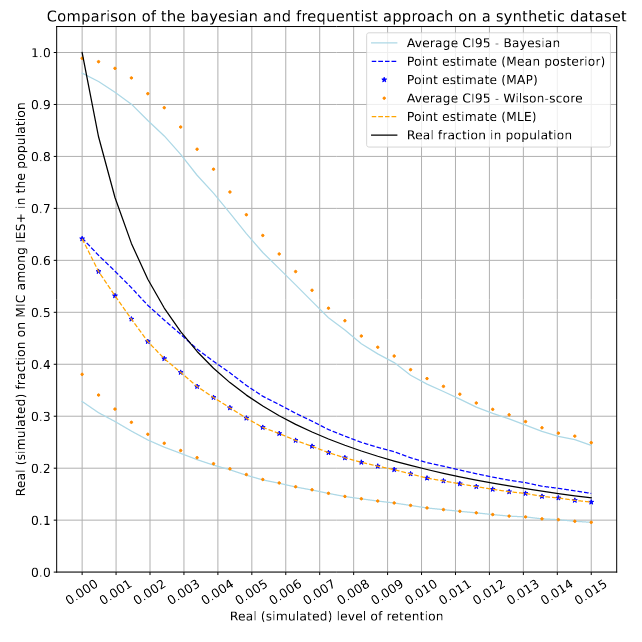
Real retention R (simulated)
P(MIC|IES+)
Summary
Can we identify the MIC sequences ?
- Very hard to have a correct estimate of P(MIC|IES+)
- When we will, it will mostly be for heavily retained IESs anyway
-
With even the slightest retention :
- Only works for IESs, not other MIC
- Only works when the ploidy is well caracterized
> We should expect lots of our MIC data to be impossible to use
$$R > 0 \implies P(MIC|IES^+) \approx 0$$
Where do IES+ molecules come from ? (2/2)
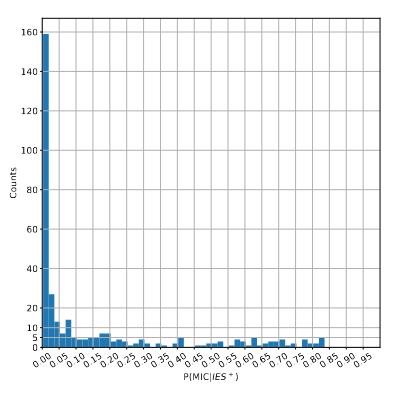
> With an updated MAC ploidy of 1600N...Most IES+ reads come from the MAC !
> Extremely problematic
for us
Results
- Small amounts of IES+ reads
-
Most IES+ reads come from... The MAC !
- The MAC ploidy was ill-estimated: we fixed a 50 years old mistake !!!
- A pipeline to analyze PacBio SMRT data
- DNA methylation in the MAC
- DNA methylation in the MIC
PB : When do we call a nucleotide methylated ?
A pipeline to analyze PacBio SMRT data
- I coded a pipeline.
- It outputs "scores". eg:
- "ModificationQv" : 0, 30, 25...
- "identificationQv": 1, 0, 50...
E.coli is used to feed paramecium (contaminants)
~100% of GATC sites = 6mA
Same for EcoK sites
>> Benchmark
- Se = 92%
- Sp = 99.9%
ipdRatio in E. coli
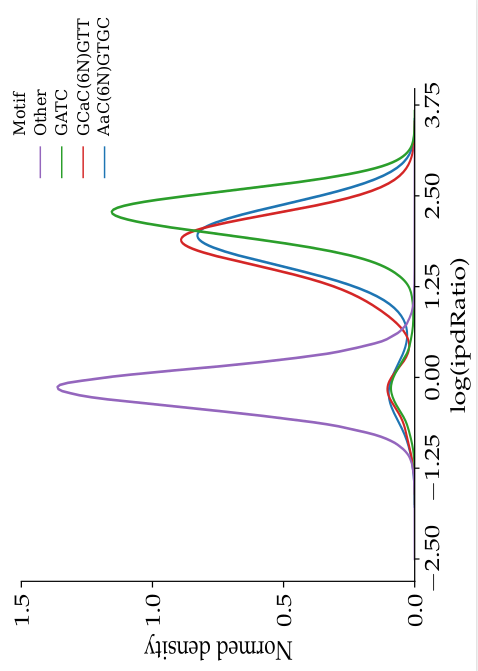
- The nucleotides we expect to be methylated have a high ipdRatio
- Slight changes between motifs
- Some exceptions : really not-methylated ?
Motif effect
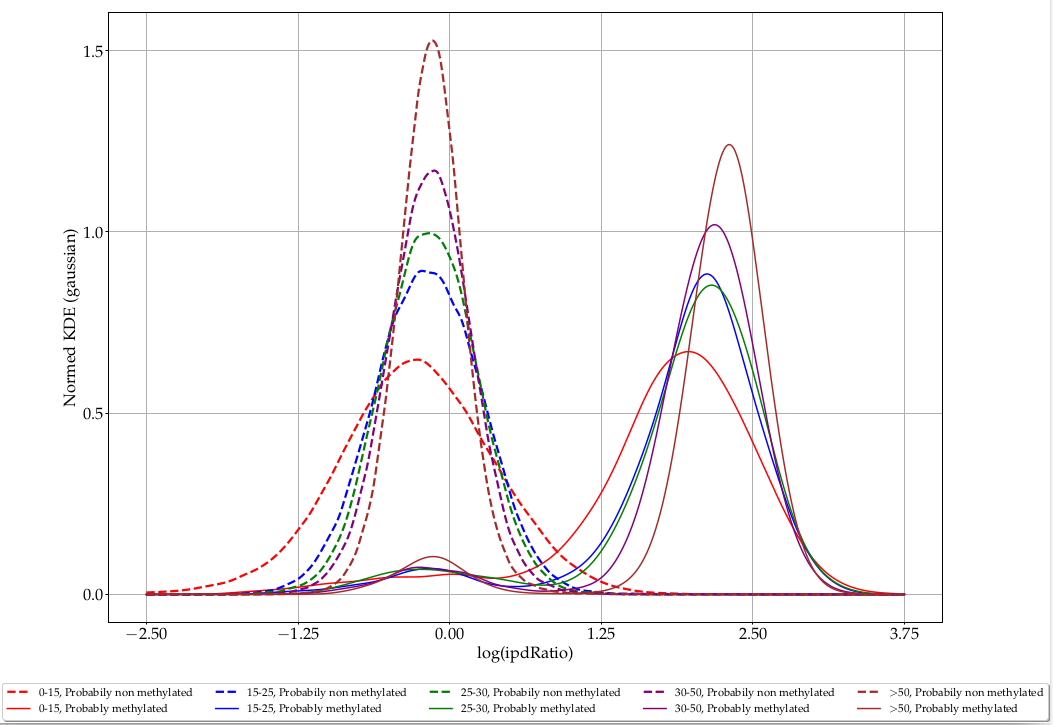
Coverage
L. methylated
L. unmethylated
> 50
35-50
25-35
15-25
0-15
ipdRatio in E. coli
Coverage effect
How to binarize the ipdRatio ?
Either a nucleotide is methylated, or it is not :
- We need to use a threshold on the ipdRatio to call modified nucleotides
- This threshold has to take account of the coverage effect
- No optimal solution anyway
Our pragmatical solution : An arbitrary linear threshold
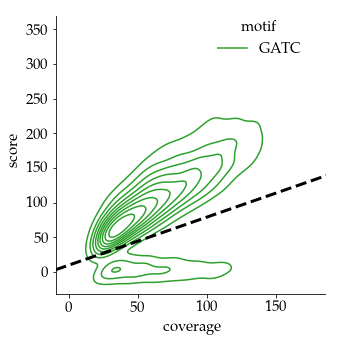
Benchmark (6mA)
- ~92% of 6mA in EcoK and GATC
- ~99.8% of non-6mA elsewhere
If we make the simplification that all GATC/EcoK sites are methylated and that 6mA is only present there :
$$Sensitivity = P(D|M)$$
$$Se = 92\%$$
But :
-
Some GATC/EcoK are unmethylated
- The real Se is actually better than 92%
-
A few amount of 6mA outside of GATC/EcoK site
- The real Sp is actually better than 99.8%
- Se = 92% and Sp = 99.8% are worst case estimates
$$Specificity = P(\overline{D}|\overline{M})$$
$$Sp = 99.8\%$$
Remark on hemi-methylation
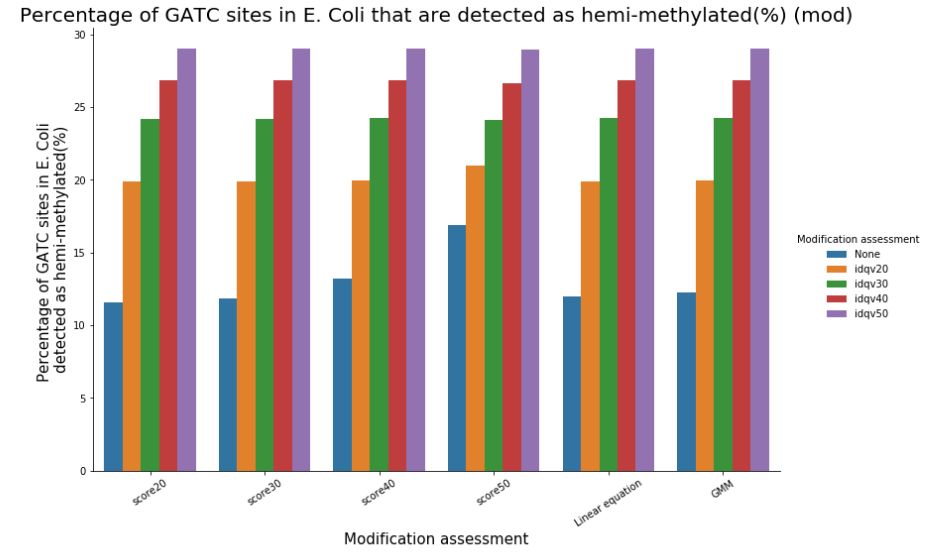
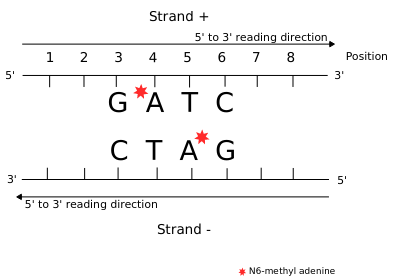
We can easily have more than 50% of so-called hemi-methylated sites that are actually not hemi-methylated
Quantifying hemi-methylation is tricky if $$Se < 100\%\ and\ Sp\ < 100\%$$
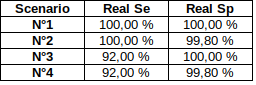
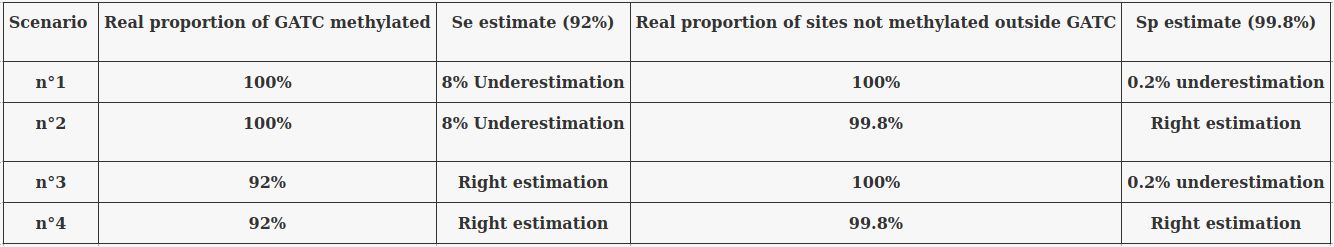
The 4 scenarii for Se and Sp
- We don't care about Se and Sp
- Only thing that matters : Does it impact the result ?
| Se | Sp | Interpretation | Scenario Number |
|---|---|---|---|
| 100% | 100% | Perfect | 1 |
| 100% | 99.8% | Sometimes it misses | 2 |
| 92% | 100% | Sometimes it invents | 3 |
| 92% | 99.8% | A few confusions here and there | 4 |
Benchmark
(other modifications)
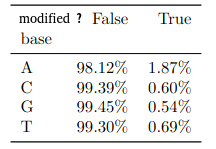
PacBio sequencing was already known for its propensity to generate false positives for 4mC (K. O’Brown et al. 2014)
Qv30
- Either false positives, or a Nature paper
- "Killer experiment": See with amplified DNA
- For now, we ignored it
Results
- Small amounts of IES+ reads
-
Most IES+ reads come from... The MAC !
- The MAC ploidy was ill-estimated: we fixed a 50 years old mistake !!!
- A pipeline to analyze PacBio SMRT data
- DNA methylation in the MAC
- DNA methylation in the MIC
DNA methylation in the MAC (1/3)
- Between 1.25% and 1.45% of 6mA in the MAC
-
97.39 to 100% of them:
- In AT sites
- Symmetrical
Depending on Se&Sp :
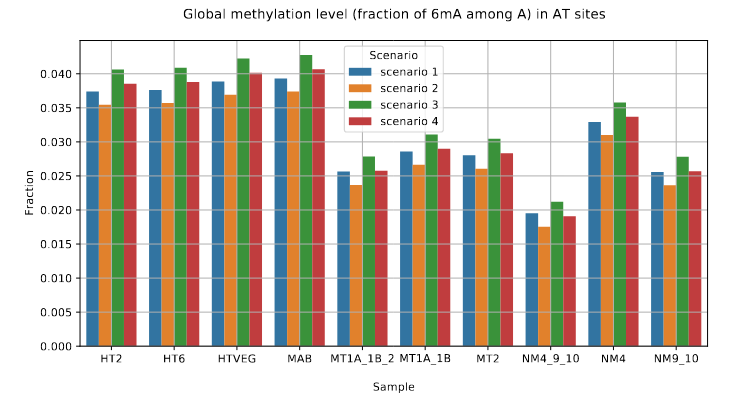
Role of our candidates
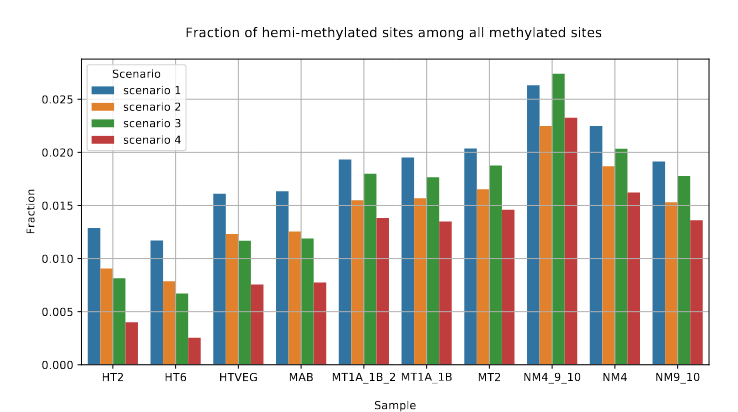
Raise of hemi-methylation, whose intensity depends importantly on how well Se and Sp are well estimated or not
Fraction of AT sites that are hemi-methylated
Role of our candidates
Role of our candidates
The capacity to make symmetrical methylation is never abolished completely
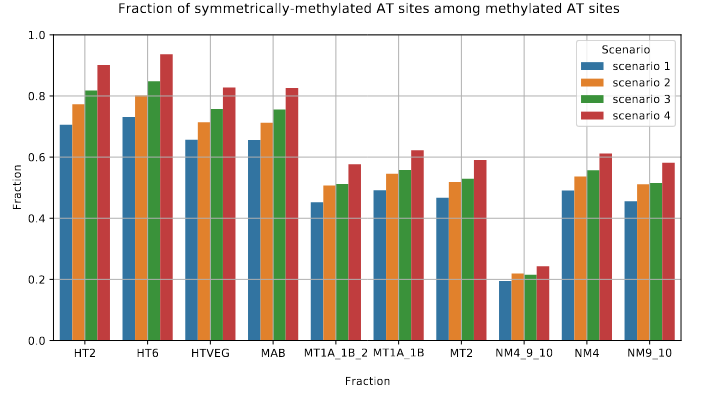
Role of our candidates
De novo methylation of unmethylated AT sites :
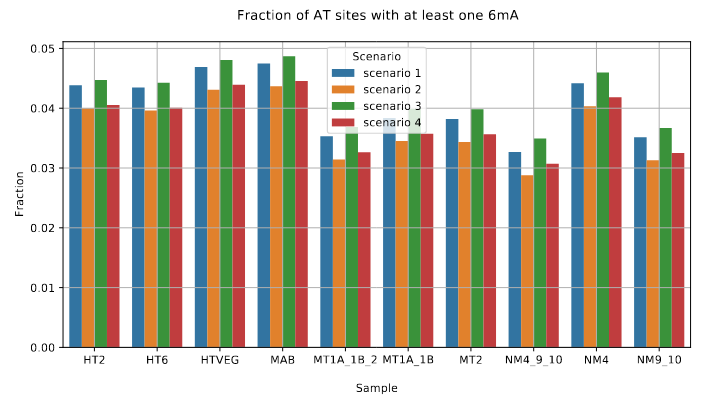
Unchanged NM4.
Drops everywhere else
Outside of AT sites
Predicted FDR : 100%
But likely detection outside of AT sites too
never erased
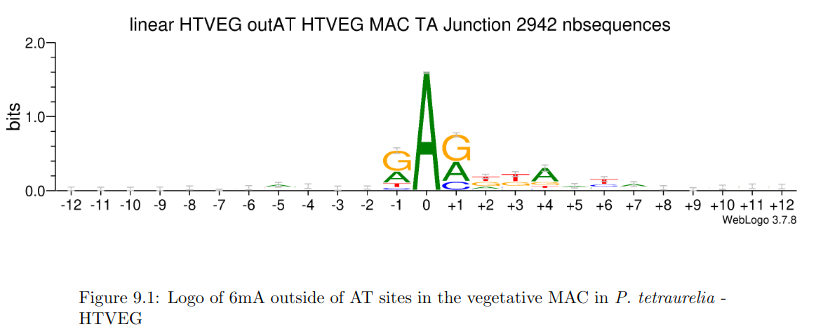
AGAA and GAGG motif
are documented as methylated sites (6mA) in C. el-
egans too (Greer et al. 2015)
Beh et al. 2019
The methylation pattern in the MAC is close to the one already observed in Oxytricha Triffalax
DNA methylation in the MAC (2/3)



