Primer Scheme Switching Considerations:
Length, and Variants, and Bears - Omicron

Date
Brad Langhorst
CDC TOAST - Brad Langhorst - NEB - 2021-12-03
NEB's Interest in SARS-CoV-2 Variants
We want be sure that our LAMP, qPCR and sequencing SARS-CoV2 kits can be updated as new variants arise
Other people probably need this too... FDA guidance

Nightly Download
Genome Alignment
Variant Calling
RelationalDatabase
Matt Campbell
Diagnostic Tests
Charité qPCR
China CDC qPCR
CDC qPCR
N2/E1 LAMP
Proprietary Assays
...
Sequencing Methods
ARTICv3
ARTICv4
ARTICv4.1
VarSkip
Midnight
...
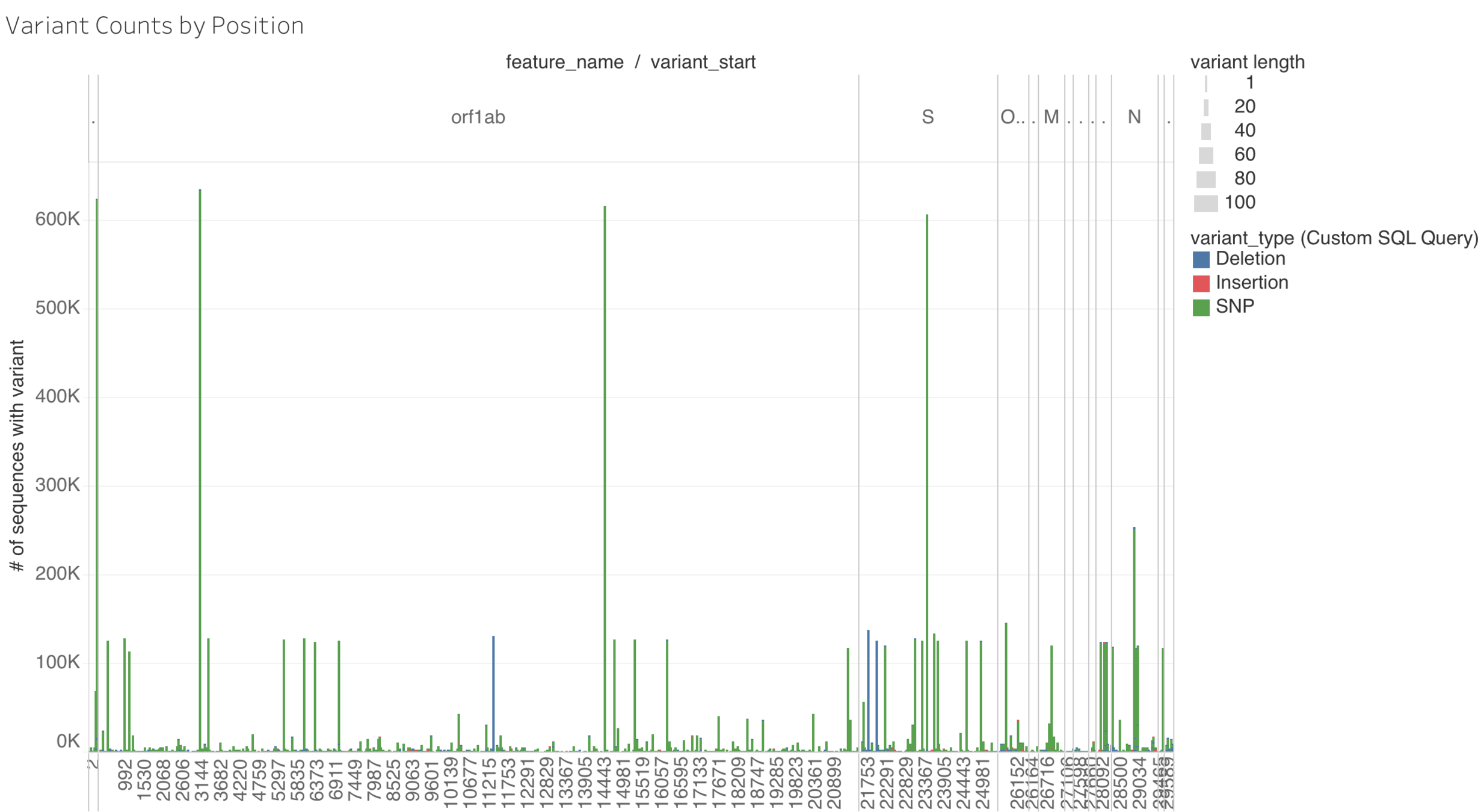
Other Variants Matter for Diagnostics
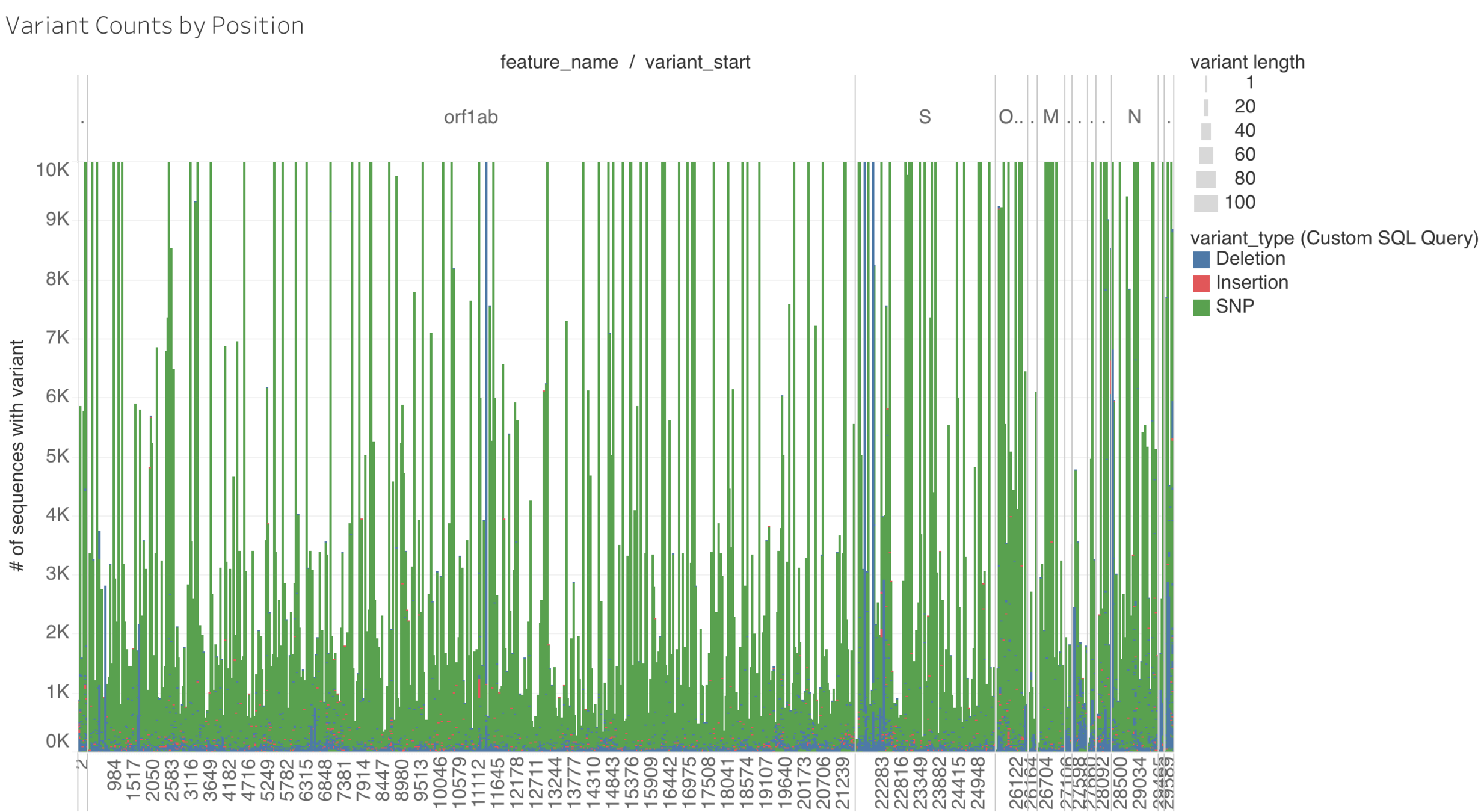
Variant Skip Design
Hypthesis: some genomic regions are more prone to variation than others.
Approach:
- Identify variants seen in GISAID sequences (651,177 sequences as of 2021-03-08) seen > 2000 times
- Mask known variant positions in reference genome with Ns
- (bedtools maskfasta)
- Design primers
- Vary lengths, overlaps, GC, and variant frequency threshold (primalscheme)
- Select best design
- Additional primers as necessary
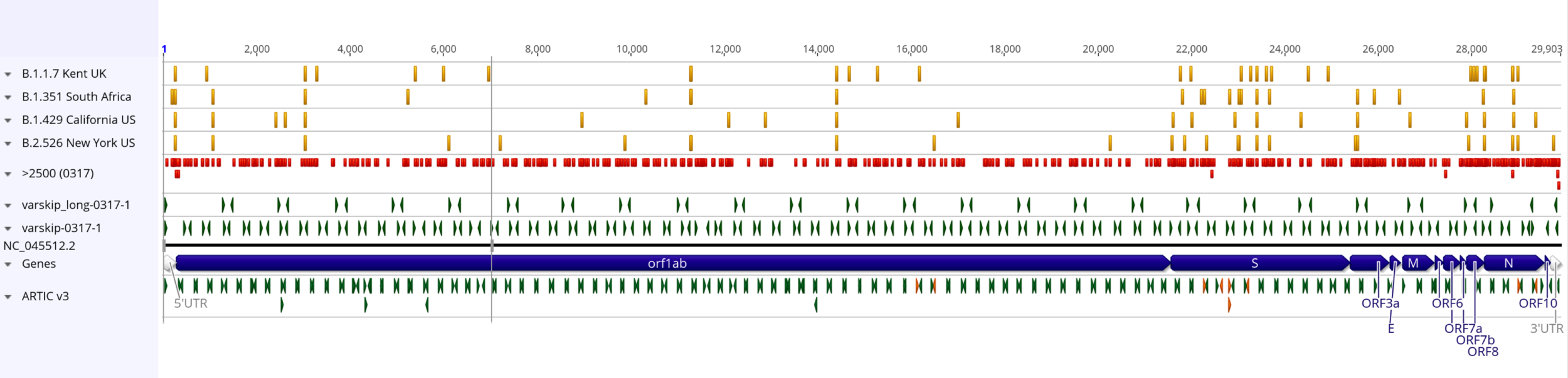
Varskip-long and varskip-short primers do not overlap >2500 variant sites (red) or variants from lineages of concern (yellow).
Orange ARTIC v3 primers overlap with a lineage of concern (yellow). Many ARTIC primers overlap sites with more than 2500 variant observations (red)
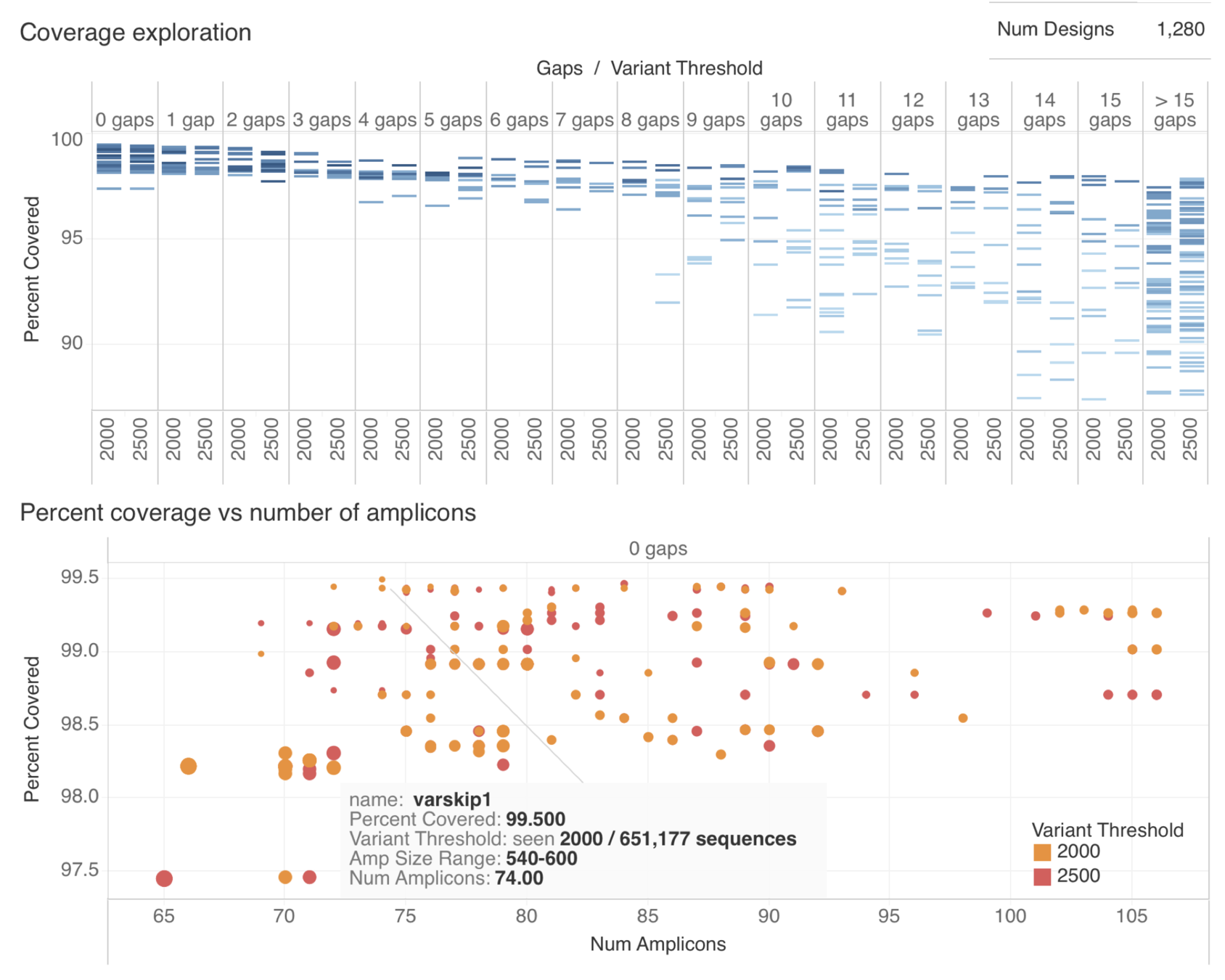
Varskip Design Summary

