
Gautam Naishadham and Brad Langhorst - New England Biolabs
Talk Goals
- Convey utility of UMIs in RNA-seq
- Examine technical factors associated with apparent transcript level differences
- Compare Transcript levels +/- UMI with expectation
- Explore correlation of transcript features with measured abundance
- For me: Hear about what other features should we consider?
What is a UMI
- Universal Molecular Identifier
- (aka MID, Molecular barcode, UID)
- Semi-random synthetic sequence of DNA bases
- Probability of any specific sequence = 1/4^n
- n = number of bases
- n=1 1/4 A,C,G, or T
- n=2 1/16 AA,AC,AG,AT,CA,CC ... GT,TA,TC,TG,TT
- n=3 1/256 ...
- We use 12 bp UMIs = 1/16,777,216
- When combined with transcript identity, probability of false collision is very low (function of transcript abundance)
- Sequencing errors might produce related UMIs
RNA-Seq Workflow Overview
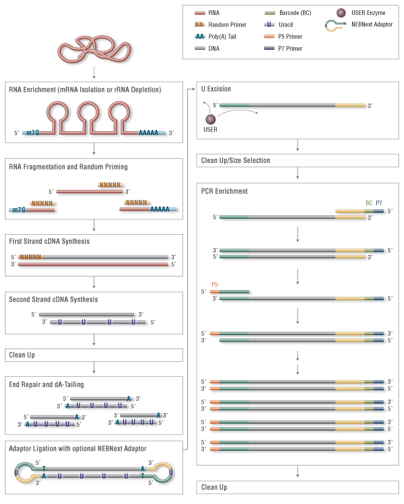
RNA-Seq Workflow Overview

UMI added here
technical factors related to observed abundance
RNA-Seq Workflow Overview
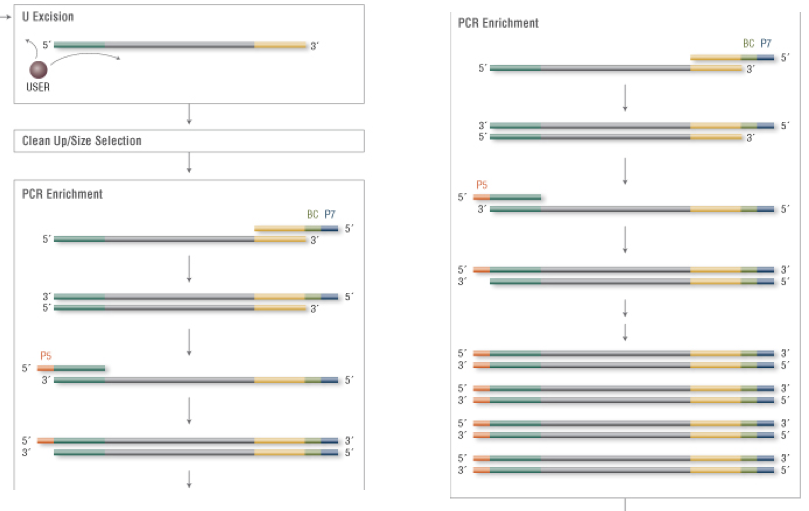
UMIs duplicated here
technical factors related to observed abundance
Technical Factors
- Pre - UMI
- Priming bias (hexamers may not be random)
- Fragmentation bias (too easy = low, too hard = low)
- A-tailing bias (lower efficiency = low)
- Post - UMI
- Size selection/cleanups (bead binding, differential elution)
- PCR (too easy = high, too hard = low)
Uneven Amplification
BRCA1
p53
Original