All weakly implied
All strongly implied
Function = Symmetry +++
- Our MTAses turn hemi-methylated sites into symetrically-methylated ones
- Total 6mA reduced by up to -50% when all MTAses are silenced
DNA methylation in the MAC (3/3)
Details on demand...
A "fun" mathematical fact about hemi-methylation
Still:
- Our pipeline outputs a correct % of 6mA
- It outputs a correct % of hemi-methylated AT sites
But: This is actually a pure coincidence
P(hemi-methylated | called hemi-methylated) ~ 20% to 50% !
> Very hard to make qualitative analysis
Reminder : Se ~ 92% and Sp ~ 99.9%
- Small amounts of IES+ reads
-
Most IES+ reads come from... The MAC !
- The MAC ploidy was ill-estimated: we fixed a 50 years old mistake !!!
- A pipeline to analyze PacBio SMRT data
- DNA methylation in the MAC
- DNA methylation in the MIC
Results
DNA methylation in putative MIC molecules
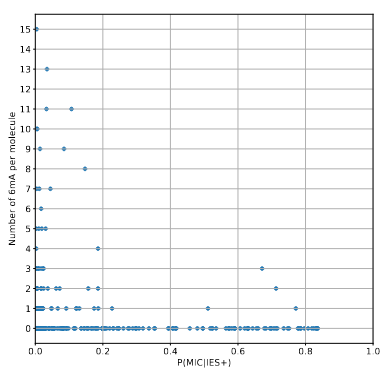
There is not a single methylated molecule for which we have a reasonable certitude that it comes from the MIC...
A ruthless data shrinkage
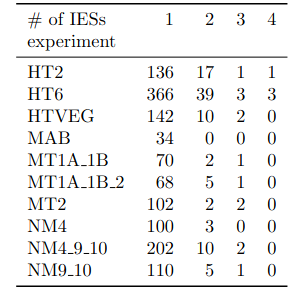
Number of molecules with at least one exploitable adenine
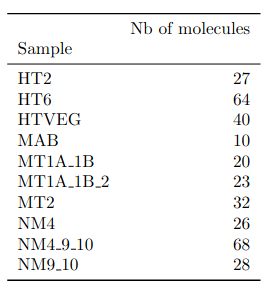
- several IESs
- variable MAC regions
- extremity outliers
Is our calculus valid for several IESs ?
Remaining with computable P(MIC|IES+)
We can never exclude the hypothesis that 6mA comes from the MAC
Summary
Methodological developments :
- Analysis pipeline for SMRT-seq
- Retention / P(MIC|IES+)
- MAC ploidy > 1600n
- [ Hemi-methylation ]
On our MAC data :
- Methylase candidates = active in the bulk of methylation in the MAC
- Symmetrical methylation of AT sites + a few de novo hemi-methylation
- ( AGAA / GAGG outside of AT sites ?)
On our MIC data:
We cannot exclude the hypothesis that all 6mA comes from the MAC

Perspectives
-
P(MIC|IES+) when > 1 IES ?
- Could invalidate the hypothesis definitely
-
Deeper analysis of HT2 and HT6
- P(MIC|IES+) is irrelevant here
- Still possible to find 6mA or other modifications around the IES very transiently
- Check 6mA / TSS
-
Hemi-methylation :
- TSS ?
- Maintained through replication ?
On these data :
For the future :
- MIC purification +++
- Nanopore ?
- Partial purification ?
- The random sampling is doomed to fail in P. tetraurelia
CONCLUSIONS
- Fixed a 50 years old mistake in the literature :
- K(MAC) > 1600N !
- A pipeline to analyze SMRT-seq data
- A new method to quantify IES retention
- Methylases = Bulk of 6mA in the MAC
-
Impossible to tell about the MIC
- Further experiments required !
Interesting discussion :
- Is hemi-methylation maintained through DNA replication ?
- If yes: How ?!?
PharmD thesis
Hematopoietic Stem Cell transplants (HSCT)

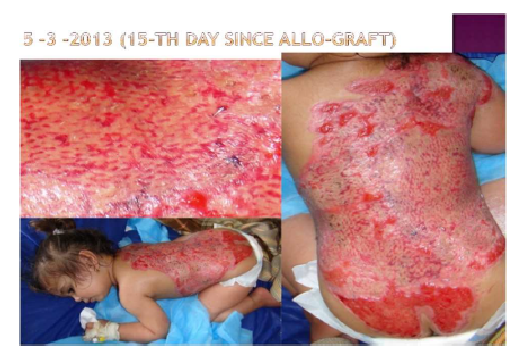
The Idea: Predict mortality or GVHD events with machine-learning
- My own project from the beginning
- Worked with a national consortium and 2 clinical experts
Preliminary results +/- encouraging
Still ongoing
More about me

Dire Straits <3

4X - Strategy games

Chess

Memes

Climbing

lolcats
who doesn't ?

Our supreme leader
Guido Van Rossum
What is he ? A man ? A God ?
We'll never know.
(when I win)


Playing music
Internet
Hiking


Thanks :)
Methylase candidates in Paramecium tetraurelia
Candidates identified:
- NM4, NM9, NM10
- Another family: MT1a, MT1b, MT2
2) PacBio sequencing with short inserts | 6mA ++
1) Grouped silencings by sequence homology
PacBio SMRT (short inserts)


Inserts up to 350bp
Sequencing : unique molecule & both strands
Polymerase slowing ~ methylated adenine
The random sampling strategy
- Purify the MIC: Impossible
- Sequencing whole MIC from whole DNA : Impossible
-
Solution: Random sampling
- 1 among 200 molecules
- Not all with cary an IES at all
- ~20 to 100 IES only per experiment
Problem n°2
IESs are sometimes retained in the MAC
What is P(MIC|IES+) ?
Other analysis challenges
(skipped)
Details that I spare you (but don't hesitate to ask) :
- Random sampling of the MIC
- Sorting MIC and MAC
- Sorting IES specific and MIC non-IES sequences
- Filter IES retention
- Estimate Se and Sp
- Retro-ingineering PacBio's formats
- Recoding parts of the softwares for single molecules
- etc.
Detecting n6mA
Imperfect detections
No scientific method is perfectly reliable
In our case:
- Sensitivity = P(D | M) > 92%
- Specificity = P(ND | NM) > 99.8%
> Expect False Positives and False Negatives
Imperfect detections (2/2)
- Sensitivity = P(D | M) > 92%
- Specificity = P(ND | NM) > 99.8%
We can use 4 extreme scenarii :
... And see what is consistent no matter the scenario we place ourselves into
Finding unbiased estimators for and FDR
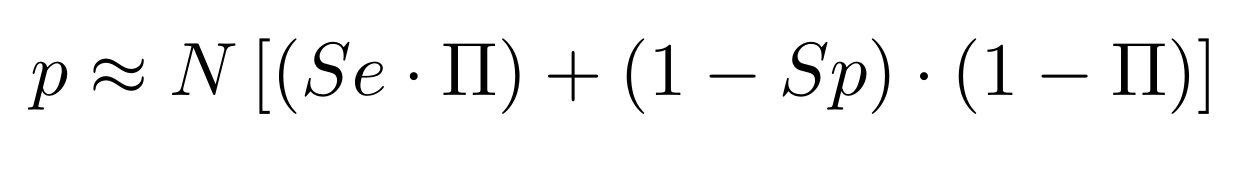
If p number of positive detections among N tests:
p = FP + TP
$$\pi$$
So,
Which means
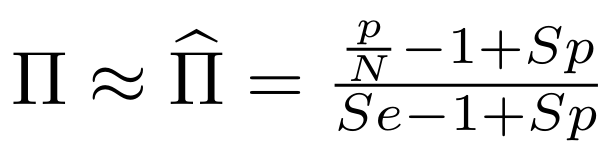
with fraction of 6mA
$$\pi =$$
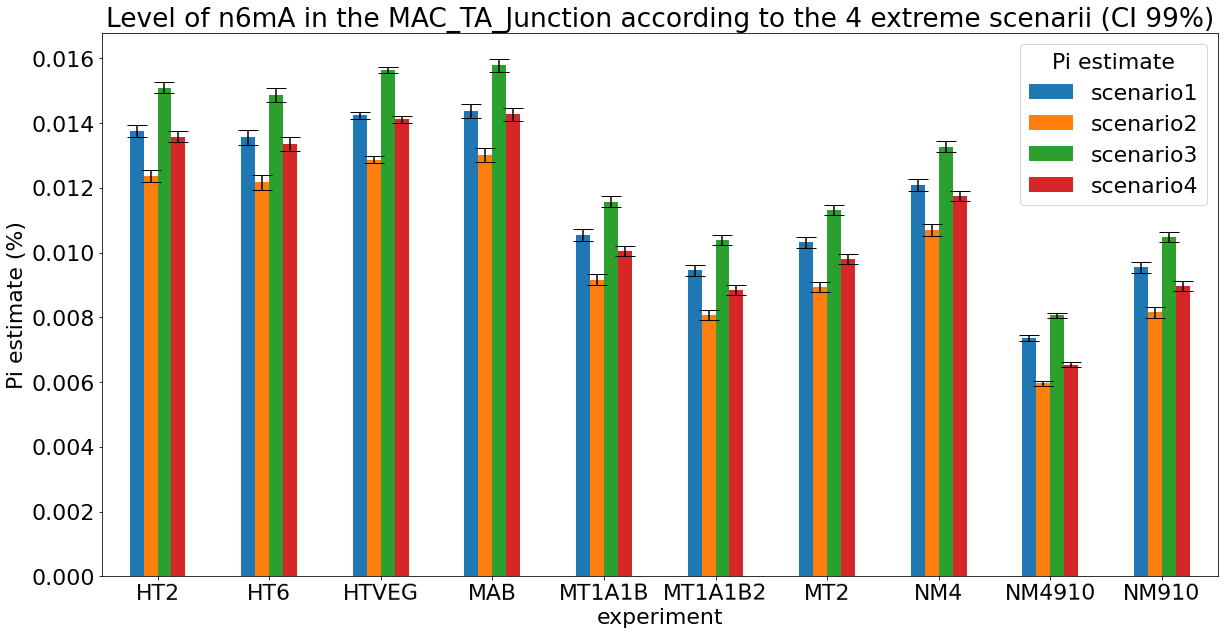
Debiased levels of m6A in the MAC
> 95% n6mA are located in AT sites
DNA n6-mA around/in the IESs
Results
Methodological development to correct hemi-methylation detection
Let FD1 and FD2 be resp:
- Fraction of AT sites detected hemi-methylated
-
Fraction of AT sites detected symmetrically methylated
PZ0, PZ1, PZ2: unbiased estimators of non, hémi, symetrically methylated AT sites
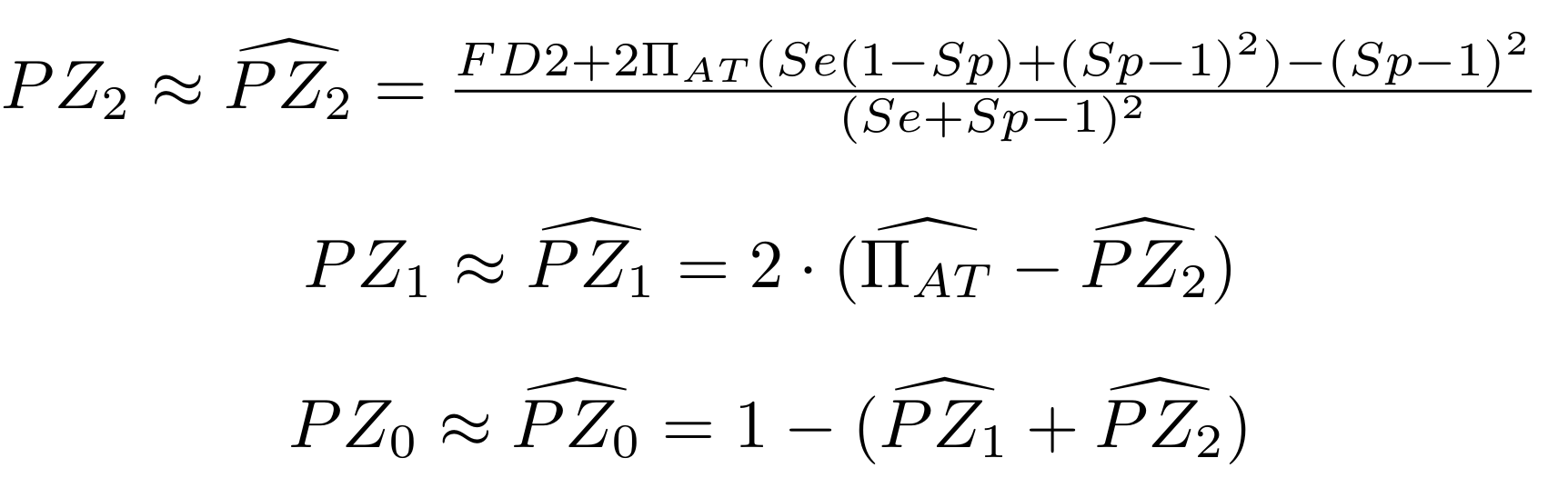
Then:
With
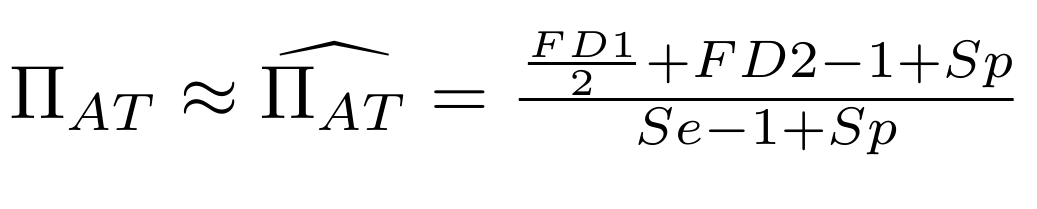
Debiased Hemi-methylation in MAC