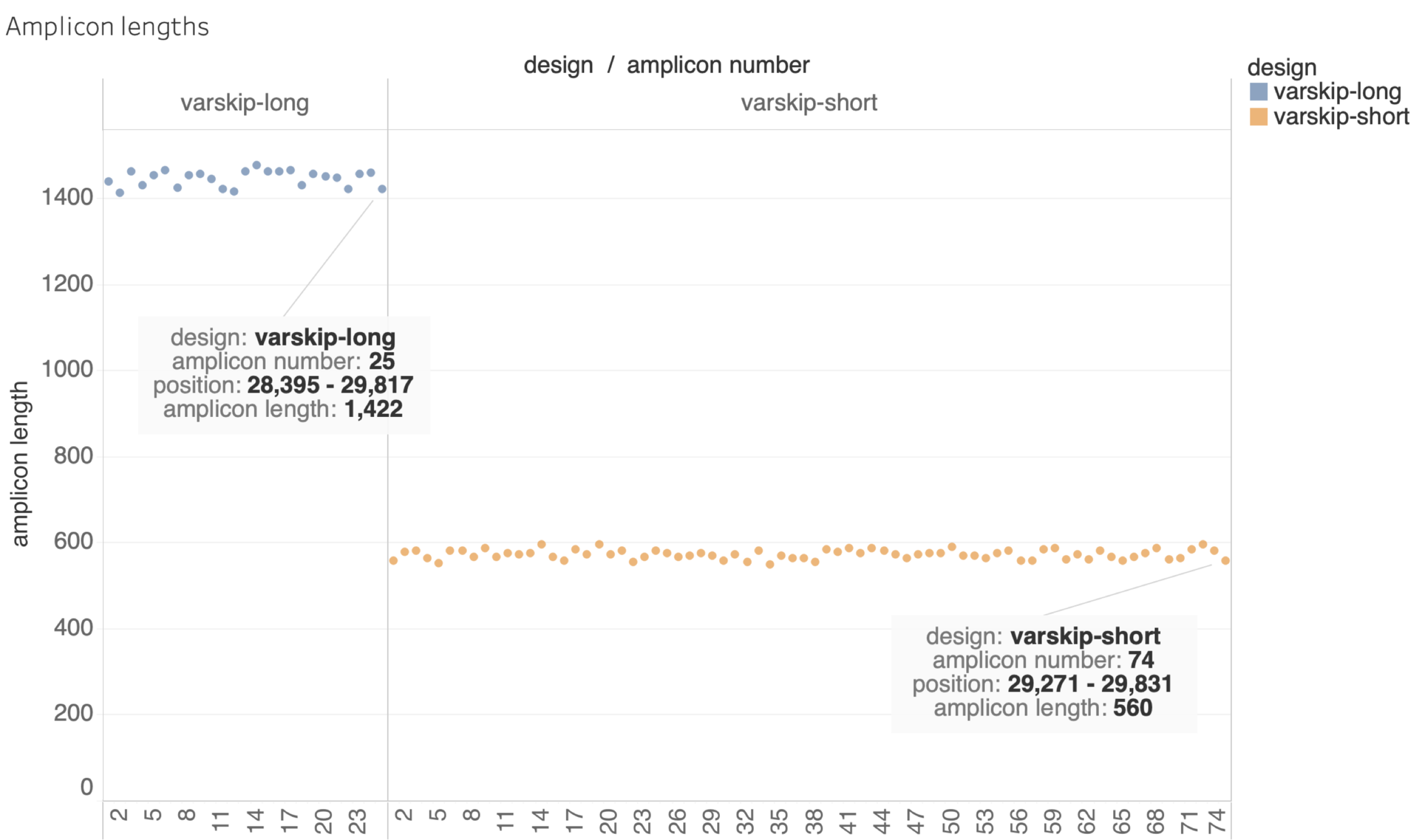
Amplicon Summary
Selected VarSkip Short
- Long vs. Short
- Long : Lower "surface area", but requires more intact RNA. Each dropout has a bigger impact on coverage.
- Beta testing indicated more robust performance for short version, especially for marginal samples
- Big Question
- Will VarSkip avoid future variants?
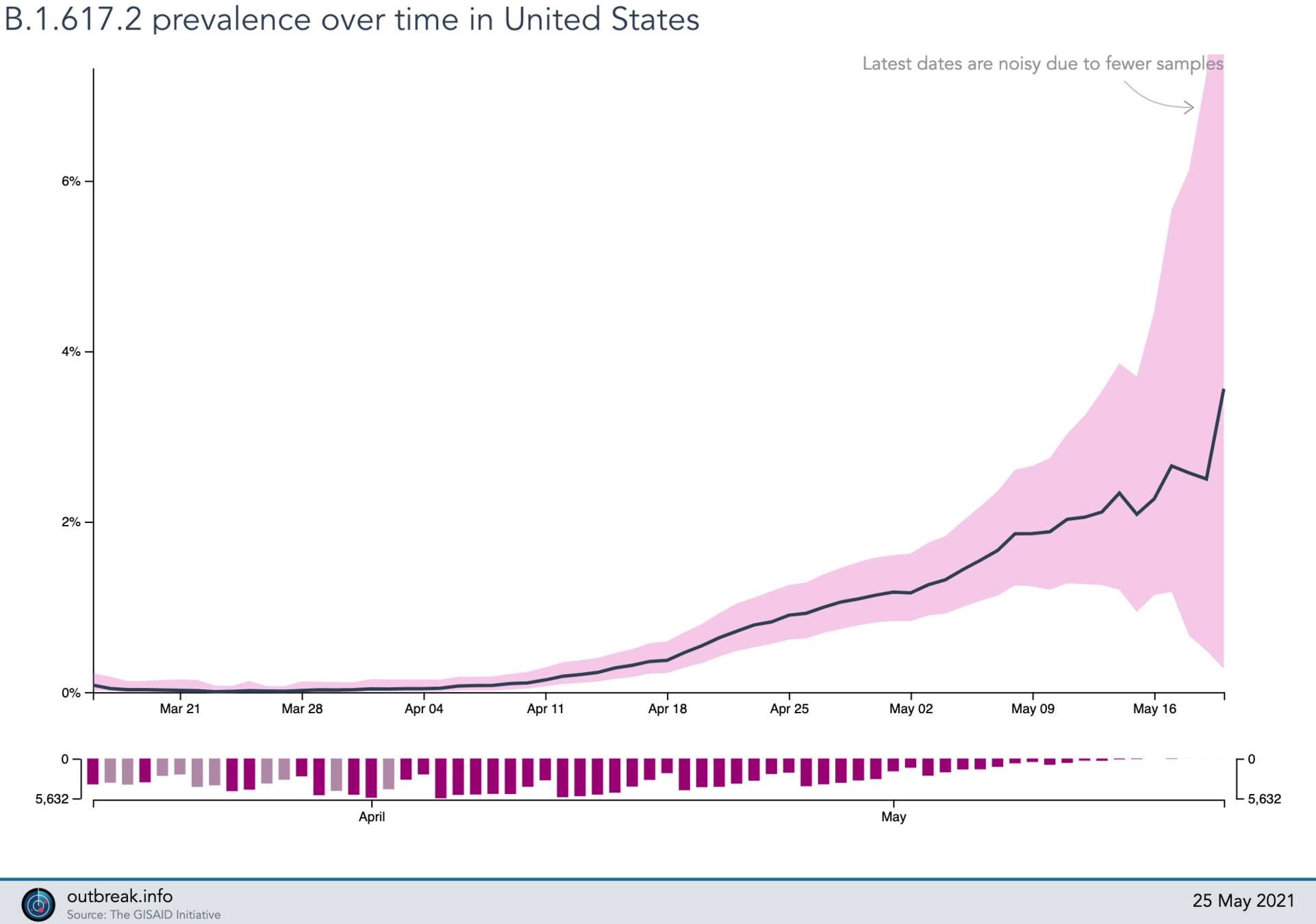
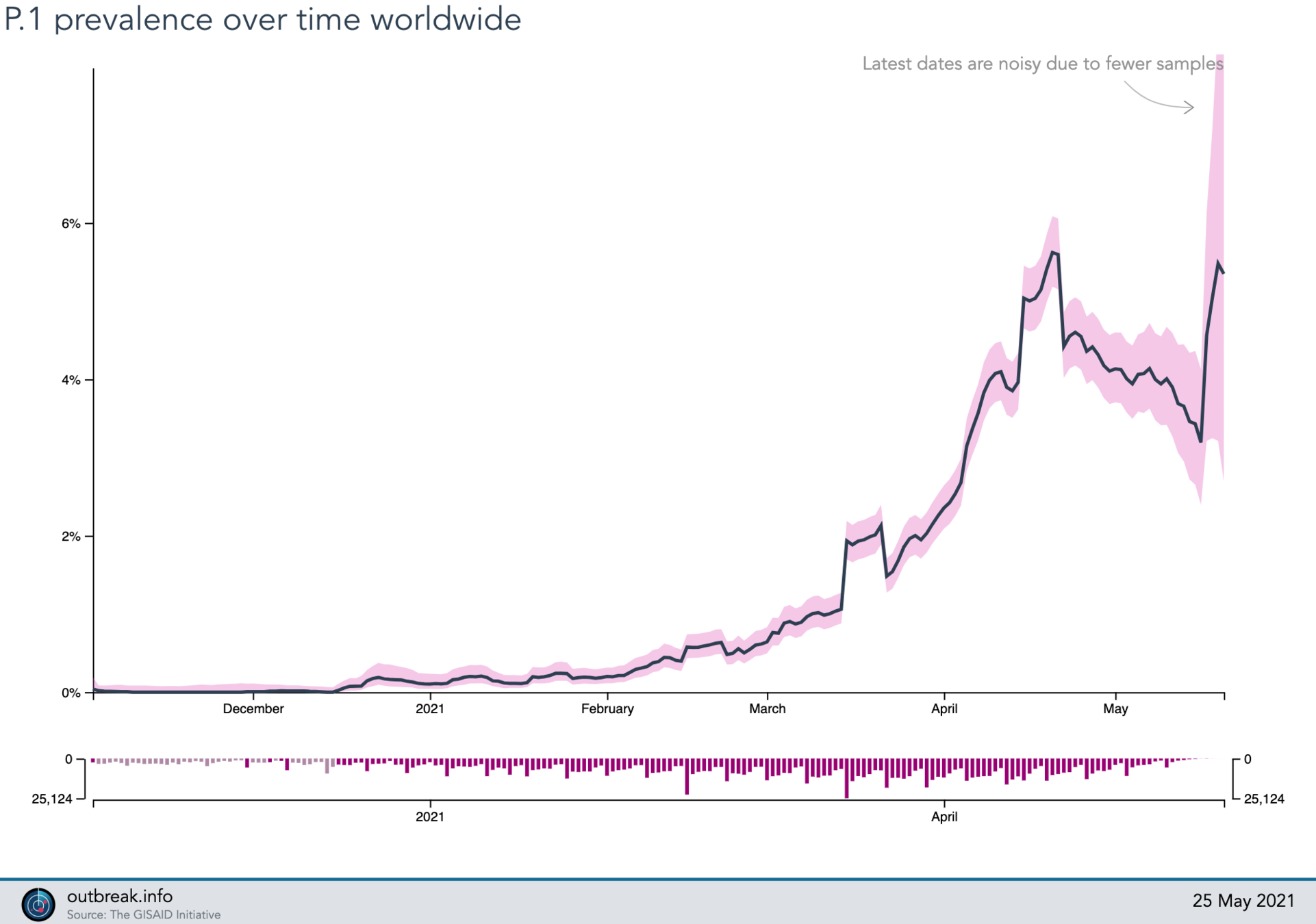
Lineage Summary
