PyGPlates workshop
Deployment
with Docker image
docker pull brmather/pygplates-course
docker run --name pygplates -p 8888:8888 brmather/pygplates-courseExample 1
- Using rotation models
- Accessing GPlates features
- Reconstructing features back in time
- Mapping with Basemap
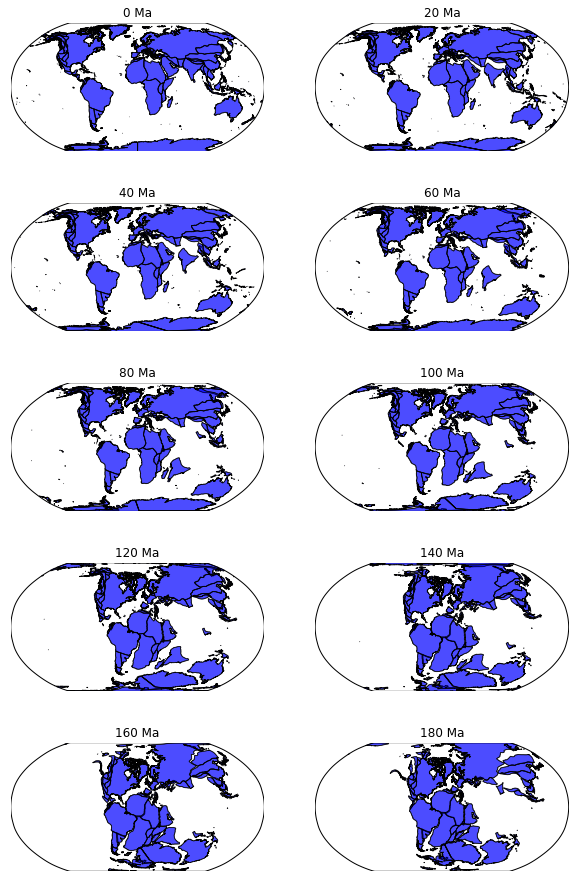

# Names of input files
input_feature_filename = './Data/Seton_etal_ESR2012_Coastlines_2012.1_Polygon.gpmlz'
input_rotation_filename = './Data/Seton_etal_ESR2012_2012.1.rot'
# Input parameters to define how the reconstruction is made
reconstruction_time = 120.6
anchor_plate = 0
# Name of ouput file
output_reconstructed_feature_filename = './tmp/tmp.shp'
# Use pygplates to carry out the reconstruction
pygplates.reconstruct(input_feature_filename,
input_rotation_filename,
output_reconstructed_feature_filename,
reconstruction_time,
anchor_plate)
Code snippet for making a reconstruction
Example 2
- Create motion paths
- Compare rotation models
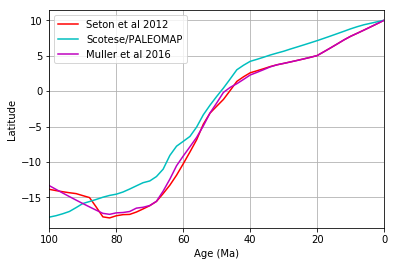
# Plot curves for three seed points on Africa
rotation_filename = 'Data/Seton_etal_ESR2012_2012.1.rot'