2) Transcient in the new forming MAC
1) Constant pattern in the MIC
Transcient ?
Our hypothesis:
Analysis showed that:
~2.5% of n6mA in Paramecium (MIC) : Cummings 1974
?
> Actually way lower (if any)
> Unexpectedly, NM and MT proteins play a whole another role in the MAC,
not the MIC
Conclusion
IGEM2014
MT and NM families :
In the MAC
Convertase activity DNMT1-like
But: Hemi-methylated AT sites kept after many mitosis
New hypothesis:
Symmetrical methylation of AT sites = Mitotic clock or maturity indicator for the cell to be allowed to go enter meiosis
A bit more about me...
I don't like

Red socks

Fun

Windows 10 update of May 2021
Some keywords for my PhD
1) Paramecium tetraurelia
3) DNA methylation


4) PacBio sequencing
2) Transposable elements / IESs
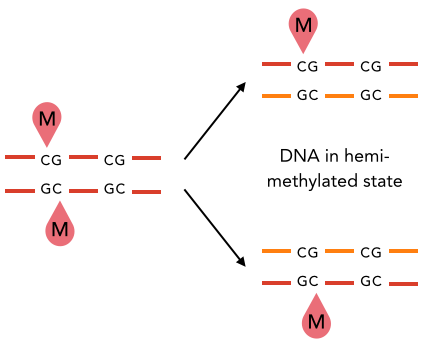
5) hemi-methylation of
palindromic motifs
(AT sites)

P. tetraurelia

Unicellular eucaryote with 3 nuclei:
- 1xMAC nucleus (up to 800n)
- 2xMIC nuclei (2n)
DNA ratio: 1 MIC for 200 MAC
The difference ?
MAC genome compared to MIC =
- Amplified+++
- Free from TEs
- Transcriptionnally active
- Not transmitted to progeny
Genome invaders in Paramecium tetraurelia
In the MIC:
- 45.000 unique sequences
- Small (<27bp), Non-coding
- Lots present in the CDS
- Remnants of TE ?
- 100% TA-bounded


--> How ?
Present in the MIC
Absent in the MAC
= Excised generation after generation




2) Transcient in the new forming MAC
1) Constant pattern in the MIC
Transcient ?
Recognition of IESs: Our hypothesis
Our hypothesis:
~2.5% of n6mA in Paramecium (MIC&MAC) : Cummings 1974
Methylase candidates
Candidates:
- NM4, NM9, NM10
- Another family: MT1a, MT1b, MT2
2) PacBio sequencing with short inserts | 6mA ++
1) Grouped silencings by sequence homology
PacBio SMRT principle


Reads up to 80 kbp
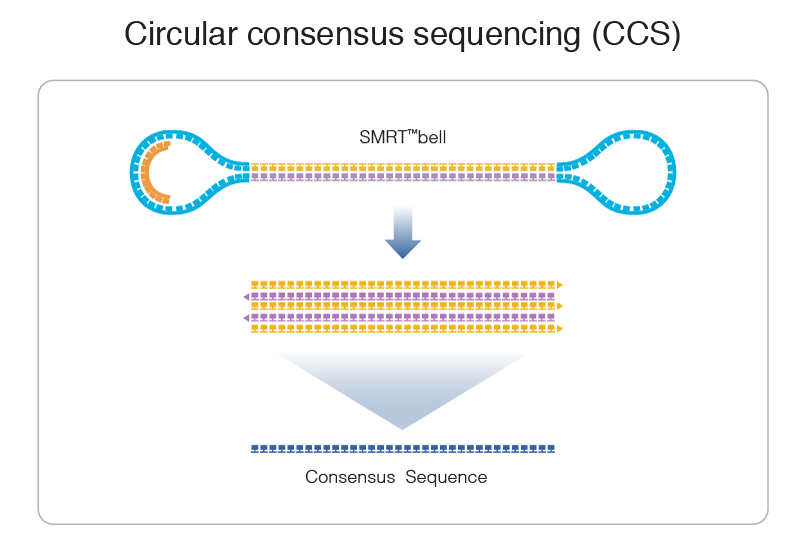
1) Sequencing (unique molecule)
2) High fidelity consensus
3) DNA methylation analysis
What are the results ?




2) Crosstalks in the new forming MAC
1) Constant pattern in the MIC
Transcient ?
Our hypothesis:
Prelimary analysis shows that:
~2.5% of n6mA in Paramecium (MIC&MAC) : Cummings 1974
Details invegetative MAC
-
~95% of n6mA locates in AT dinucleotides in the MAC
-
75% of the methylation in an AT dinucleotide is actually symetrically modified
Bulk of methylation:
Total = 1.2 to 1.5% of adenines
0.6% of the adenines outside AT sites:
Now a bit more details...
Imperfect detections
No scientific method is perfectly reliable
- We don't care about Se and Sp
- We care about the fact that eventual mis-estimations of them doesn't really change anything
In our case:
- Sensitivity = P(D | M) > 92%
- Specificity = P(ND | NM) > 99.8%
Finding unbiased estimators for and FDR
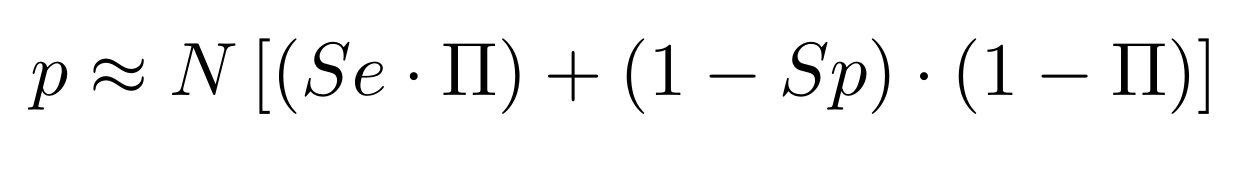
If p number of positive detections among N tests:
p = FP + TP
$$\pi$$
So,
Which means
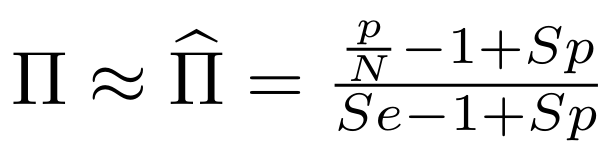
with fraction of 6mA
$$\pi =$$

Debiased levels of m6A in the MAC_TA inserts
Methodological development to correct hemi-methylation detection
Let FD1 and FD2 be resp:
- Fraction of AT sites detected hemi-methylated
-
Fraction of AT sites detected symmetrically methylated
PZ0, PZ1, PZ2: unbiased estimators of non, hémi, symetrically methylated AT sites
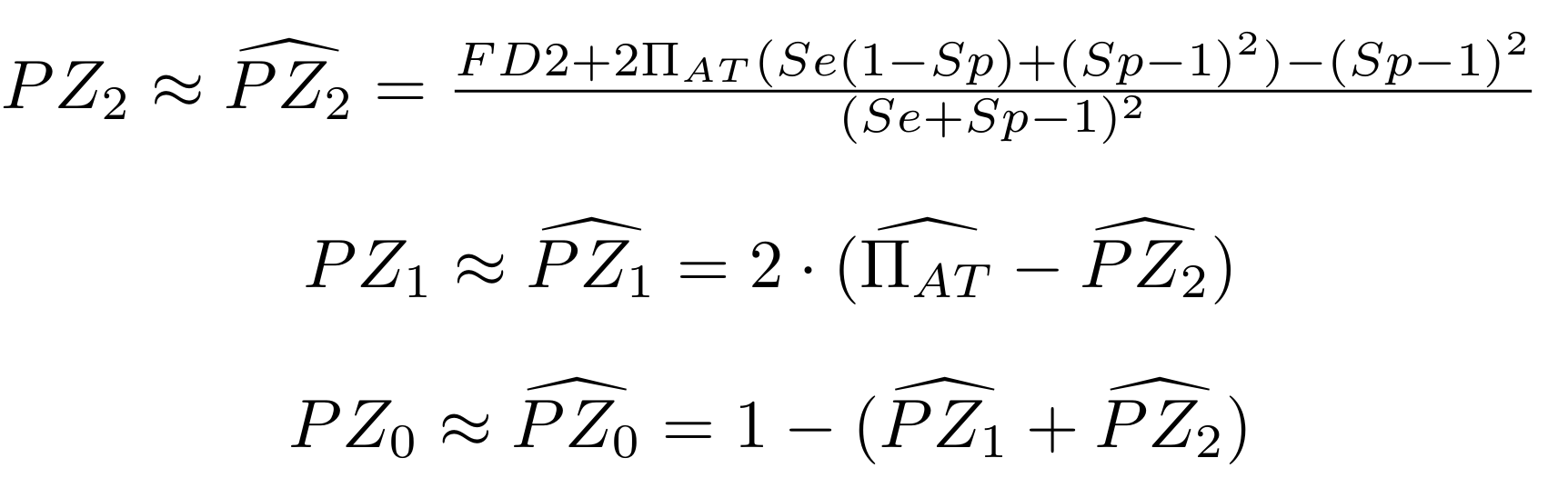
Then:
With
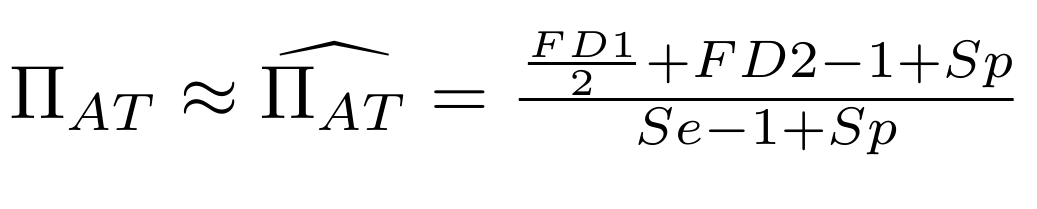
Debiased Hemi-methylation in MAC_TA

Conclusion
- In the MIC:
- Probably no m6A after all
- Hard to imagine a role in TE/IES excision
-
In the MAC
-
Our 6 genes are implied in the bulk of the MAC m6A
- Sym-methylated -> Hemi-methylated
-
MT and NM families: Functional analogs of DNMT1 ?
- Not really: Not kept after replication
-
Lots of questions raised by the MAC methylation:
- TSS ?
- IES excision Junctions ?
- Nucleosome positionning ?
-
Our 6 genes are implied in the bulk of the MAC m6A
-
One important phenotype:
- NM4-9-10 somehow makes the cell unable to go into autogamy

Thanks :)
Role of DNA-6mA in Paramecium tetraurelia






Guillaume DELEVOYE
3rd Year PhD student
Bioinformatics
P. tetraurelia: Genomic architecture

Unicellular eucaryote with 3 nuclei:
-
2xMIC nuclei (2n)
- Germline nucleus
-
Contains: TE & IES
- No transcription outside meiosis
-
1xMAC nucleus (up to 800n)
- Somatic nucleus
- Amplified and "fixed" version of the MIC
- Free from TE and IES
- Transcriptionnally active
DNA ratio: 1 MIC for 200 MAC
+ DIfficult to purify the MIC DNA
Profiling the IESs
- Non-coding
- Excised after sexual processes
- Remnant of TE ?
- Most recent IES are very short (< 27bp), and are the majority of IESs
45.000 Unique sequences

How are IESs recognized ?
(1/2)

- 100% TA-Bounded
-
Weak consensus TAYAG
- Degenerated TC1-Mariner TE insertion site
- Not sufficient
- Periodic size distribution
30% of IESs only are small-ncRNA dependant (shown by DICER-like2-3 silencing)
- What about the majority remaining ?