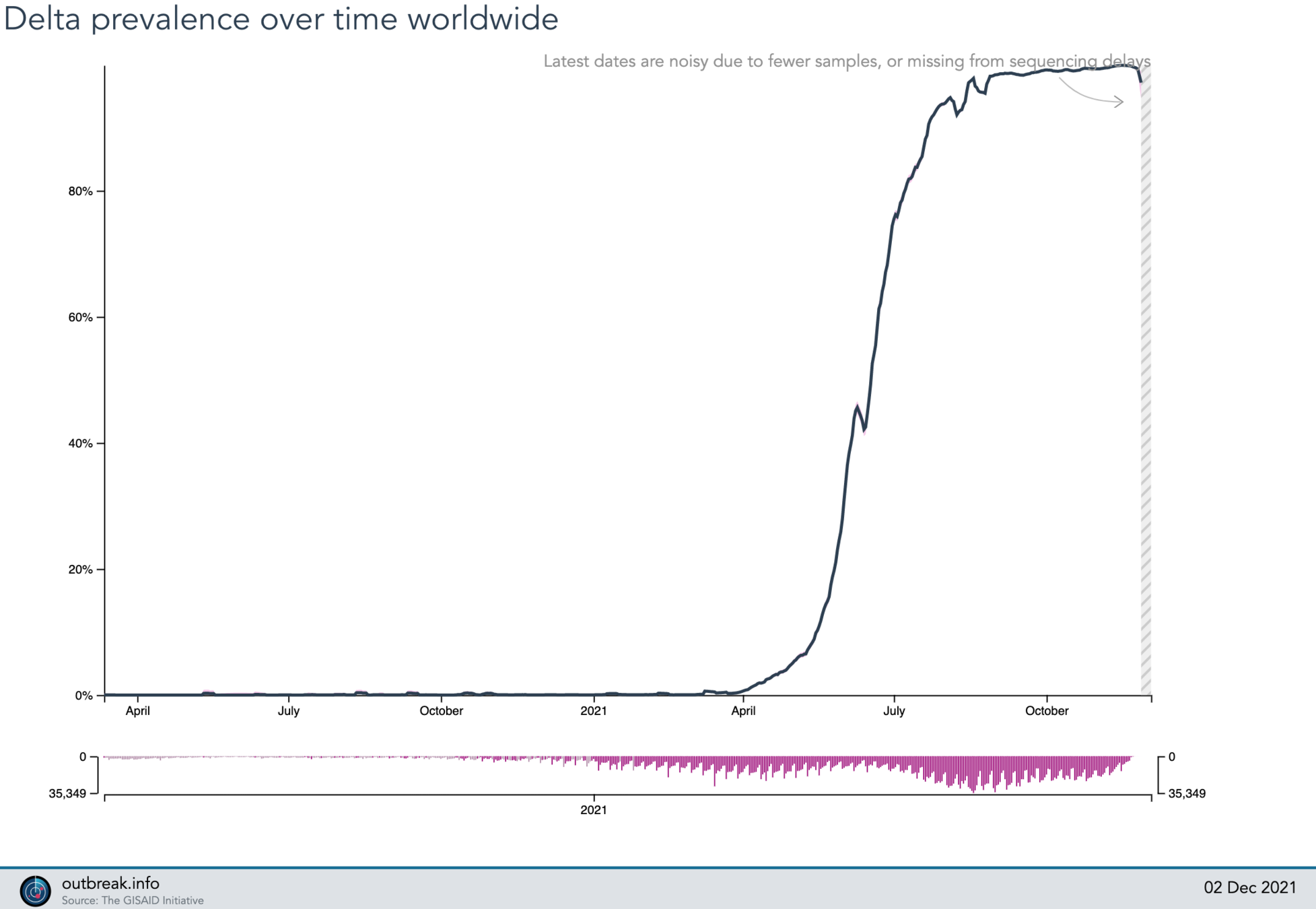
2021-05-25
6%
6%
P.1 (gamma)
B.1.617.2 (delta)
Julia L. Mullen, Ginger Tsueng, Alaa Abdel Latif, Manar Alkuzweny, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Kristian G. Andersen, Chunlei Wu, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology outbreak.info. Available online: https://outbreak.info/ (2020)
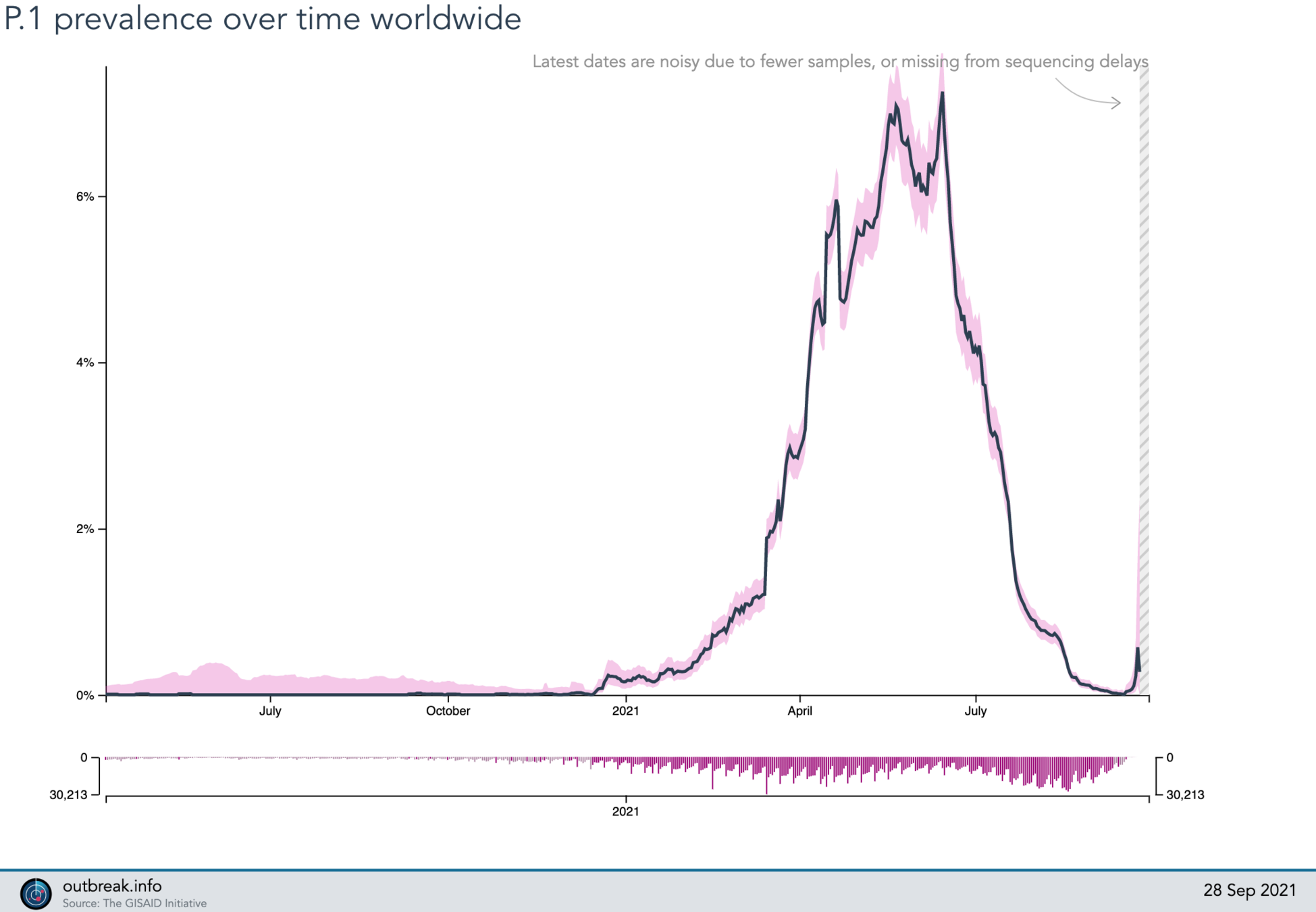
2021-09-28
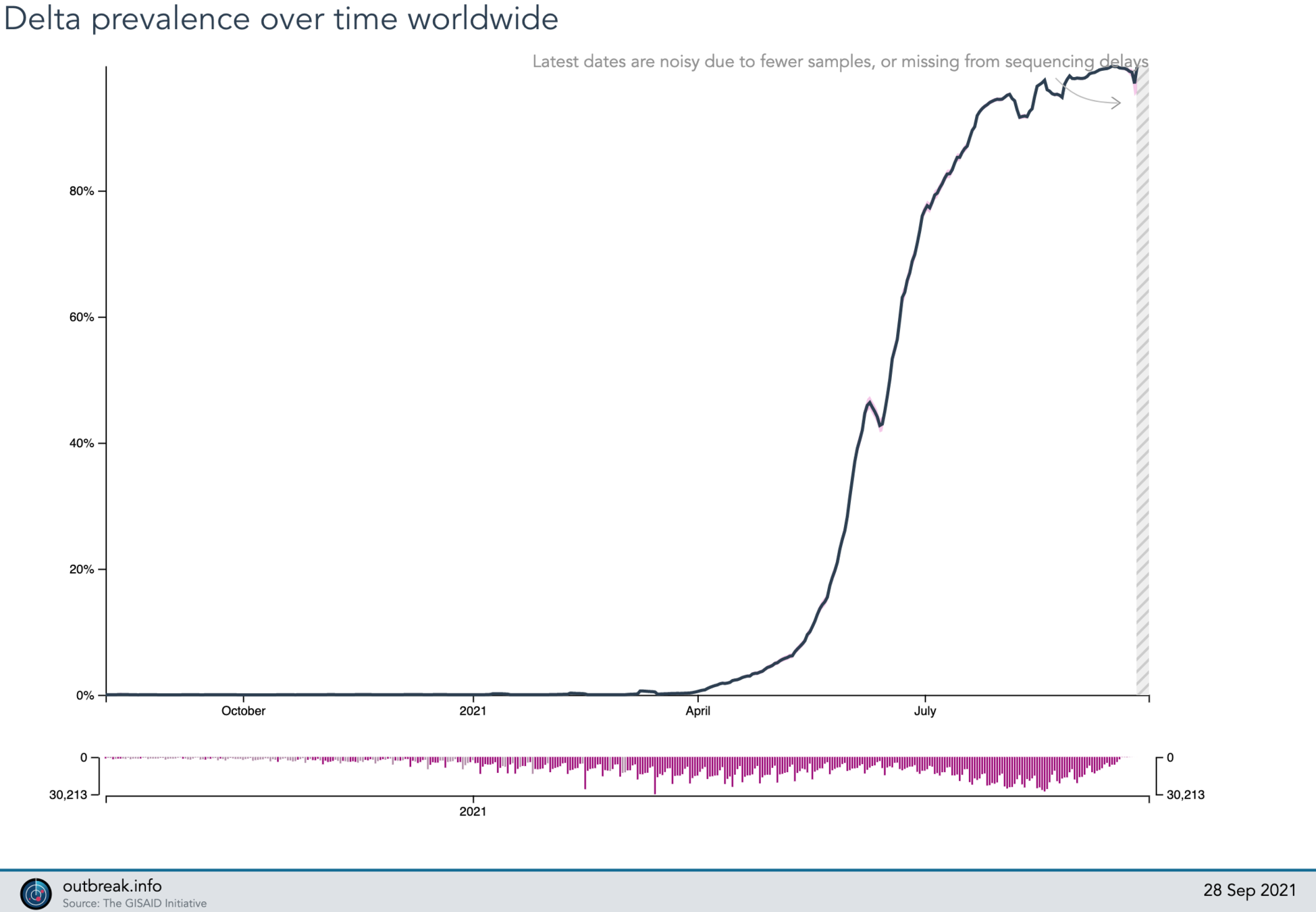