MovingPlate = 701
RelativePlate = 0
times = np.arange(0,130,10)
# get motion paths for three different points in Africa, at the same latitude but
# different longitudes
trail_1 = get_motion_path(rotation_filename,
(-20,0),
MovingPlate,
RelativePlate,
times)Retrieving a motion path
Example 3
- Downloading palaeodata
- Filtering specific keywords
- Export to GPlates feature
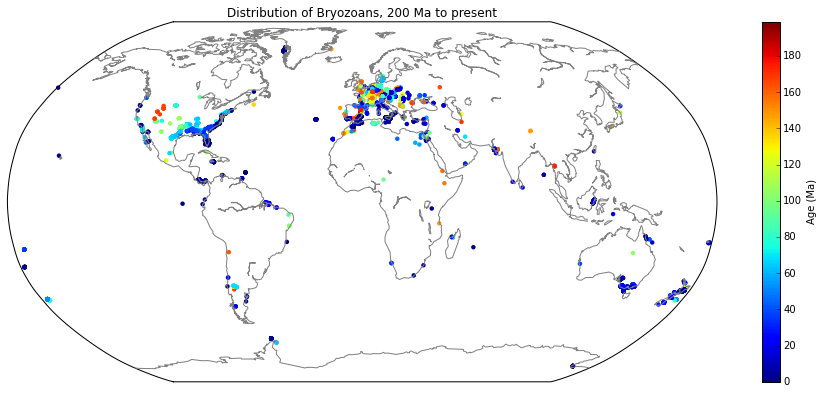
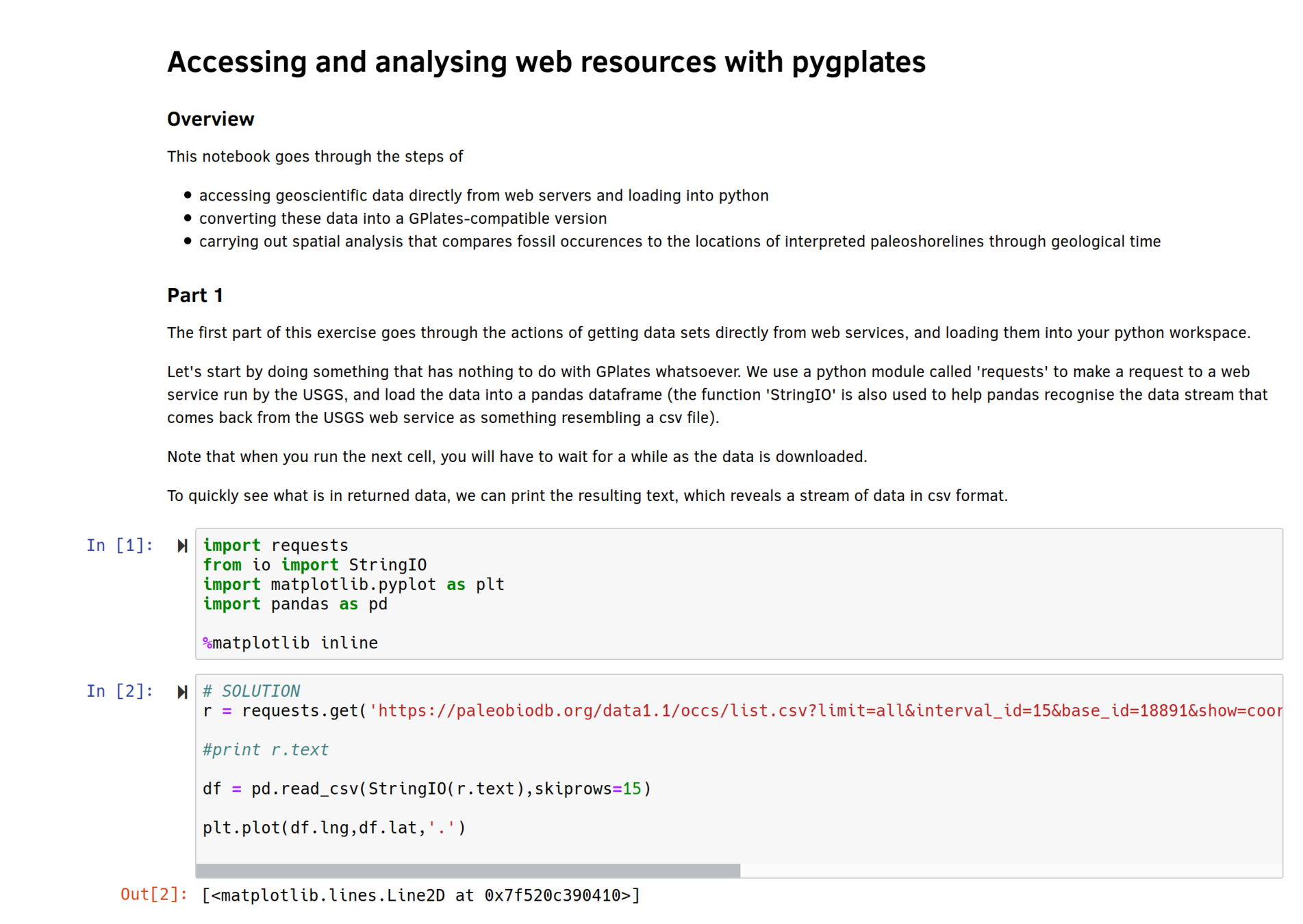
import pygplates
fossil_point_features = []
for index,row in df.iterrows():
point = pygplates.PointOnSphere(float(row.lat),float(row.lng))
point_feature = pygplates.Feature()
point_feature.set_geometry(point)
fossil_point_features.append(point_feature)
# Save a feature collection to file
# can be *.gpml, *.gmt or *.shp
output_feature_collection = pygplates.FeatureCollection(fossil_point_features)
output_feature_collection.write('my_fossils.gpml')Exporting feature collections
Roadmap
- Deforming plate margins
- Move from Python2 to Python3
- Replace Basemap (depreciated) with Cartopy
- GitHub repository