How are IES recognized ? (2/2)
The sc-RNA pathway
The DNA methylation hypothesis
6mA likely to be abundant in Paramecium:
-
Suspected 2.5% in the MAC of P. aurelia by Cummings et Al (1975)
-
Also documented 6mA in the MIC
-
-
-
Detected methylation by SMRT in the MAC in our lab (unpublished data)
-
Detection by SMRT in the MAC by Sandra Duharcourt's lab (unpublished)
- Detection in Oxytrichia by L. Landweber et Al (2019)
- no 5mC, 4mC in the MAC a priori





2) Crosstalks in the new forming MAC
1) Constant pattern in the MIC
Transcient ?
Methylase candidates
Candidates:
- NM4, NM9, NM10
- Another family: MT1a, MT1b, MT2
2) PacBio sequencing with short inserts | 6mA ++
1) Grouped silencings by sequence homology
PacBio
TL;DR overview
PacBio SMRT principle


Reads up to 80 kbp

1) Sequencing (unique molecule)
2) High fidelity consensus
3) DNA methylation analysis
PacBio SMRT principle (2)

- Strategy 1: Long inserts (long reads)
- Ideal for assembly of long repeated sequences
-
Poor resolution for DNA methylation analysis
-
Strategy 2 : Short inserts (long reads)
- Much higher resolution for DNA methylation analysis
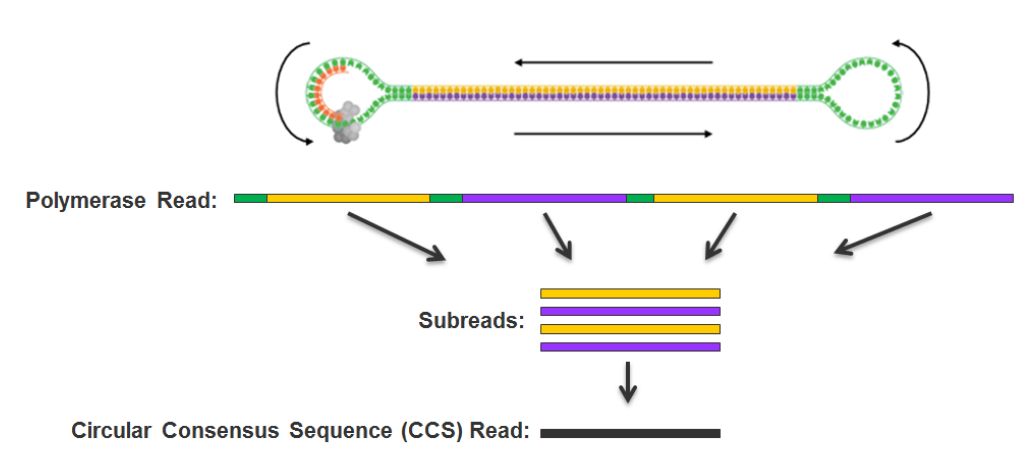
Step 1: Consensus

99% accuracy
Max
75% accuracy

Because our inserts are circular and shorts, we can make CCS of high accuracy despite a 15% error-rate
Step 3 : DNA methylation analysis
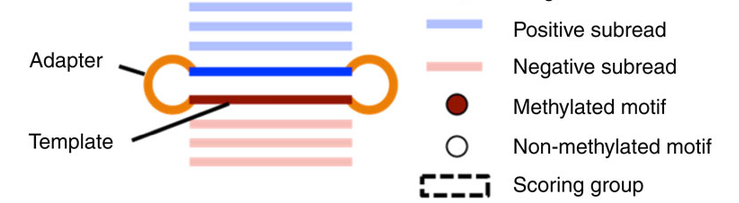
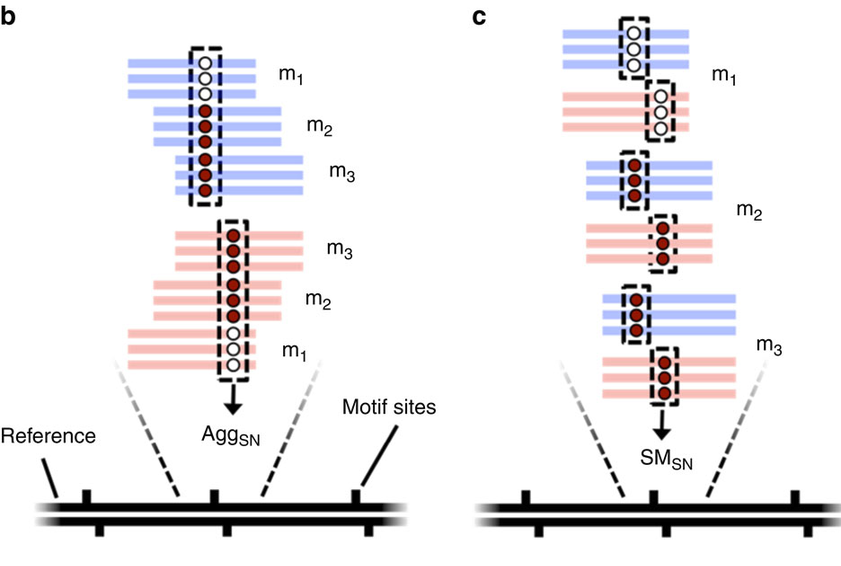
- Single-molecule
- Single-nucleotide resolution
- Independant yet pairable analysis on both strands
Some more details:
the random sampling approach
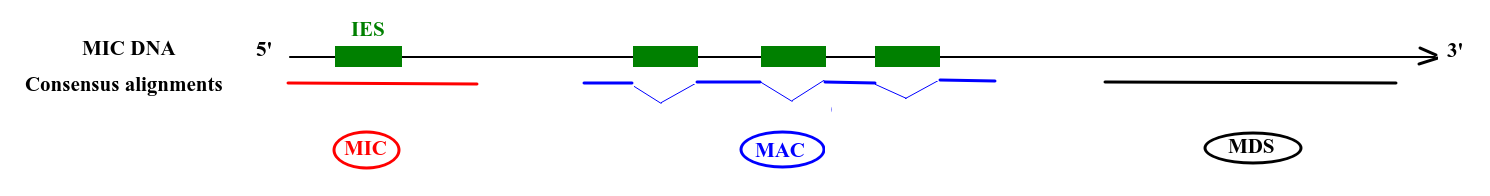
Deduced origin
MIC DNA
Alignment of consensus
Only a few remaining: ~ 10 to ~200 sequences
100% should carry a methylation pattern
-
1 out of 200 comes from the MIC
-
1/6 of MIC inserts will carry an IES
-
For 50% of IESs, we cannot be sure whether it come from the MIC or from the MAC
- 30% of the remnants are scanRNA dependant
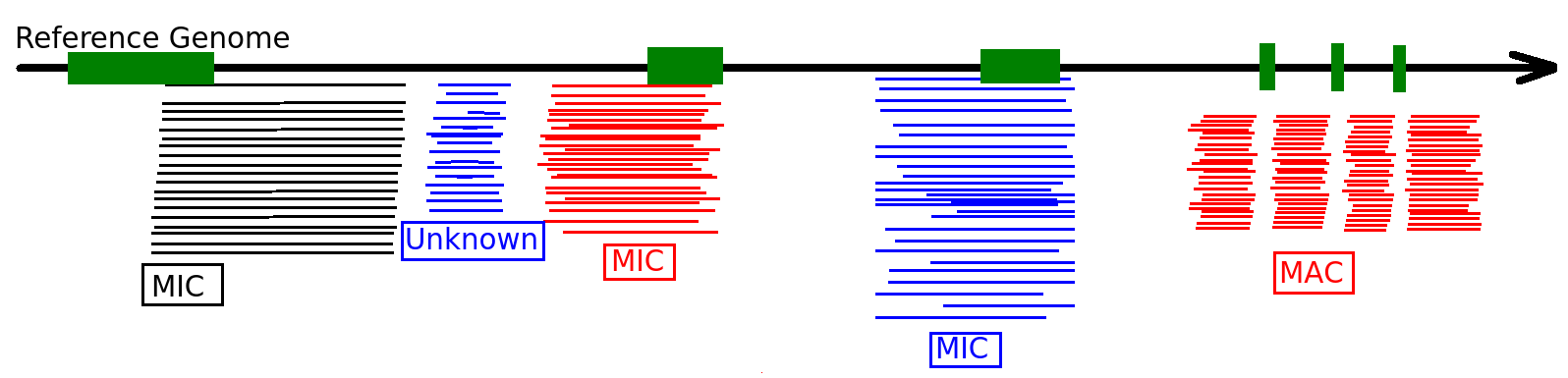
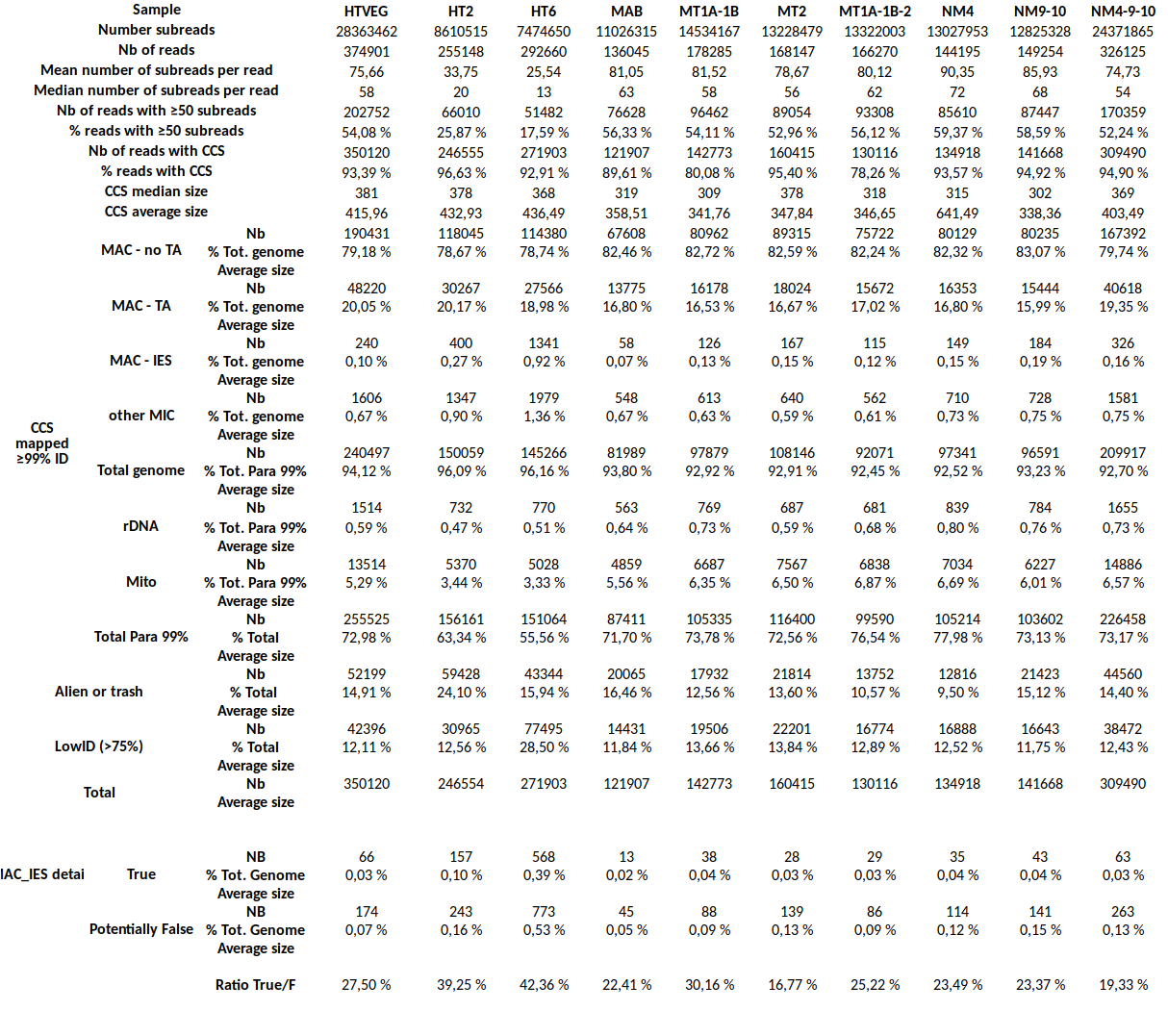
Final categories
- MDS
- = "MAC" for 99% of sequences
- MAC_IES
- "true" MAC_IES (never seen retained)
- other MAC_IES (sometimes retained)
- MIC
- Other MIC specific (TE, repeated sequences)
- MAC (TA Junction)
- Overlap a TA junction of excision
- rDNA
- mtDNA
-
Other:
- Contaminants
- Alternative excision boundaries ("LOWID")
- low identity consensus ("Trash")
"genome"
Total Paramecium
Total sequencing
Little bug to be corrected
30%
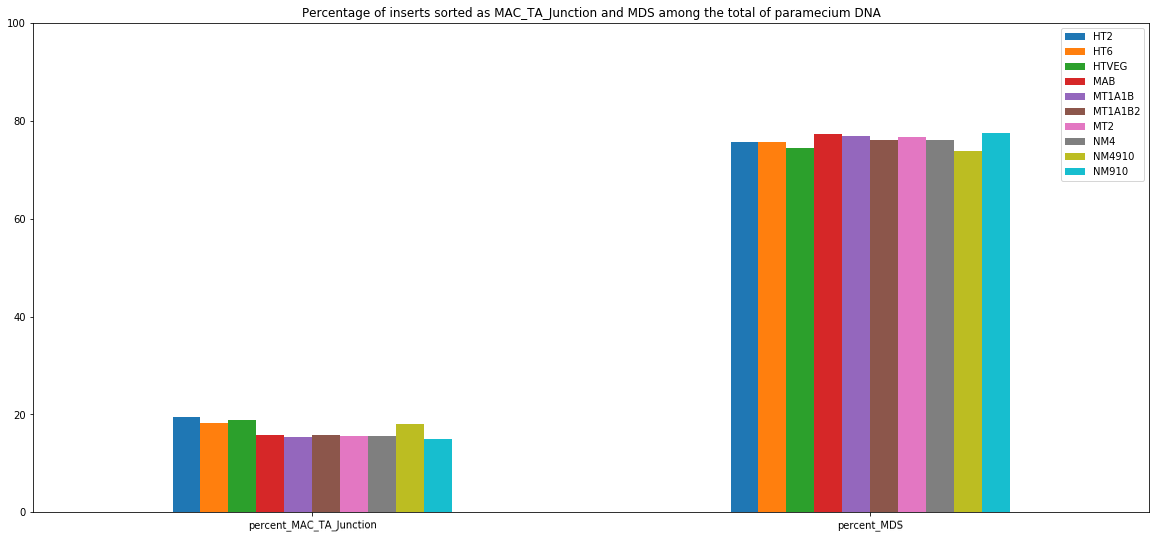
MDS and MAC_TA_Junction represent a vast majority
MIC specific and IES are as rare as expected
+ Applying the retention filter divides the number of "MAC_IES" approximately by 50%
Detecting m6A in details
Detecting m6A optimally
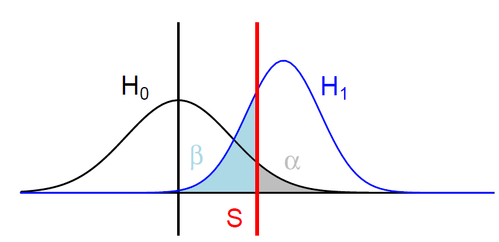
H0: ipdRatio of umethylated-Adenines
H1: ipdRatio of 6mA
S: Threshold on the pvalue
--> Specificity
--> Sensitivity
$$\alpha$$
$$1 - \beta $$
Adenine
6mA
ln(ipdRatio) ~ N(0,1)
log(ipdRatio)
A pvalue is just the probability that log(ipdRatio) in the tail of H0
Using E.coli as ground truth