?
?

Lineage Summary

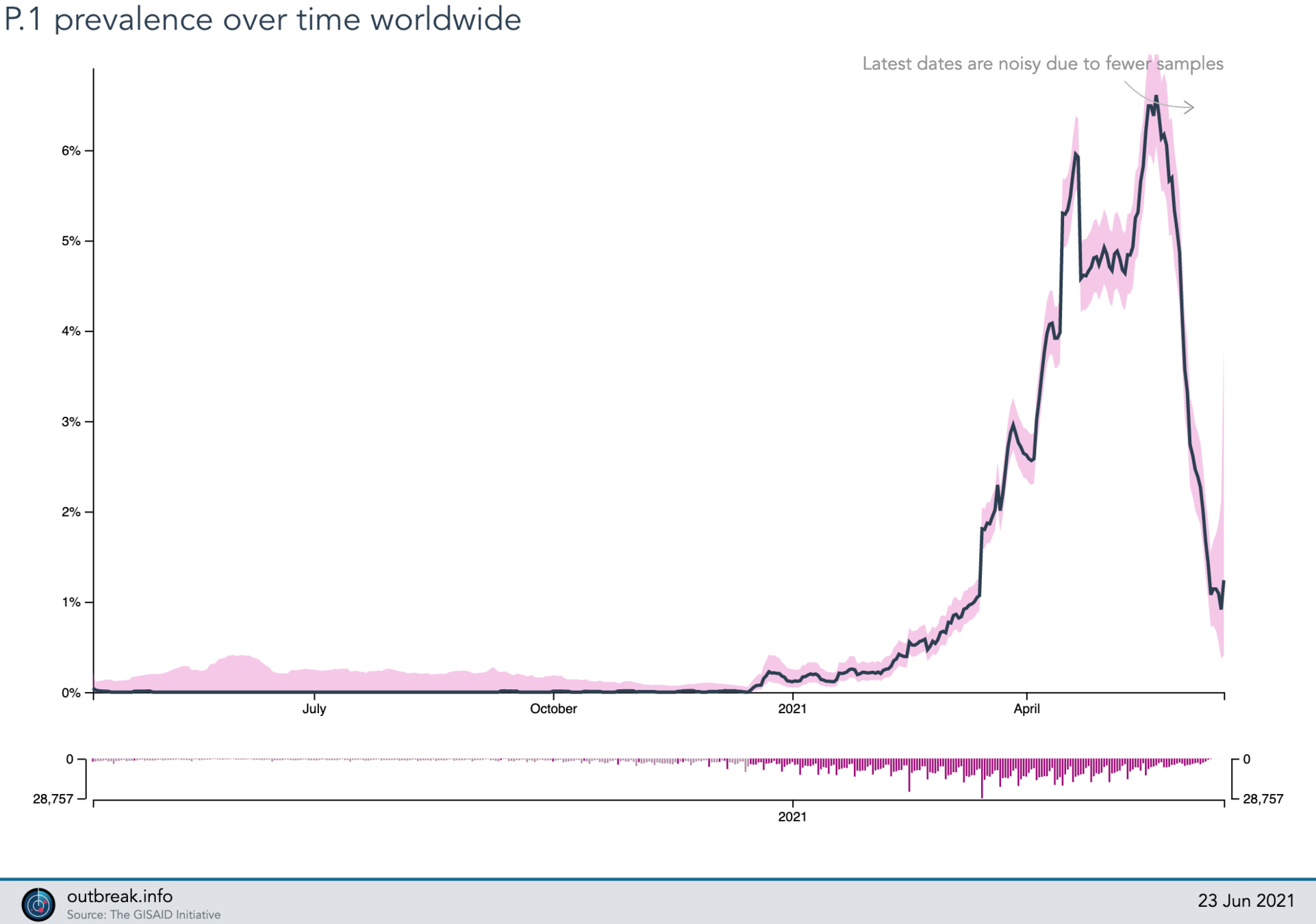
2021-05-25
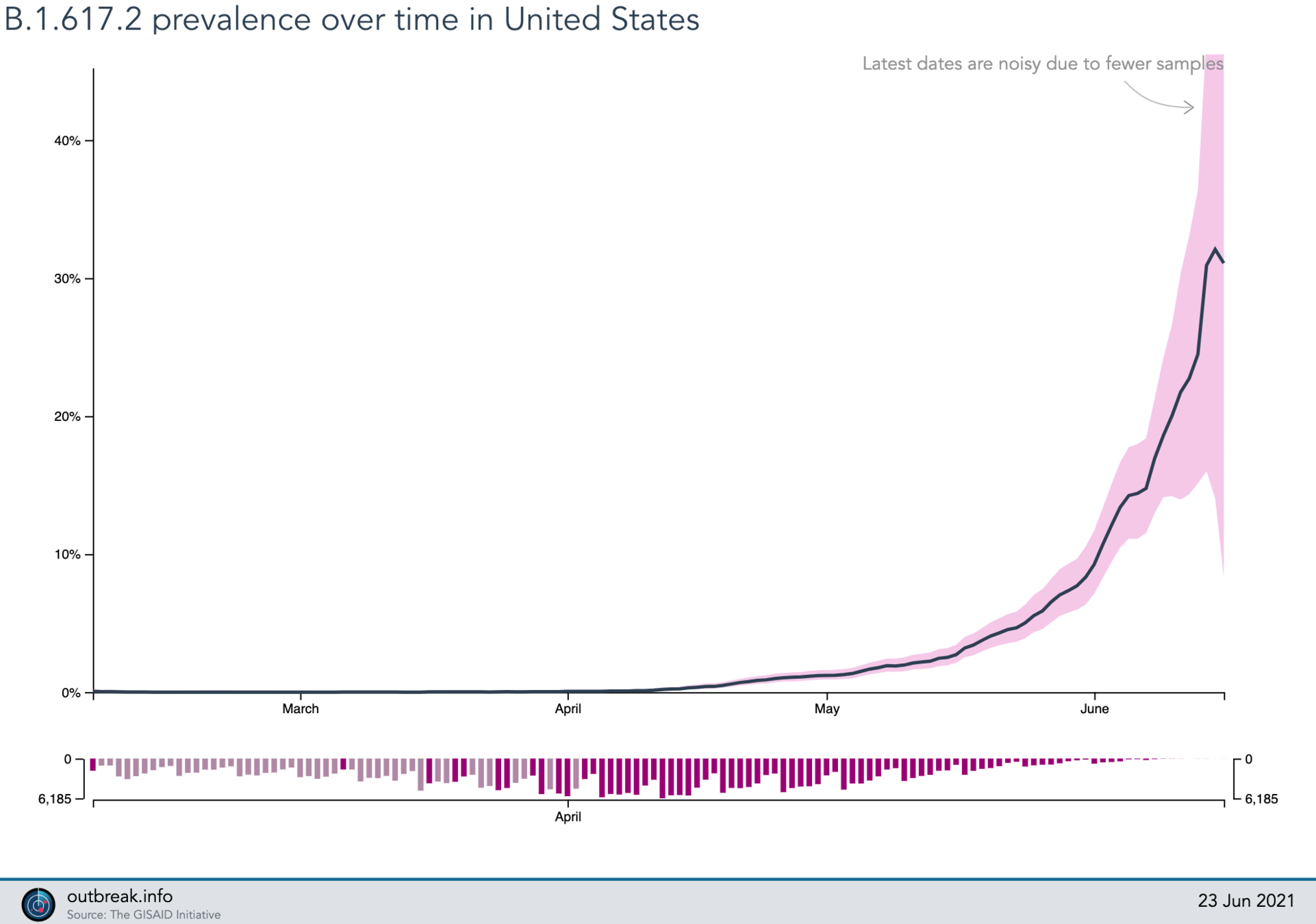
2021-06-23
6%
6%
6%
40%
P.1 (gamma)
B.1.617.2 (delta)
Julia L. Mullen, Ginger Tsueng, Alaa Abdel Latif, Manar Alkuzweny, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Kristian G. Andersen, Chunlei Wu, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology outbreak.info. Available online: https://outbreak.info/ (2020)