Library
+ UMI
Fragments
2
1
3
4
5
6
7
9
1
2
3
4
5
PCR
1
1
1
2
2
3
4
4
4
4
4
4
5
6
7
8
9
5
6
7
8
9
1
2
3
4
5
1
2
3
4
5
2
3
4
5
4
4
4
5
9
9
8
8
25/18 = 1.8
9/5 = 1.4
BRCA1/p53 ratio
4/3 = 1.3
Identification of Duplication
Dedup
1
1
1
2
2
8
9
8
9
9
9
8
Using UMI
No UMI
BRCA1
Alignment
1
2
9
= 12
= 3
1
1
1
2
2
8
9
8
9
9
9
8
1
2
8
9
= 4
= 12
BRCA1
BRCA1
BRCA1
+/-UMI Experiment Design
C. Devoe, K. Krishnan, D. Rodriguez
Goal: Compare counts of transcripts with and without UMI
- 10 ng total RNA (~ 300 cells, human/mouse blood)
- NEBNext Ultra II RNA library prep + UMI adapters
- 12 PCR cycles
- Sequenced on Illumina NovaSeq 4000 S2, 2x75bp
Experimental Results
C. Devoe, K. Krishnan, D. Rodriguez

Experimental Results
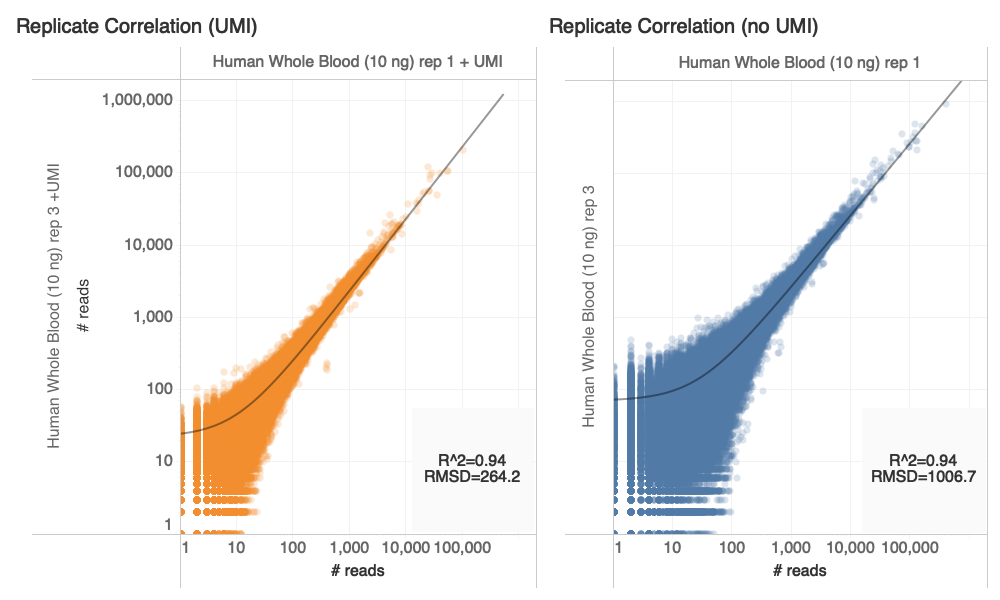

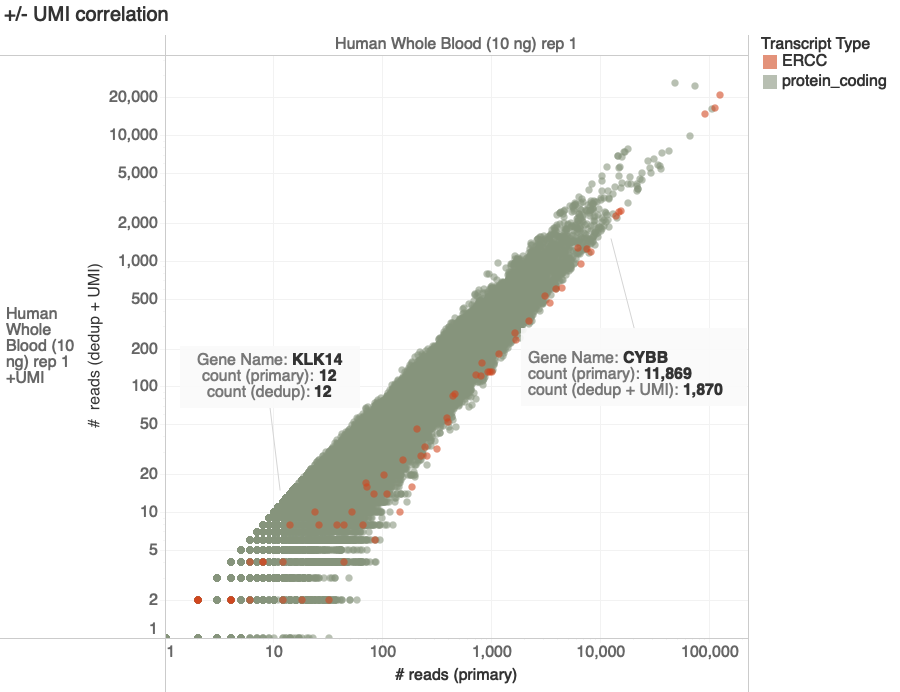
C. Devoe, K. Krishnan, D. Rodriguez
Experimental Results

Hard to Amplify
Easy to Amplify
C. Devoe, K. Krishnan, D. Rodriguez
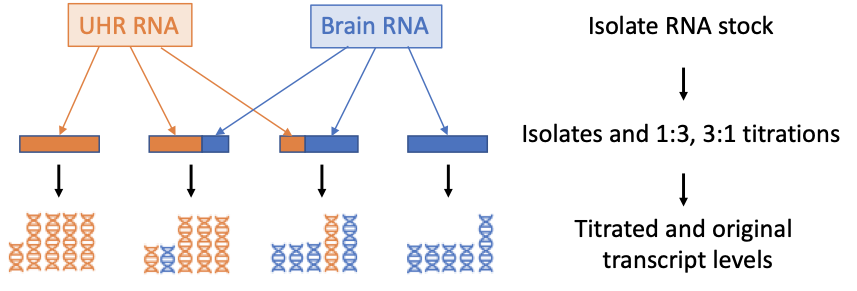
RNA Mixture Experiment
G. Naishadham
Goal: Compare counts of transcripts to expectation
- 10 ng Input (~ 300 cells, cell line Blood/Brain RNA)
- Defined mixtures 1:3 and 3:1 to generate expected values
- NEBNext Ultra II RNA library prep + UMI adapters
- 12 PCR cycles
- Sequenced on Illumina NovaSeq 4000 S2, 2x75bp
RNA Mixture Experiment
G. Naishadham
Galaxy Workflow
G. Naishadham
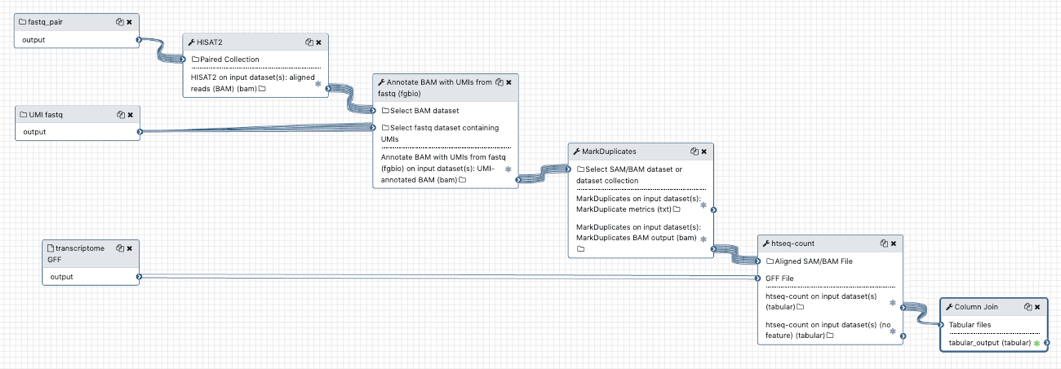
Challenges: 3 reads R1, R2 + UMI
memory = f(umilen)*num reads
Experimental Results