E.coli:
- Feeds paramecium: contaminants+++
- Nearly 100% symetrically-methylated with m6A
- GATC
- EcoK
- Few others
- Depends a lot of the strain
- Outside of GATC and EcoK: very low levels of m6A
I investigated the PacBio's output on it's GATC & EcoK VS other sites
How log(ipdRatios) look in E.coli
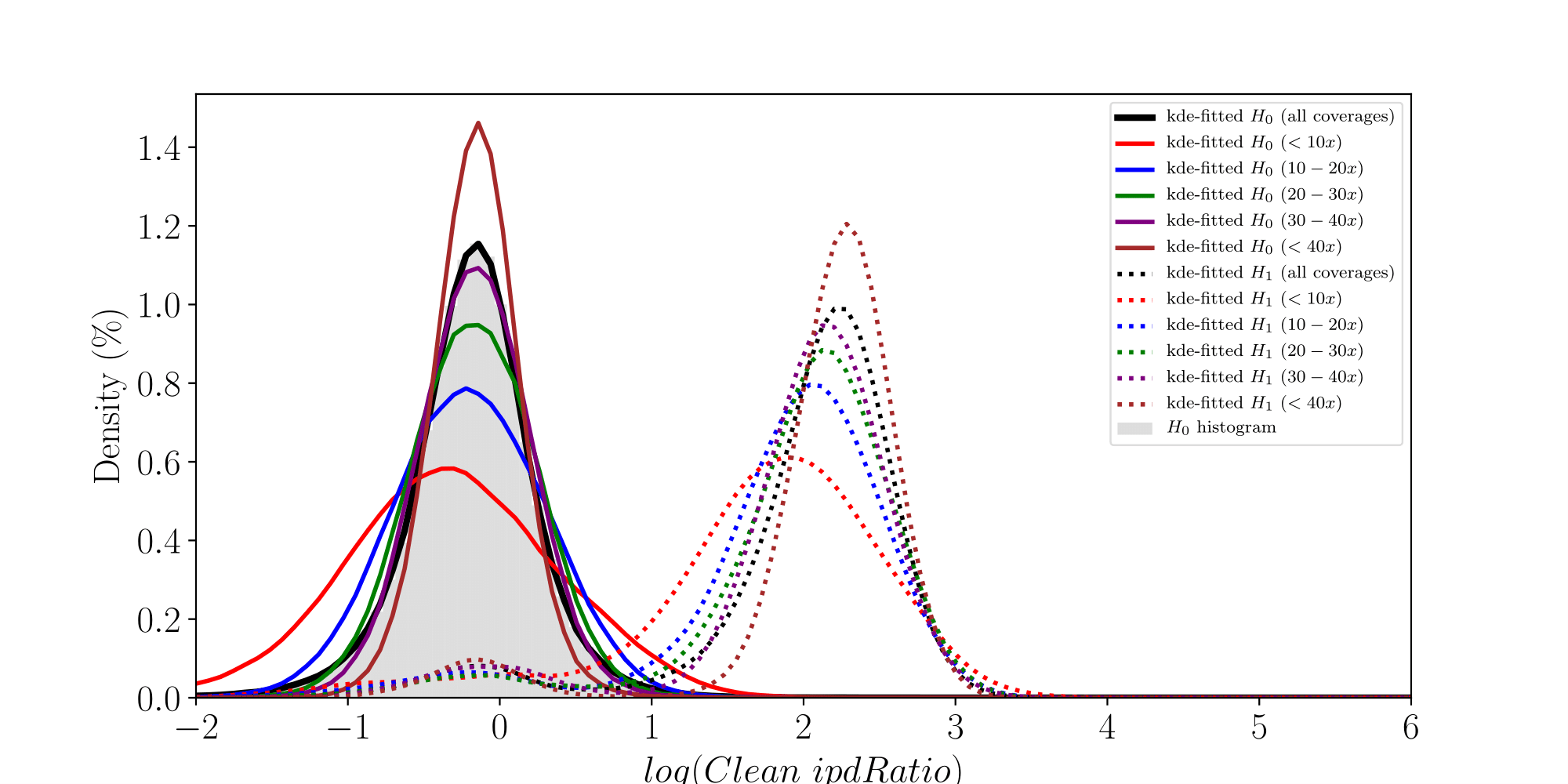
Separability raises with coverage, which is expected
PacBio pvalues do have some biological meaning
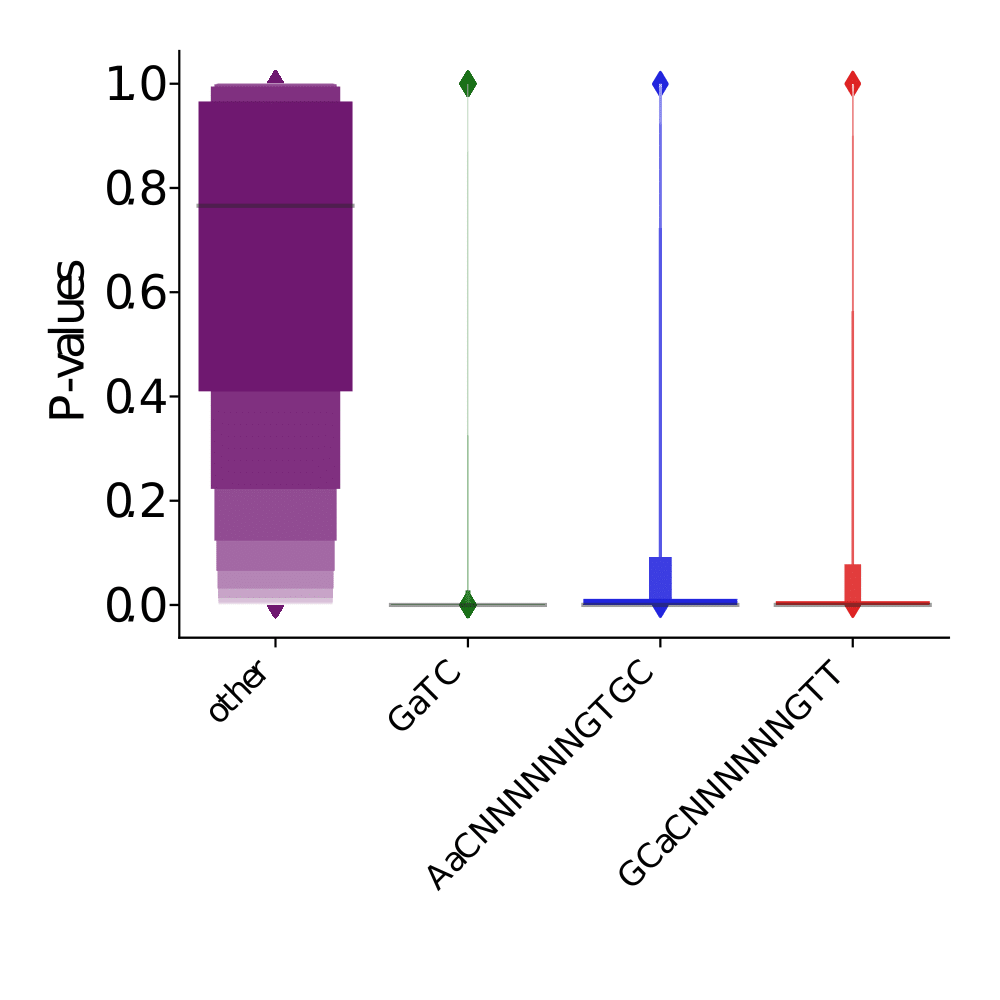
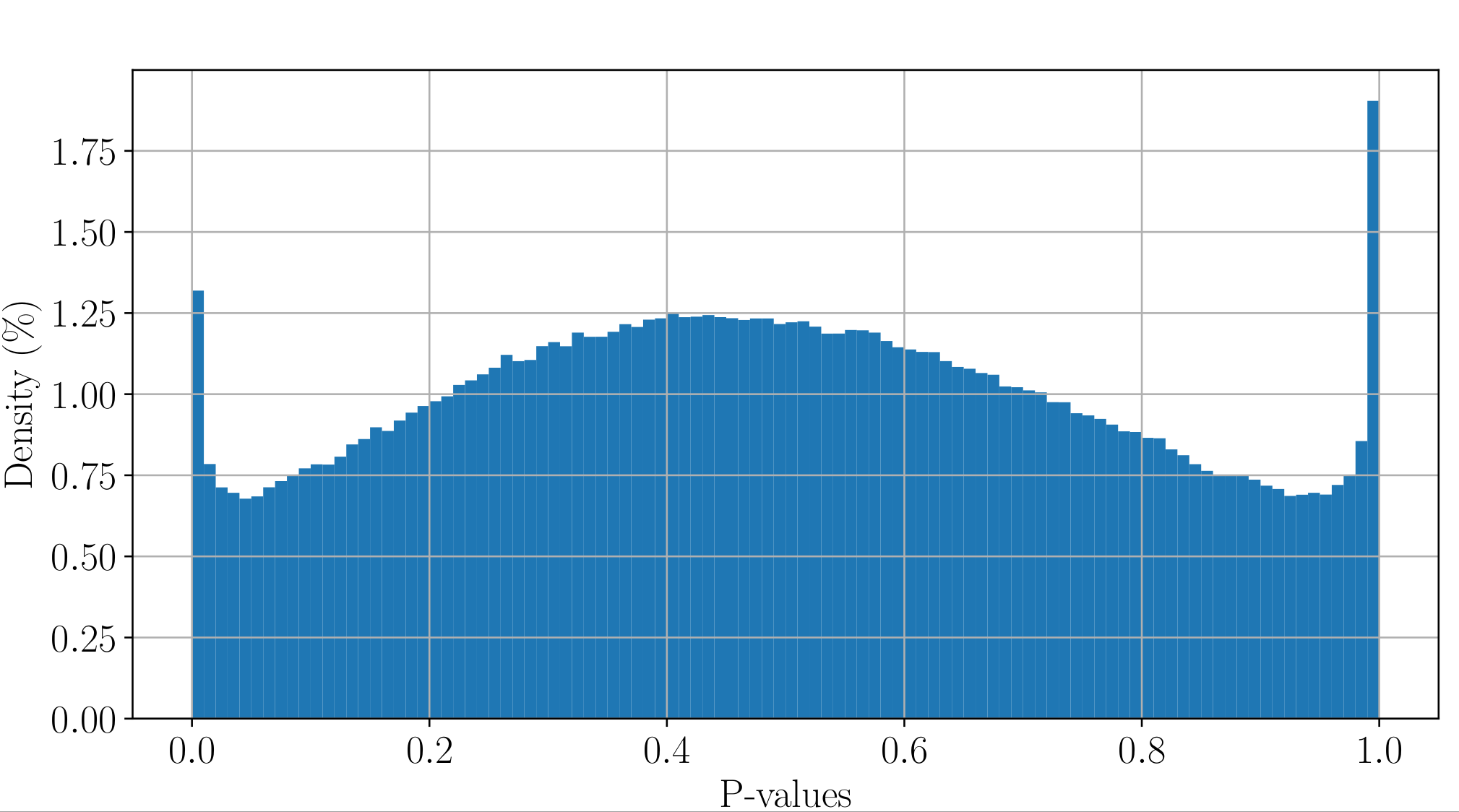
PacBio pvalues do have a meaning...
...but are not ideally distributed
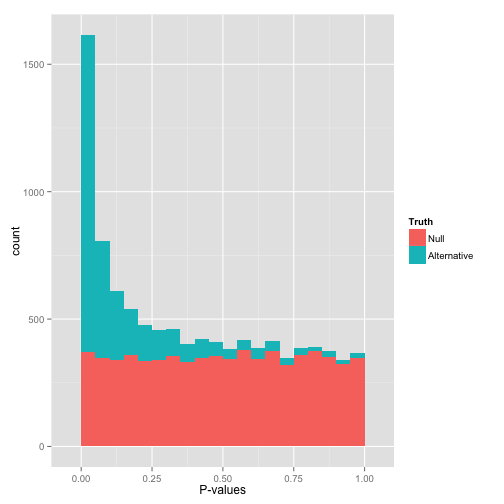
Ideal pvalues
--> Allows magic !
PacBio's
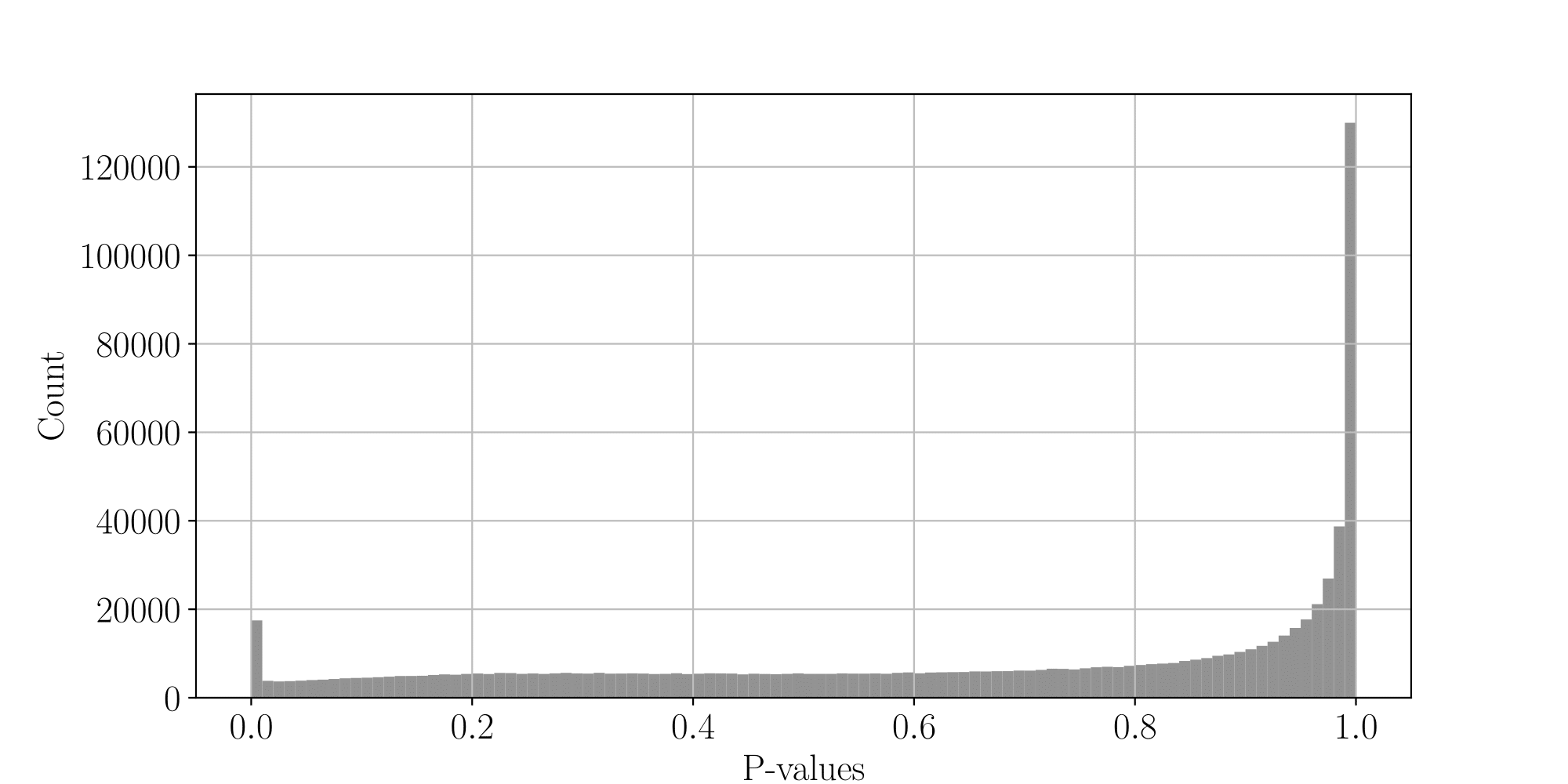
- Allow estimation of
- Allows optimal, adaptative FDR control
$$\pi_{0}, \pi_{1}$$
- Just don't
-
Obvious point n°1:
- log(ipdRatio < 1) -> -inf
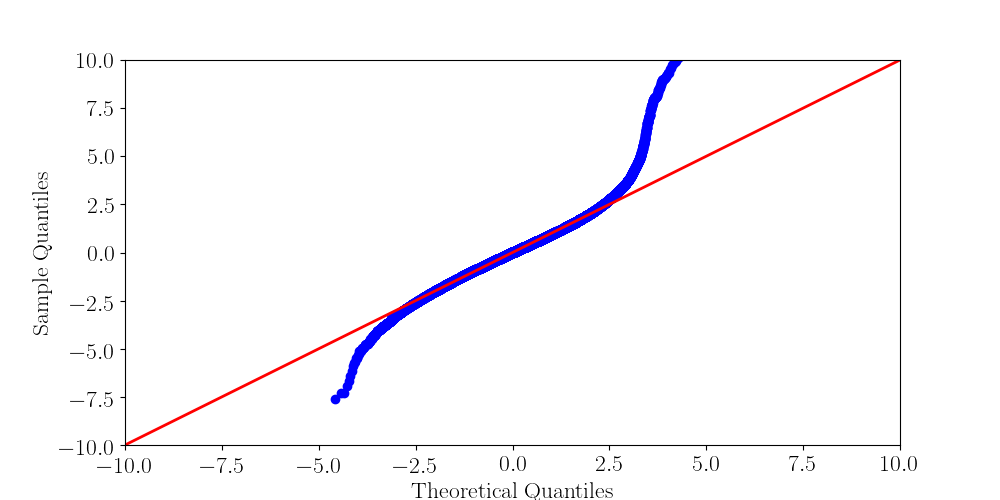
Normality assumptions under H0 are broken under high coverages
- The higher the coverage, the worse (here, >40x)
-
Will cause bell-shaped pvalues under null
- Hidden phenomena on the previous curve
Normality assumption matters
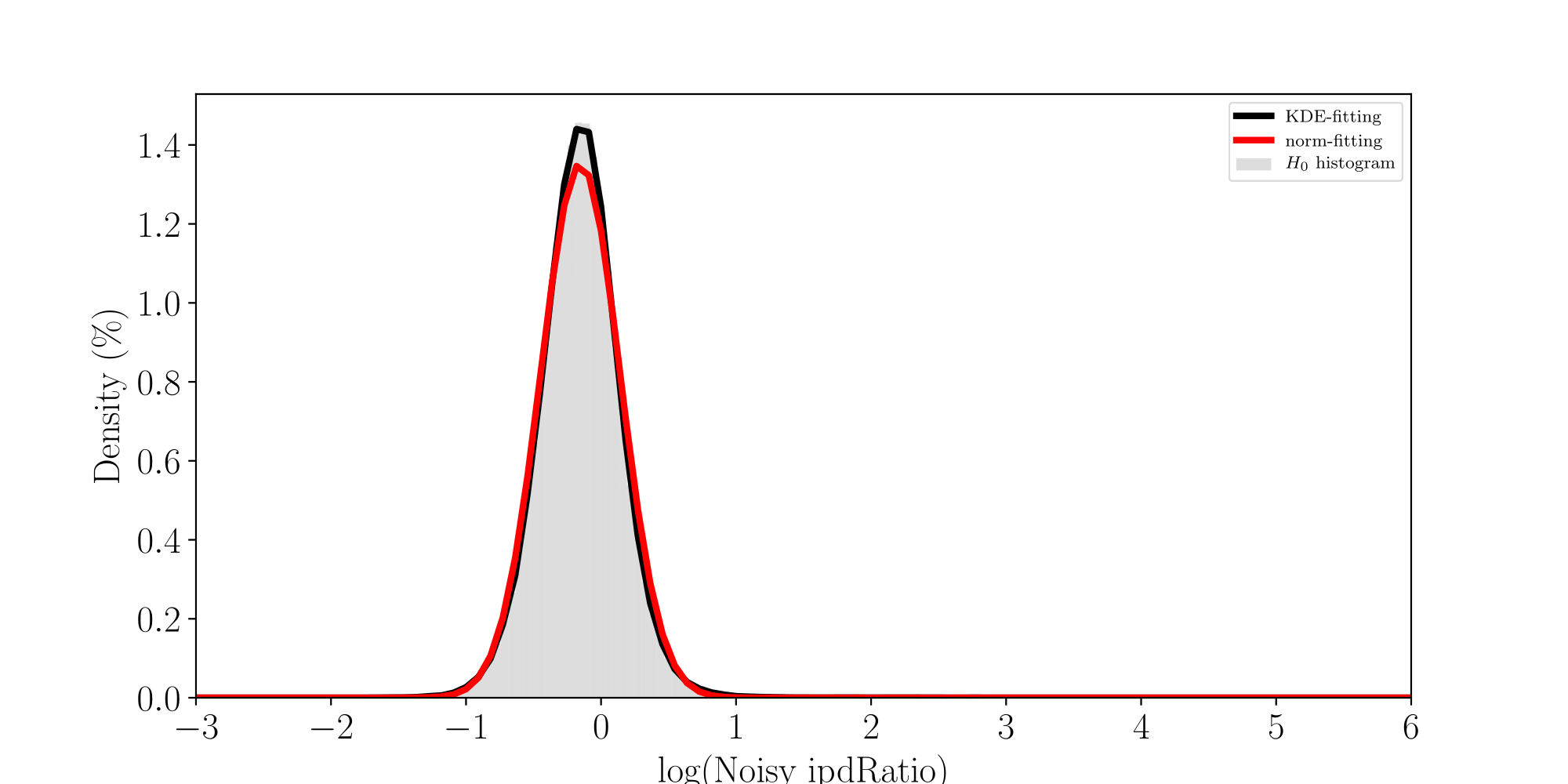
All Adenines' pvalues [E.coli] coverage > 40X
Long story short
PacBio's pvalues:
- Are biologically relevant
- We can build a reliable ad-hoc system with them
- Are produced by a linear combinations of > 150 different coverages
- Which forbids the usual statistical treatments
- Are somehow broken on a coverage-dependant manner
- Which forbids a simple fix for point 2
- We can't use the classical statistical treatments directly on pvalues
Other PBio's scores for m6A
For n6mA, PacBio produces:
-
A modification score --> Slowing of the polymerase
- pvalue against H0 only (the one we presented earlier)
-
An identification score --> Kinetic signature of a modification
-
loglikelihood between H0 and H1 (H1 = Other modifications or secondary peaks)
-
loglikelihood between H0 and H1 (H1 = Other modifications or secondary peaks)