6%
2021-09-28

100%
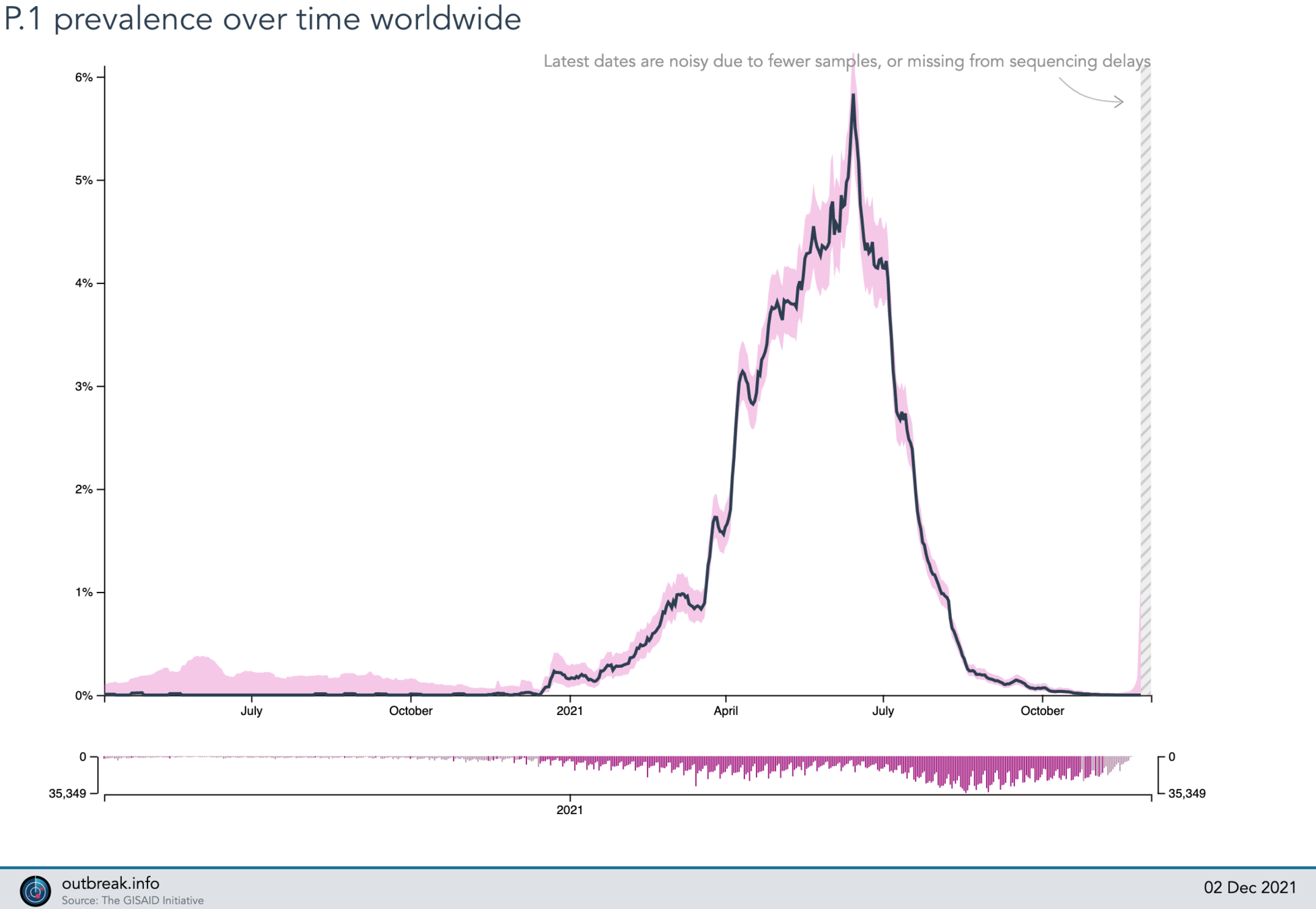
2021-12-02


VarSkip Coverage in NCBI
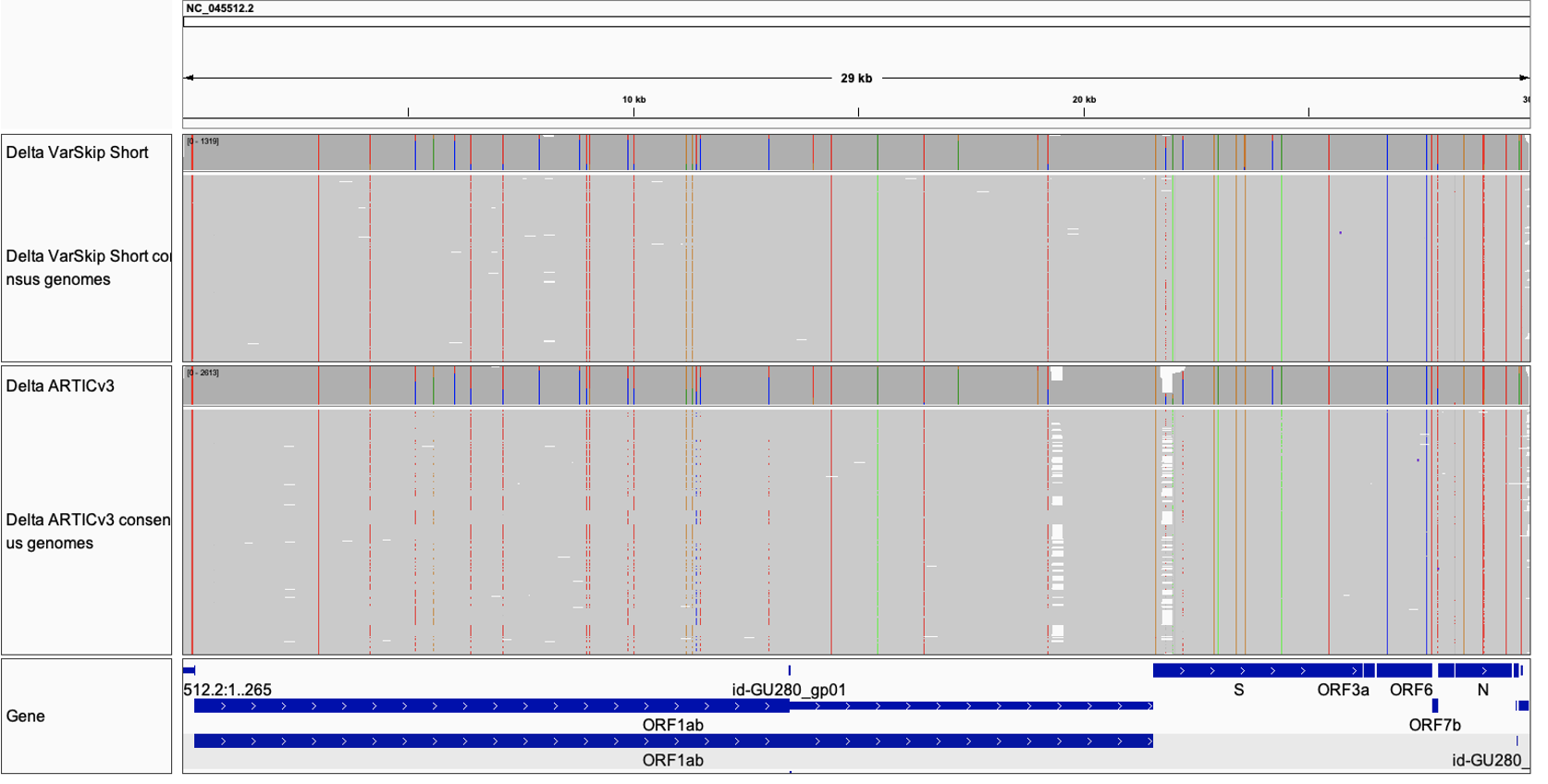
Consensus sequences classified as B.1.617.2 (Delta) in Massachusetts (where VarSkip Short was field tested) were collected from NCBI Virus and partitioned into 2613 ARTICv3 and 1319 VarSkip Short sequences using the SRA “design” field (NEB_VarSkip_v1 = VarSkip Short, others assumed to be ARTICv3). Reads were aligned to the NC_045512.2 reference using minimap2 (minimap2 -x asm5) and visualized using IGV. Consensus sequences generated from VarSkip amplicons suffered fewer dropouts (indicated by arrows).
NCBI virus link to data
VarSkip Presence in NCBI
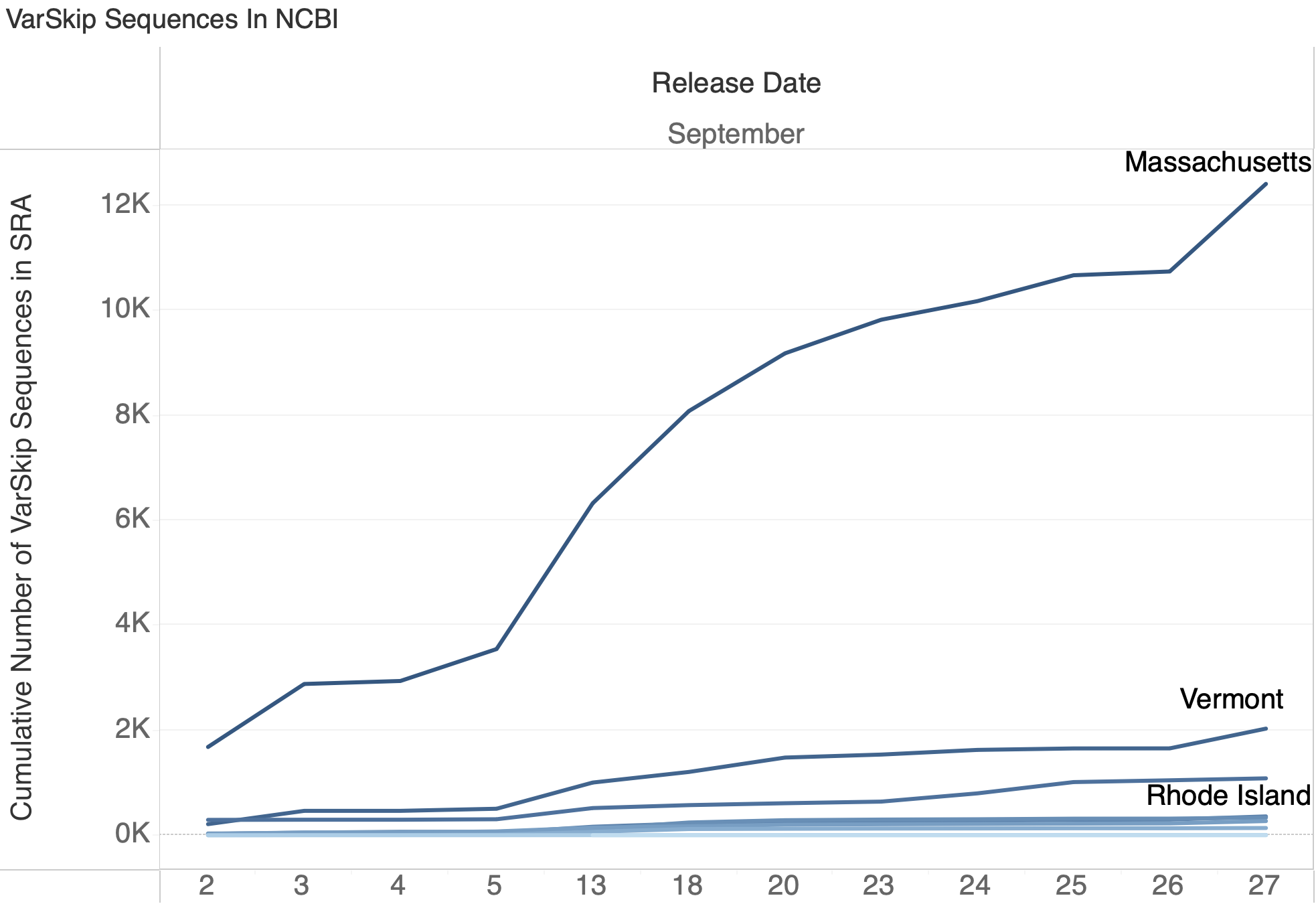
Varskip Durablity