G. Naishadham
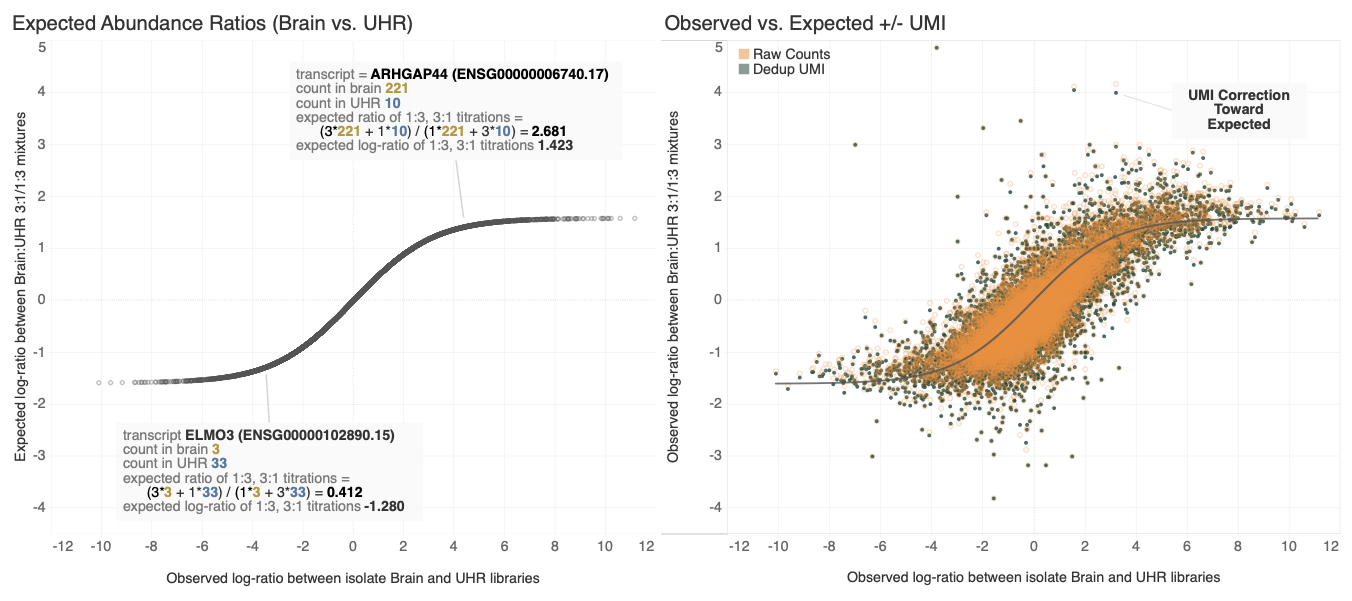
Experimental Results
G. Naishadham


Do transcript features explain variable amplification?
- GC
- Fragment Length
- RNA structure stability
- K-mer enrichment
- Others?
Transcript GC%
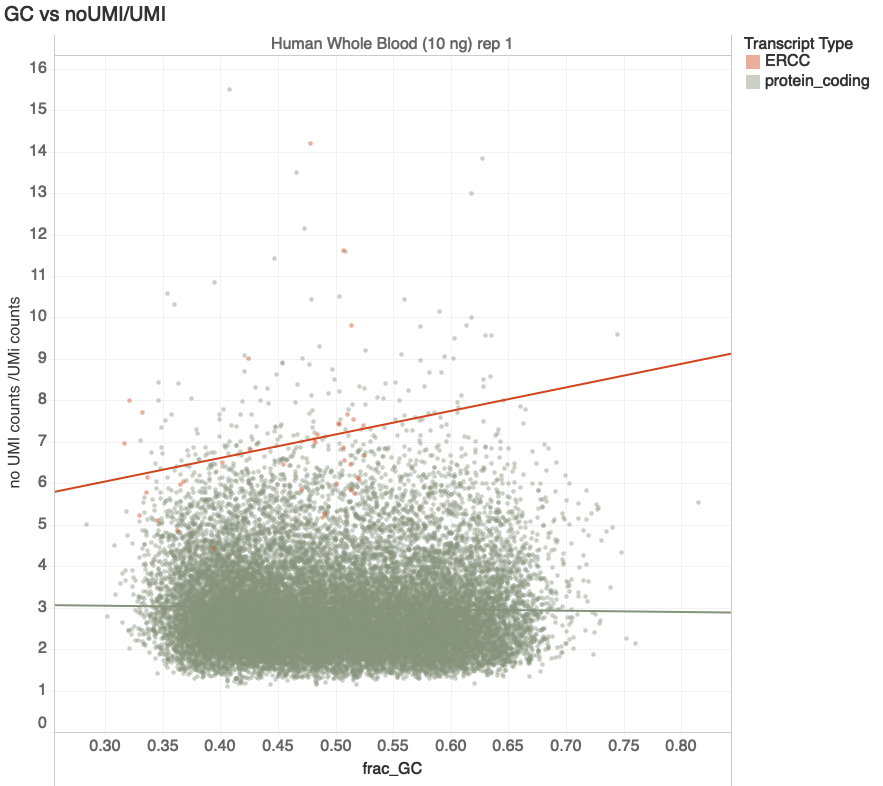
Increasing Duplication
transcripts with > 100 reads
Fragment Lengths
Increasing Duplication
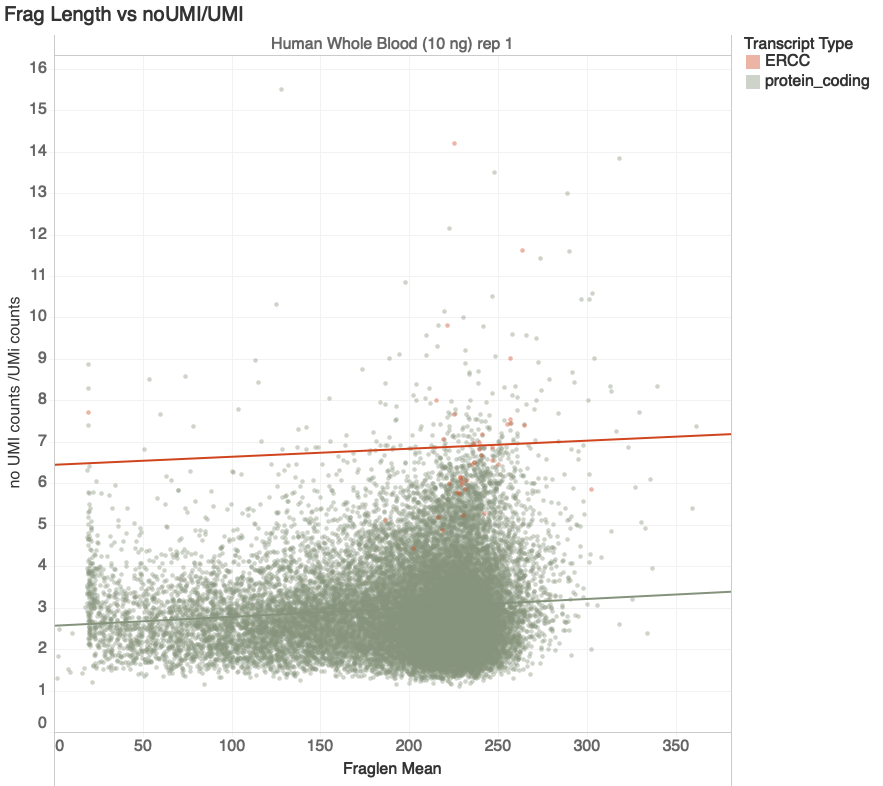
G. Naishadham
transcripts with > 100 reads
Mean Free Energy
Increasing Duplication
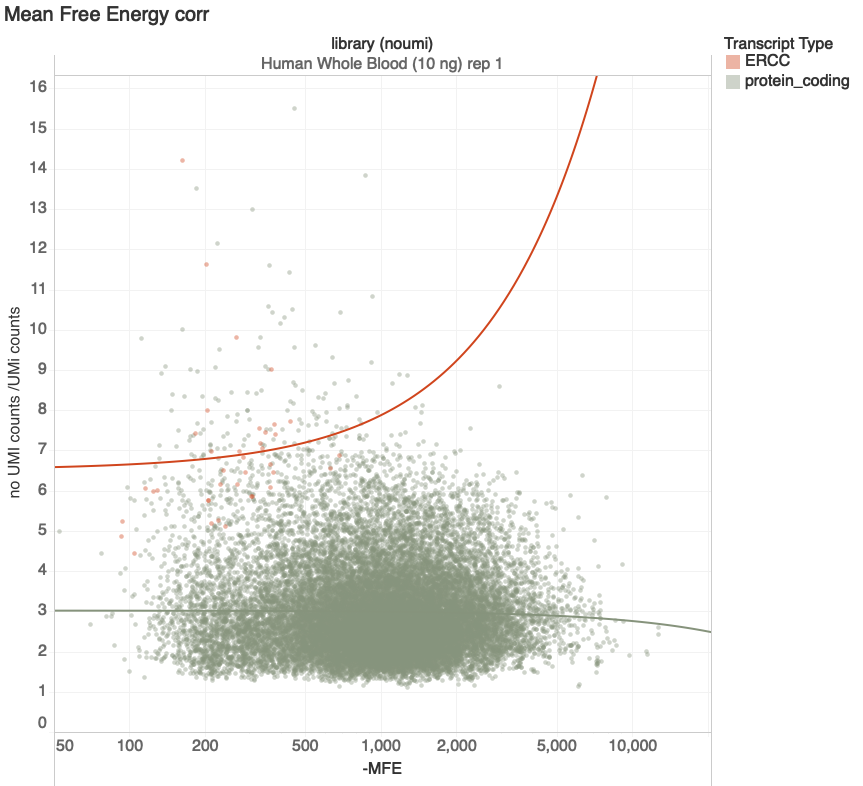
Increasing Predicted Stability
transcripts with > 100 reads
G. Naishadham
Other Factors Under Consideration
- Complexity (Shannon information content)
- Maximum 200 bp folding energy
- Other ideas ?