They are PHRED-transformed p-values of two different statistical tests, that rely on the mean of the IPDs
PHRED scores
The scores (Qv) are PHRED-transformed p-values
Typical covscore plot
Modification score / coverage
Using flat threshold on modification score = Hudge lack of power
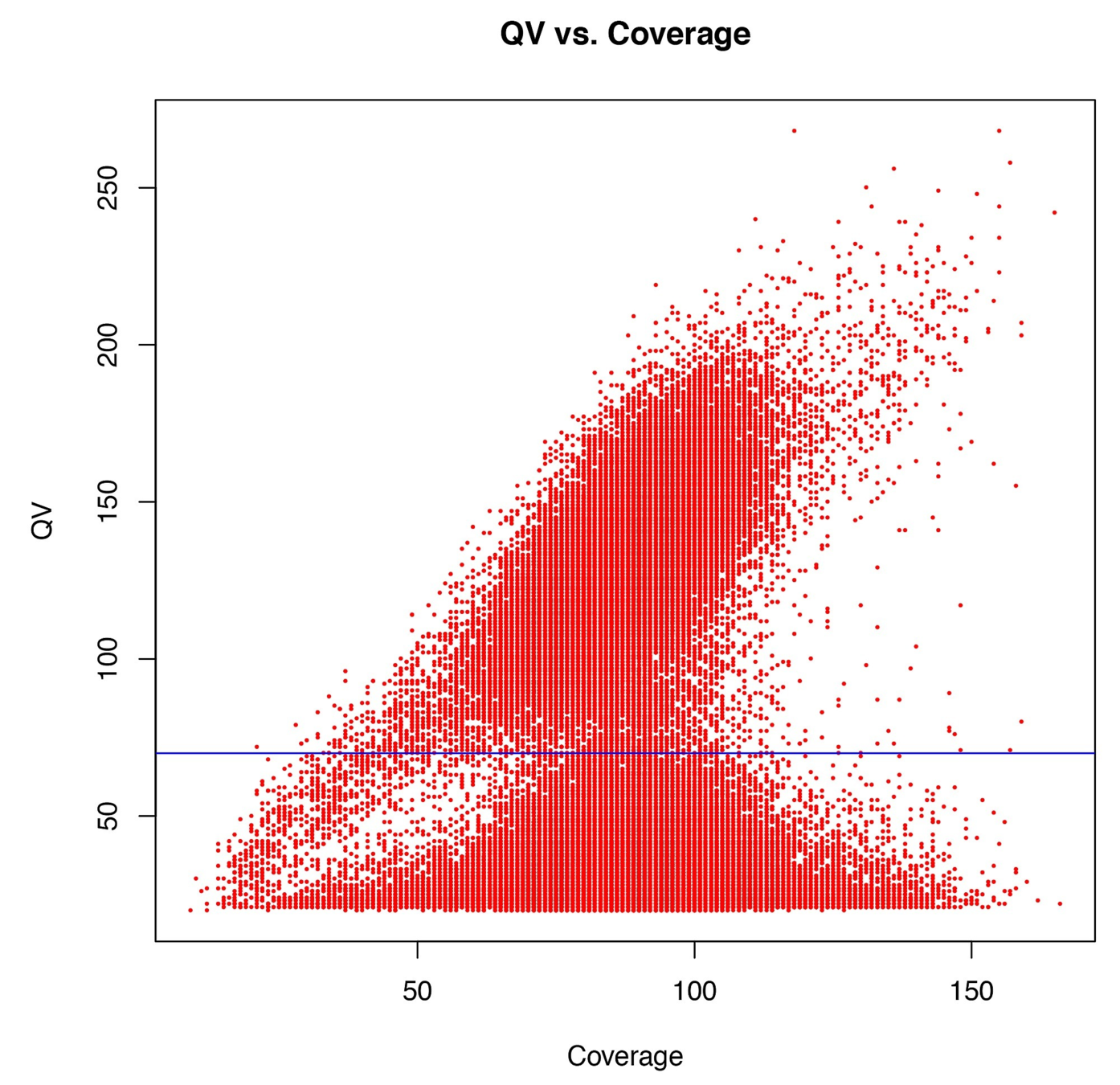
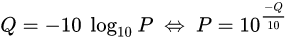
Solution : A coverage-dependant threshold on the scores
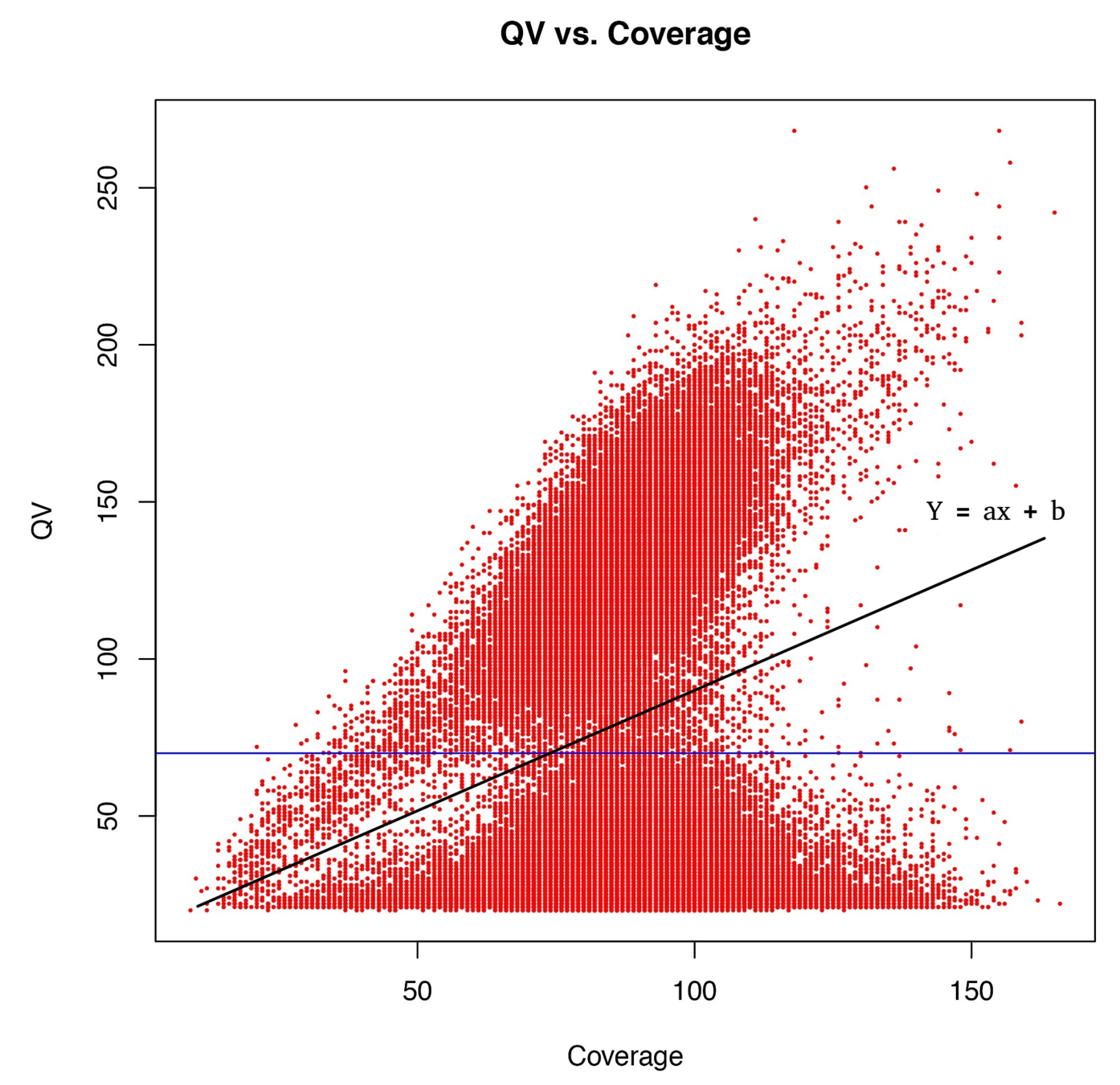
From now on
"positive detection"
=
score > linear thershold
(only >25X considered)
Benchmarking
How good (or bad) is our method ?
$$Se = P(D^+|M) ~ 92\%$$
$$Sp = P(D^-|NM) ~ 99.8\%$$
Starting from sufficient coverages (~20X to ~30X), Se and Sp don't depend on the coverage anymore
The 4 scenarii for Se and Sp
- We don't care about Se and Sp
- We care about the fact that eventual mis-estimations of them doesn't really change anything