Multiplex PCR Design
Mutations
Varskip Durablity

Multiplex PCR Design
Mutations

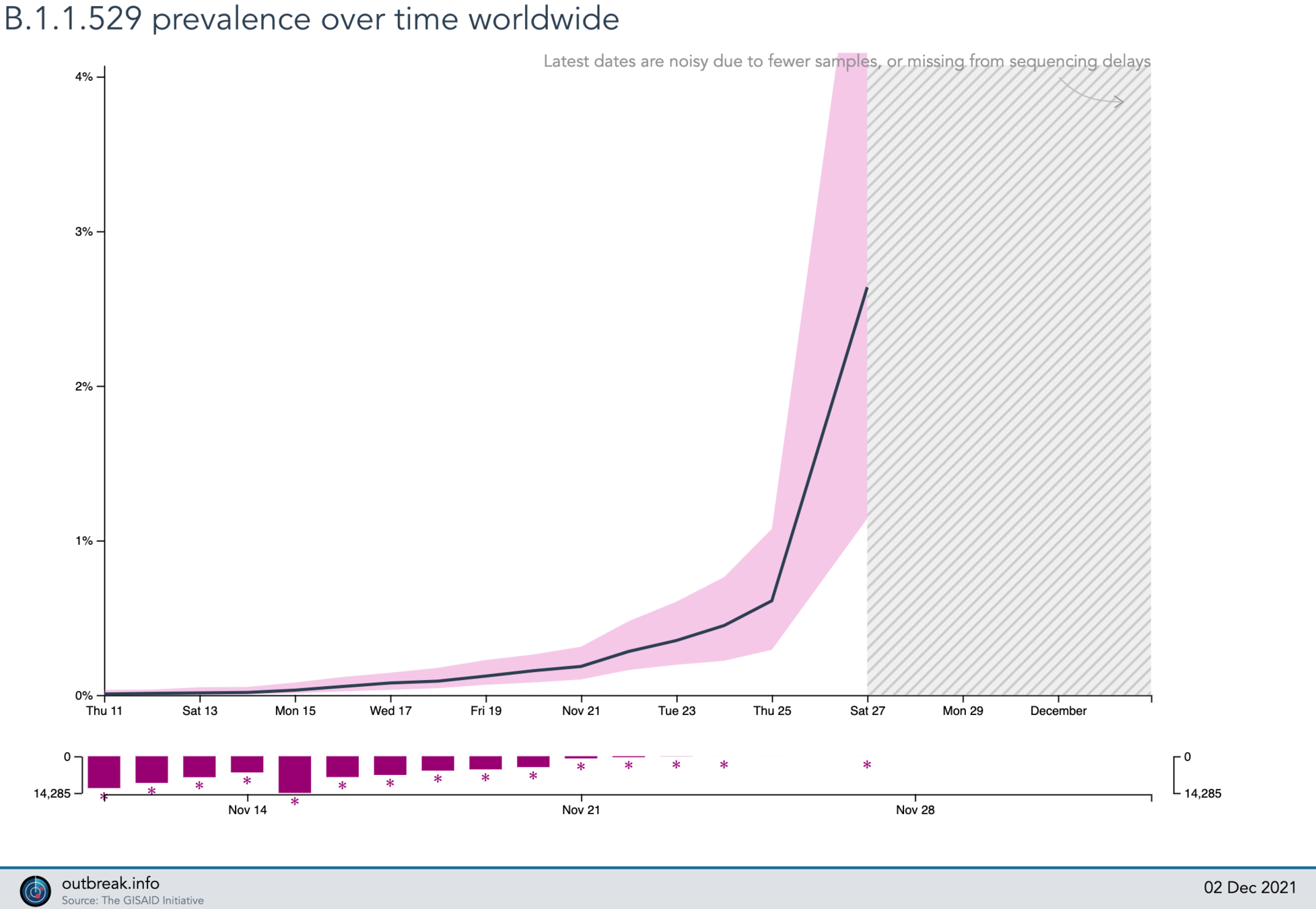
4%

?
Omicron
Lineage Variants


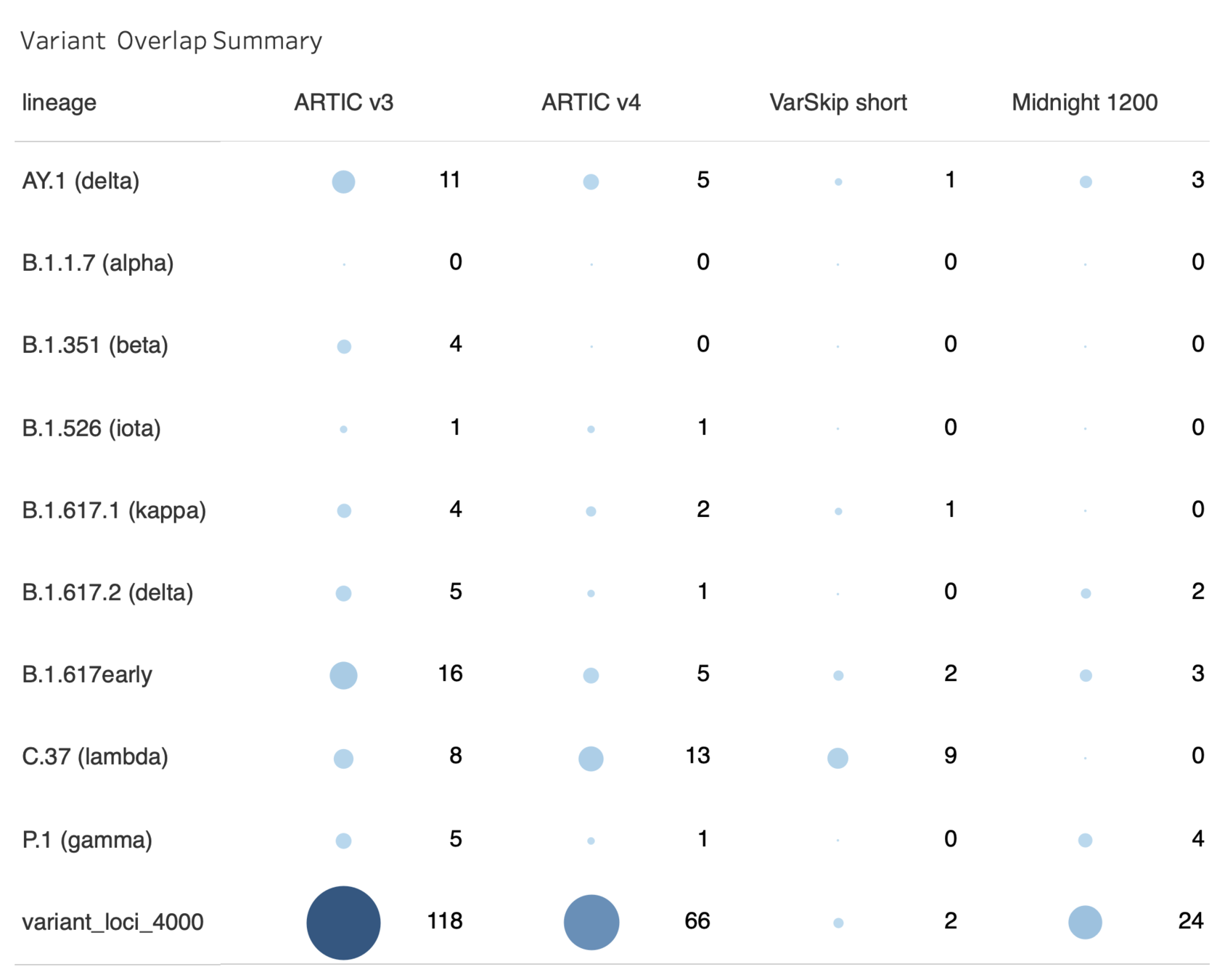
Summary of Primer Overlaps by Method
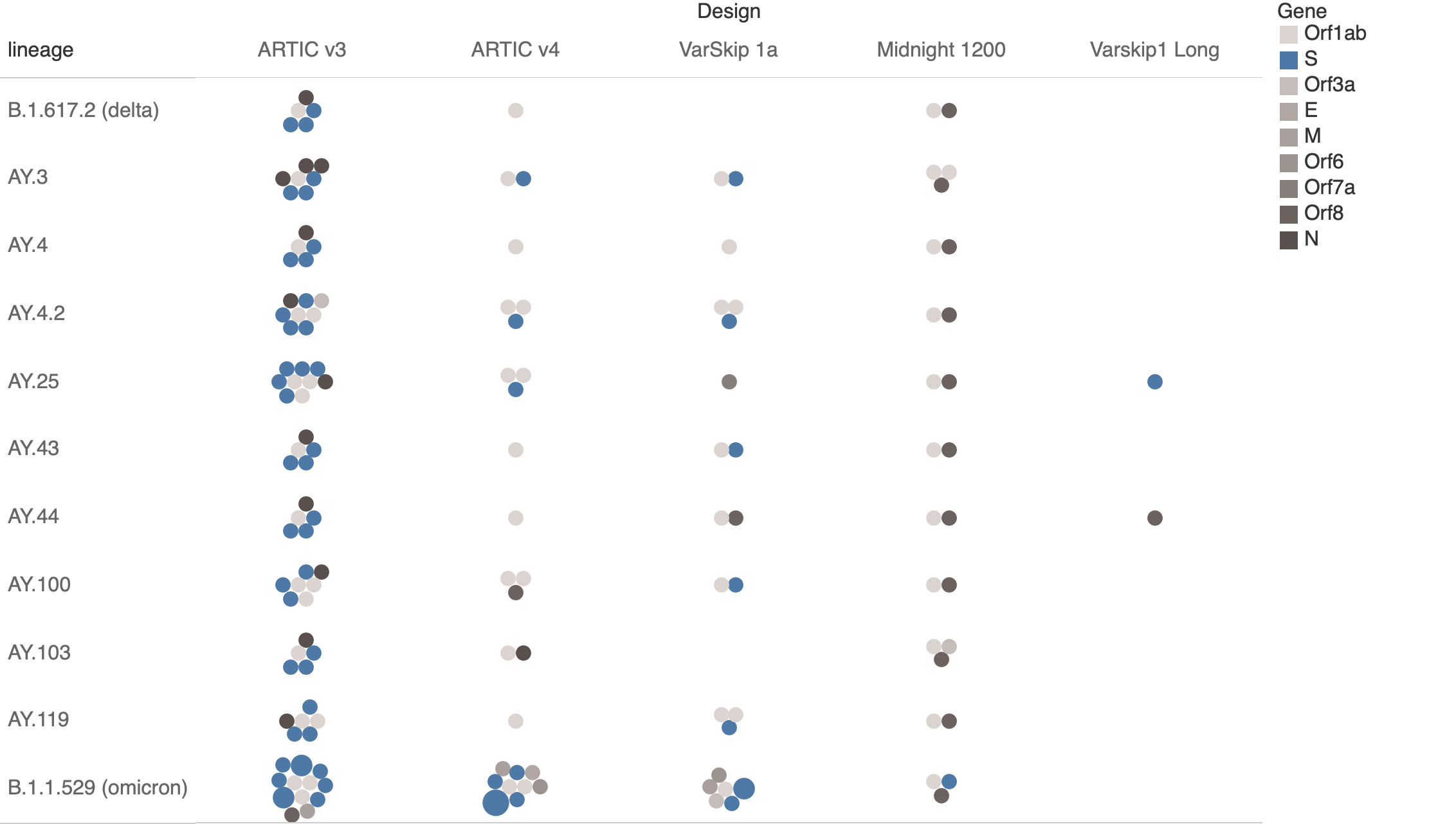
One dot per amplicon, Size=number of overlapping variants