Finding unbiased estimators for and FDR
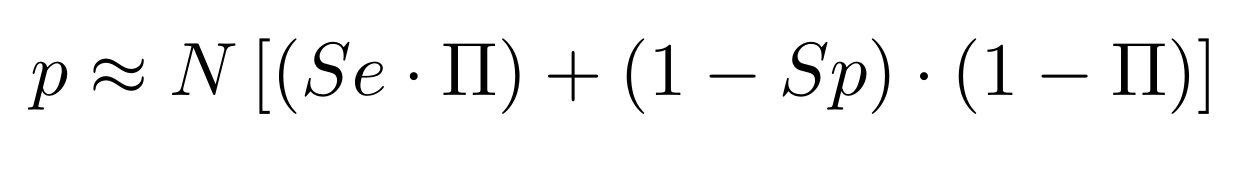
If p number of positive detections among N tests:
p = FP + TP
$$\pi$$
So,
Which means
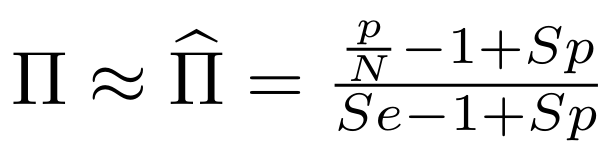
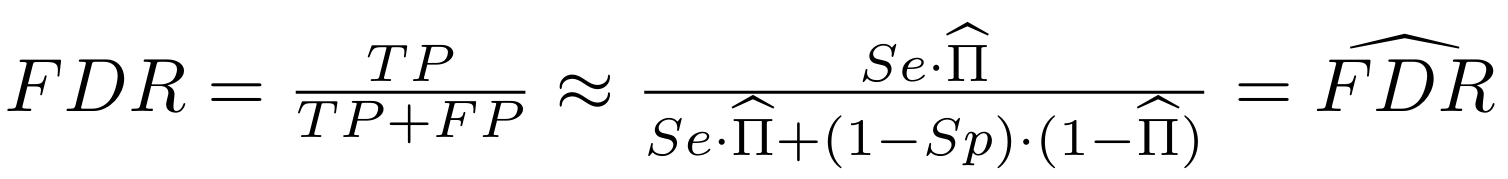
And:
What it gives in Paramecium

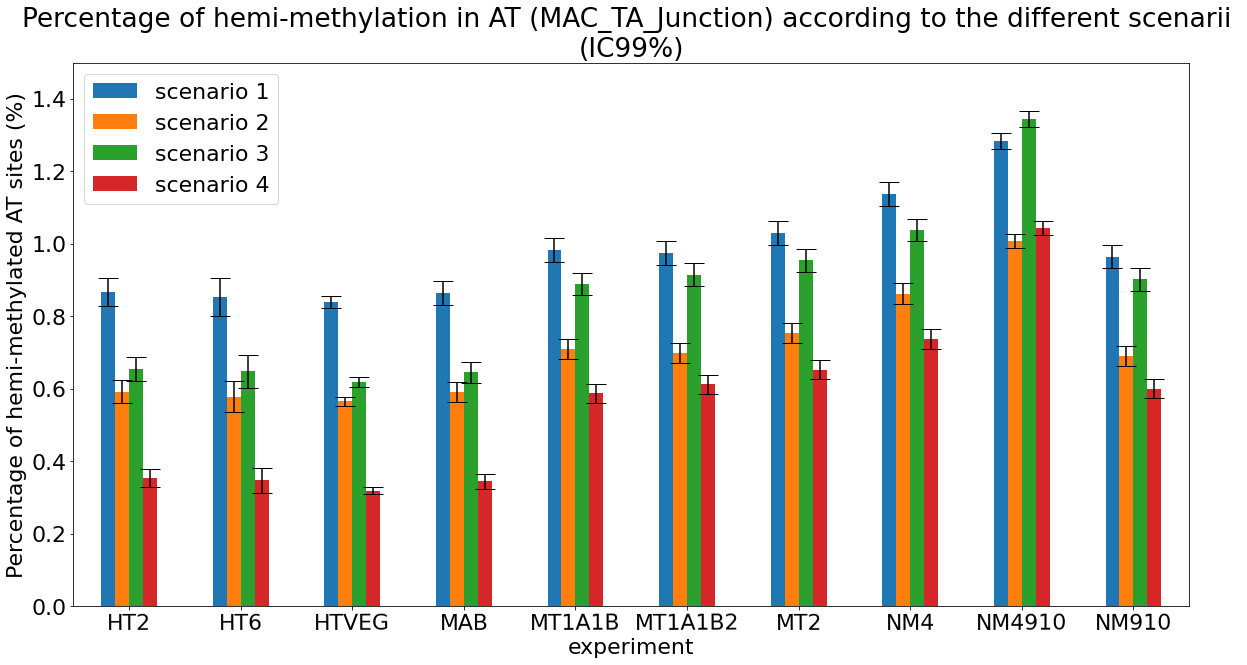
Debiased levels of m6A in the MAC_TA inserts
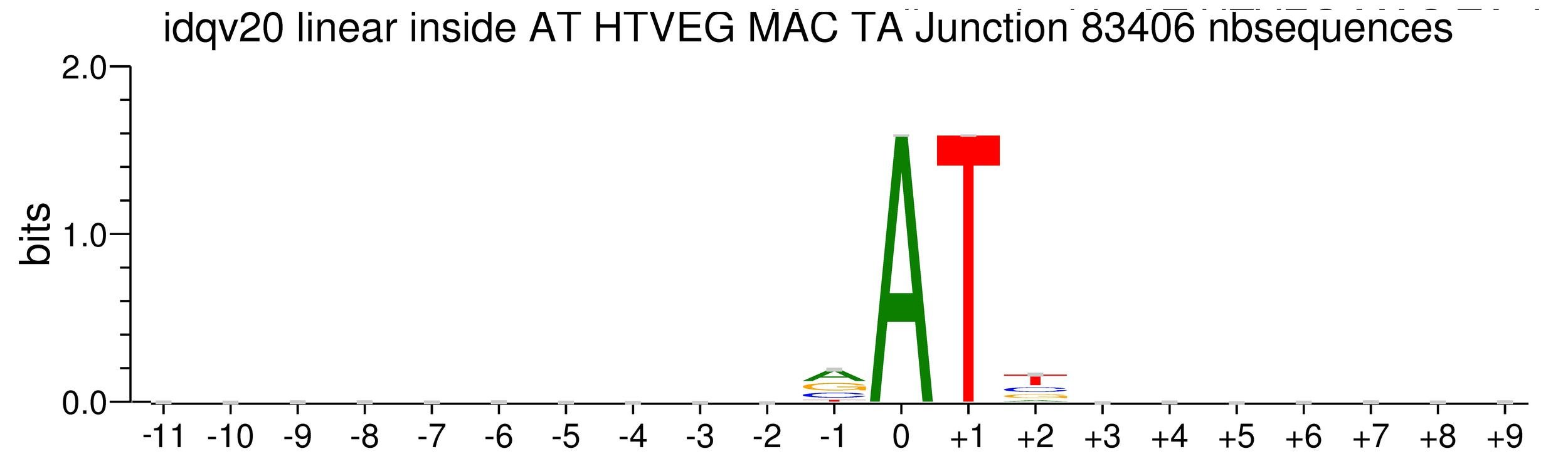
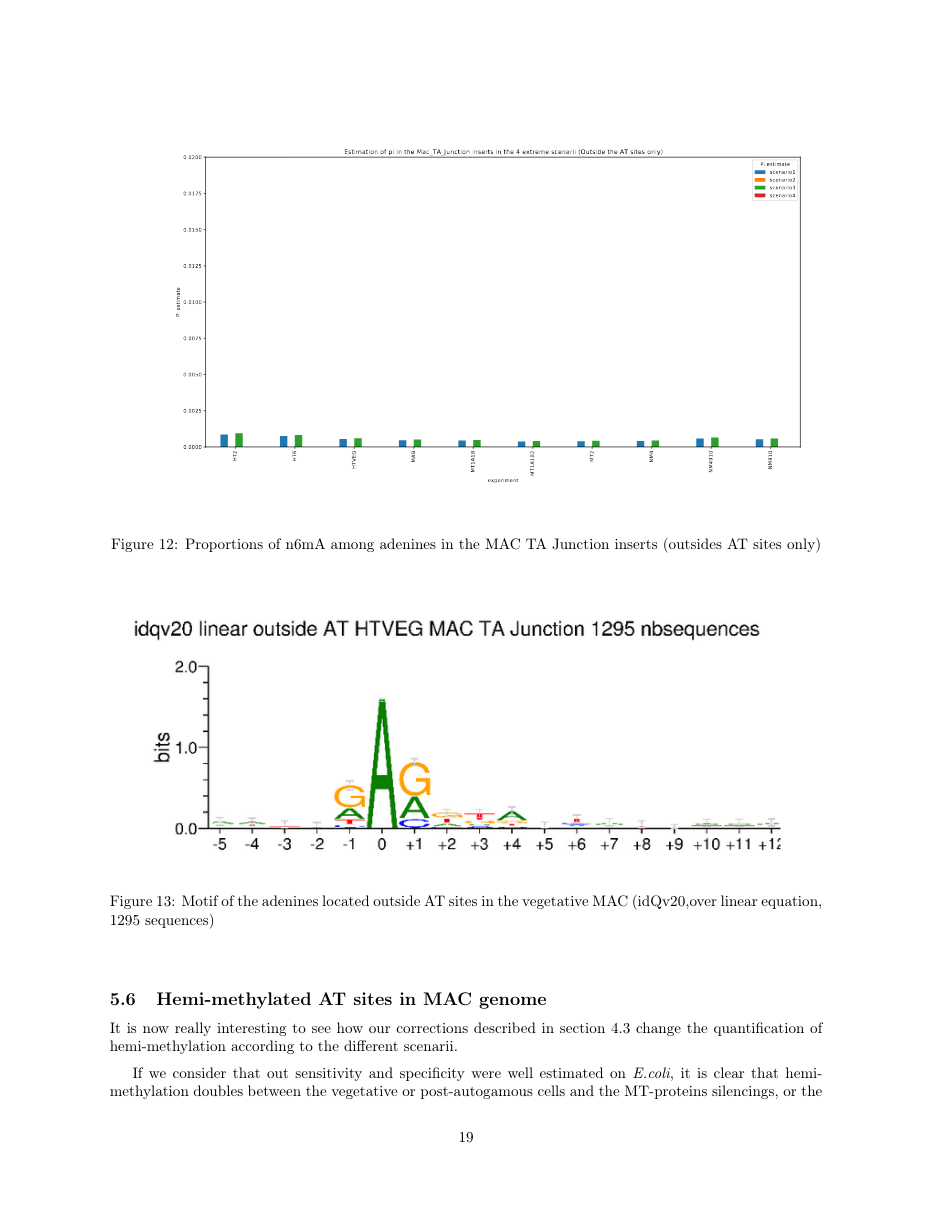
Details in MAC HTVEG
-
~95% of the methylation locates in AT dinucleotides in the MAC
-
True in any condition
-
-
75% of the methylation in an AT dinucleotide is actually symetrically modified, independantly from being in the MAC or the MIC