Variant Induced Coverage Problems
"ARTIC" from South Africa by the Kwazulu-Natal Research Innovation and Sequencing Platform (KRISP) and the Centre for Epidemic Response and Innovation (CERI).
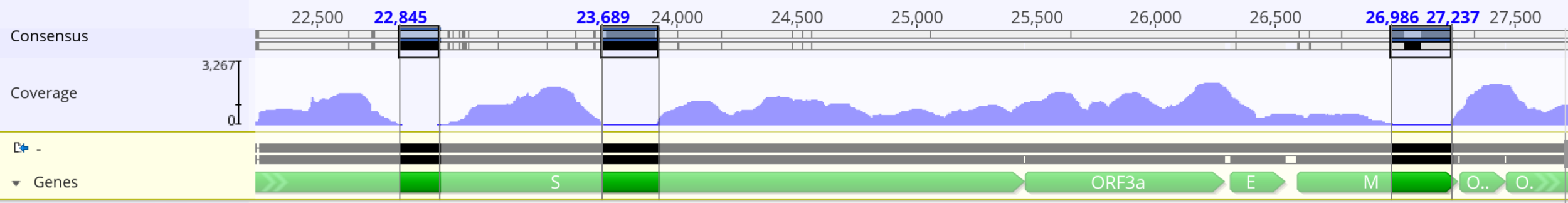
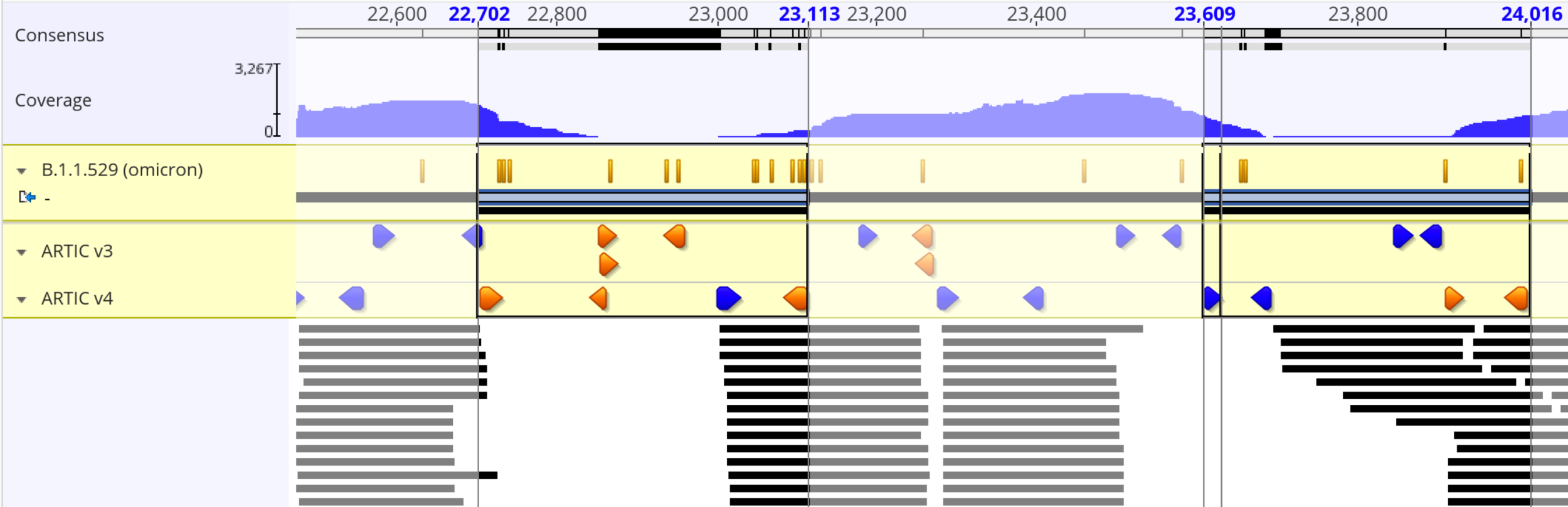
ARTICv4
Midnight