Kept in MIC and MAC
All conditions
MAC_TA outside of AT sites
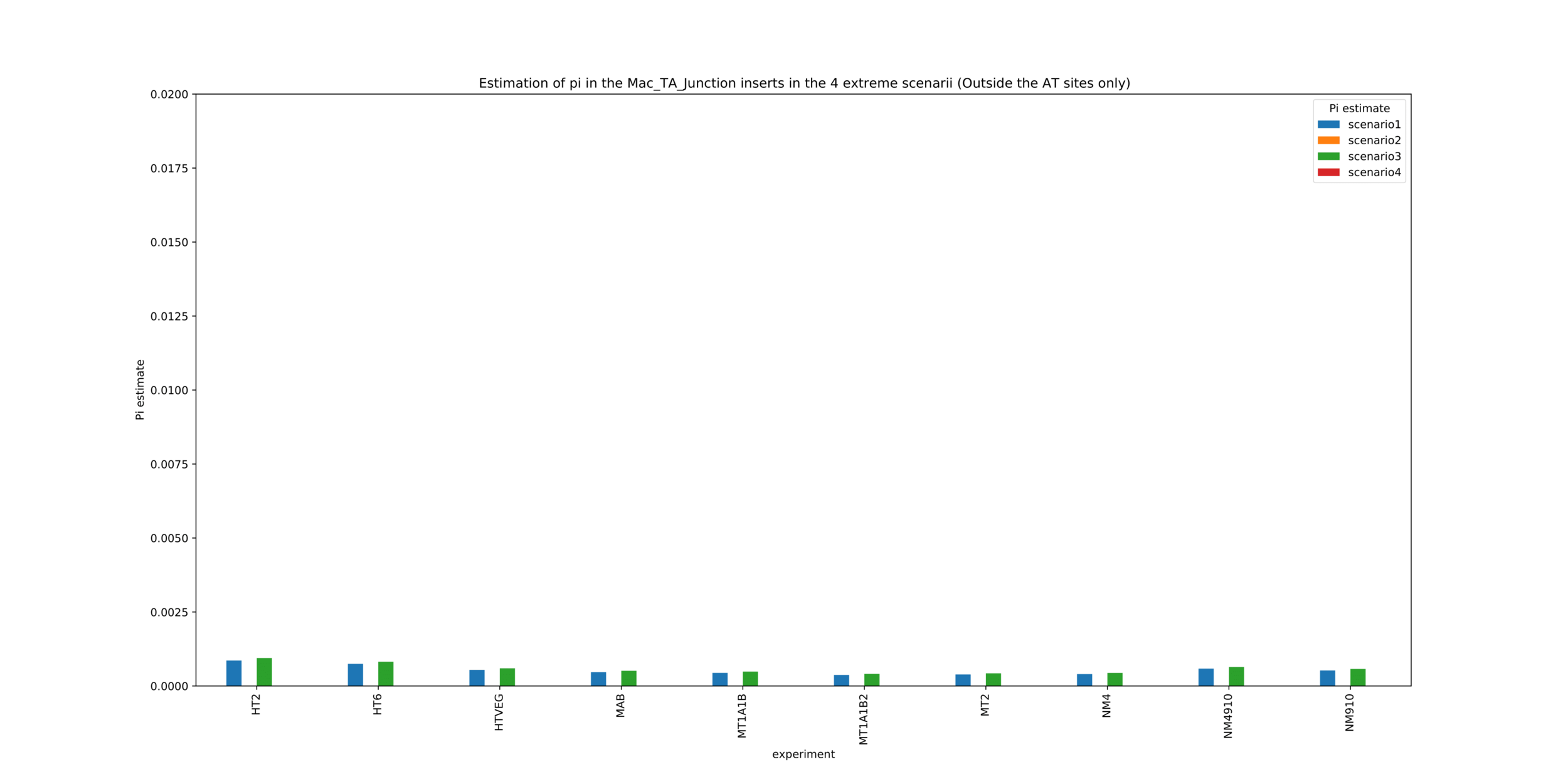
Detections outside AT sites are likely to be at least partly true positives

Present in all samples
Never erased
Conclusion: We are either in scenario 1 or 3
(Sp largely underestimated)
Was expected but confirmed
Methodological development to correct hemi-methylation detection (1/2)
Let FD1 and FD2 be resp:
- Fraction of AT sites detected hemi-methylated
-
Fraction of AT sites detected symmetrically methylated
PZ0, PZ1, PZ2: unbiased estimators of non, hémi, symetrically methylated AT sites
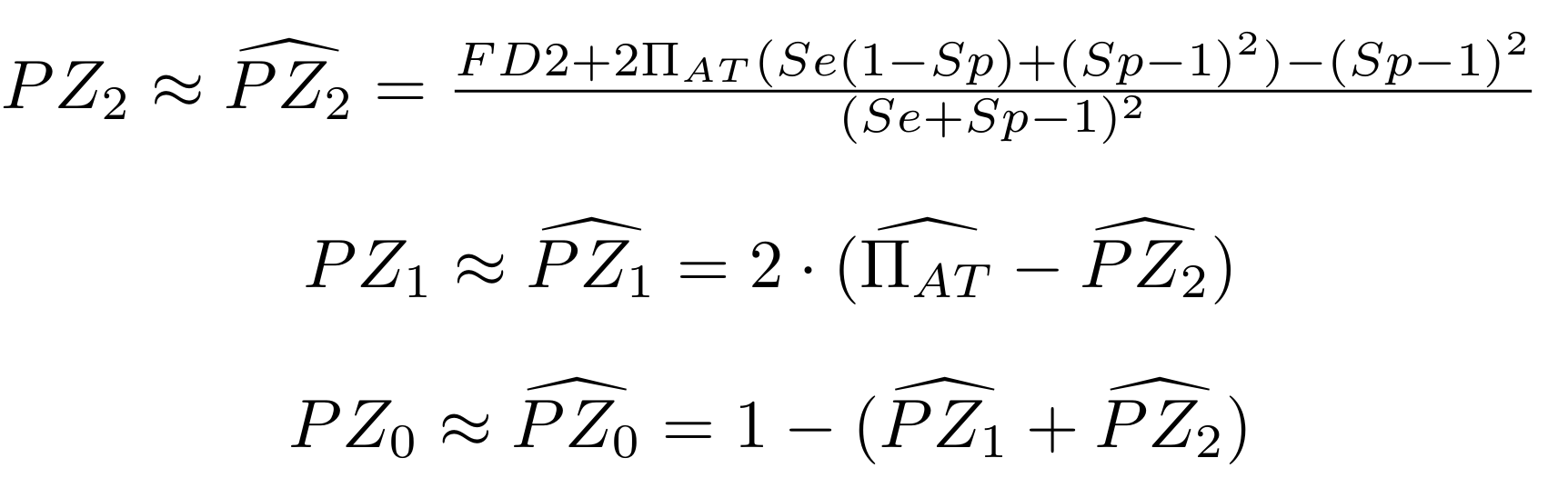
Then:
With
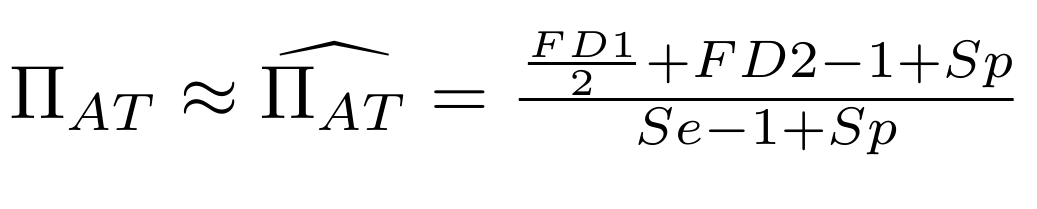
What it gives in Paramecium
Debiased Hemi-methylation in MAC_TA

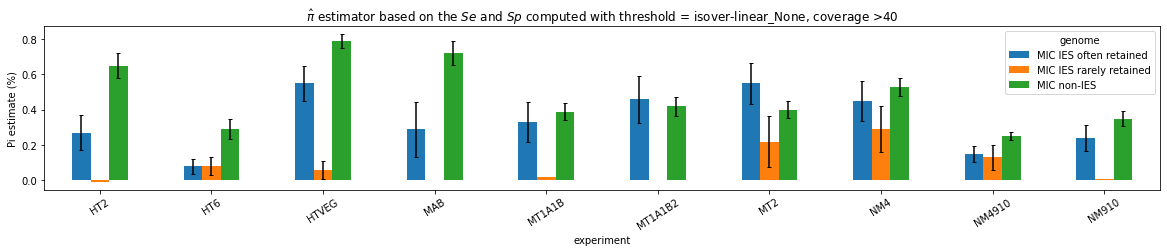
n6mA in the MIC (preliminary)
Pure MIC sequences: Cannot yet be trusted (error in the pipeline)


Coverage >= 40X made us lose too many materials, should restart with >=20X
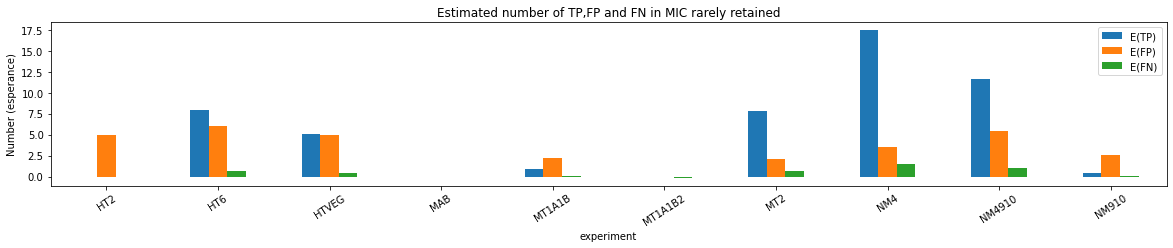
n6mA in the MAC_IES rarely retained ("TRUE MAC IES")

HTVEG

MT2
--> Some molecules carry all the detections, in sym-A*T
Very likely to be sequences comming from the MAC to my opinion
At first look, seems like the same in all samples
Conclusion
- Lots of sweat spent on methods : Now we start the cool things
-
In the MAC
- Quantification in the MAC is well-characterized in all the samples
- Our 6 genes are implied in the bulk of the MAC m6A
- MT and NM families: Functional analogs of DNMT1 ?
- Lots of questions raised by the MAC methylation: TSS, IES Junctions, nucleosome positionning...
-
In the MAC IES
- Very preliminary analysis between TRUE mac IES and other MAC IES tends to show that all detections could come from accidental retention in the MAC, and the MIC could actually carry 0% m6A or very lower levels than 1%. Really to premature to be sure
- HT2 and HT6 are still potentially full of surprises
- TE in the MIC: No idea yet
- It's just a question of time now ! Which we asked (prolongation LABEX memolife)

Sorry for the headaches !! Thanks for your time :)
Ciliates are great model organisms

1862 - Pasteur: Refutation of the spontaneous generation theory with infusoria


1937 - Sonneborn: Non-mendelian inheritance of sexual type in paramecium


Elizabeth blackburn & Carol Greider: 1985 - Telomeres and telomerases in Tetrahymena (Nobel Prize 2009)

Eric Meyer, Sandra Duharcourt
(IBENS, I. Jacques Monod 2014)
Sexual type in paramecium are transmitted by maternal RNAs, not by DNA
2.1 Retroingineering
The capping of IPDs
-
modelPrediction is the predicted IPD value by the model in a given context of nucleotides at this position
-
globalIPD is the mean of all the IPD values of the read.
-
localIPD represents all IPDs that have been mapped at a given position in the genome, including those from other sequences
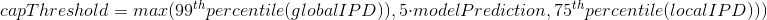
Conclusion on the capping
- Isn't coded as advertized by PacBio
- The way it's implemented for AggSN is problematic and doesn't really make sense
- Paradoxally, it should be more relevant for our approach than for the default one
- We expect no methylation to be undetected due to the capping
Laura landwebehr 2020
Oxytrichia trifallax
p values (A)
p-values (A score >20)
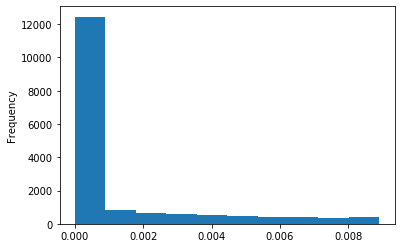
Out GATC score < 20
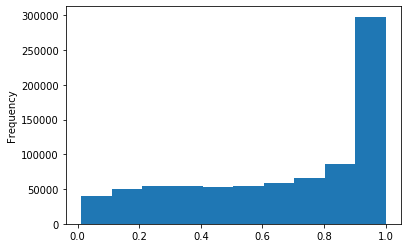
Out GATC
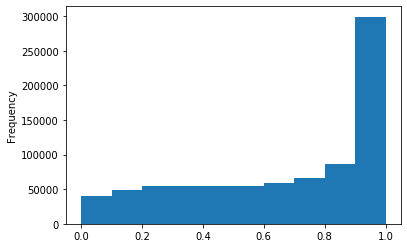
ipdRatio score 20
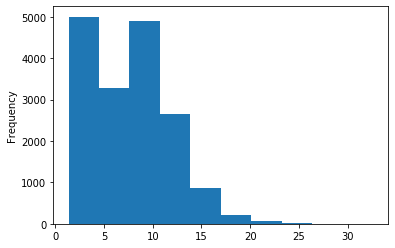
ipdRatio idv20/score20
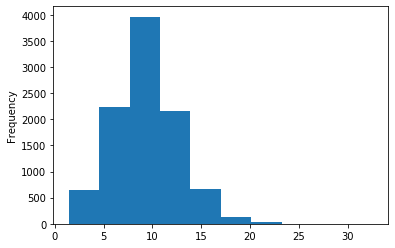


A outAT score 20 isQv20 (812 seq)
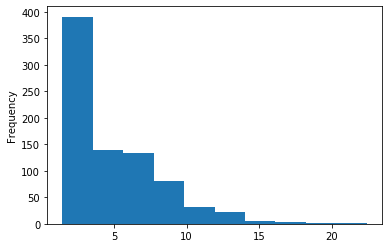
A outAT score20 idQv20 + Strong BH correction (176 seq)
ipdRatio out GATC before filtering BH vs after (qv20/idqv20)
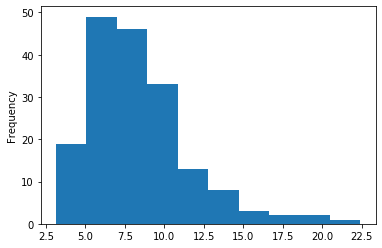
In GATC