VarSkip
?
I have the first data as of 12PM EST...
We will reformulate as quickly as possible - meanwhile VarSkip Long is a potential one library solution.
When to Switch Primer Schemes?
| Wait |
|---|
| Lab procedure changes |
| Analysis update |
| Validation study |

| Switch |
|---|
| Coverage improvement |
| Better lab workflow |
| Turnaround time |
| Material availability |
Log scale coverage
VarSkip Clinical Results
Delta Lineage Saliva Sample By Primer Set
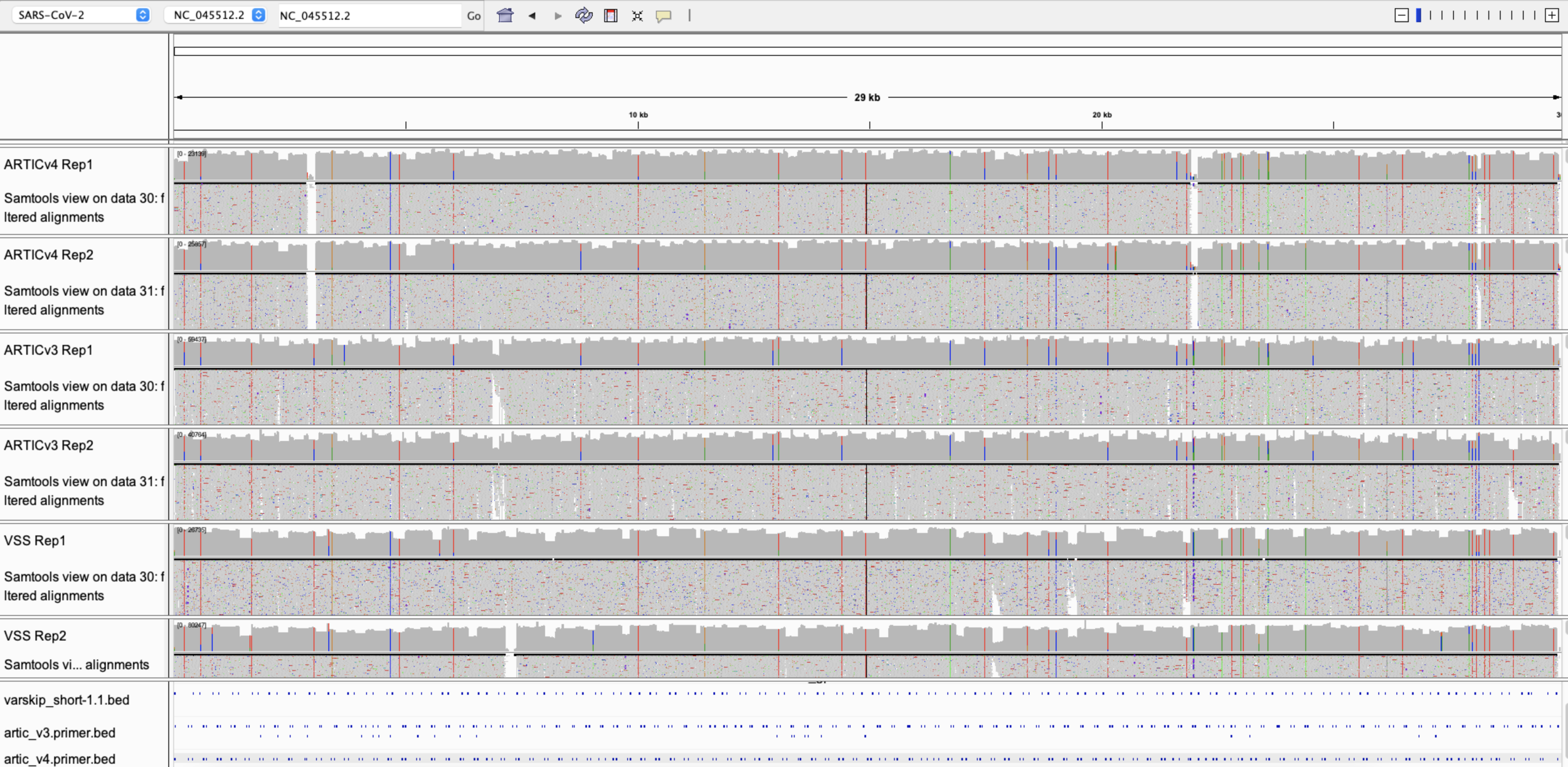
ARTICv3
ARTICv4


VarSkip
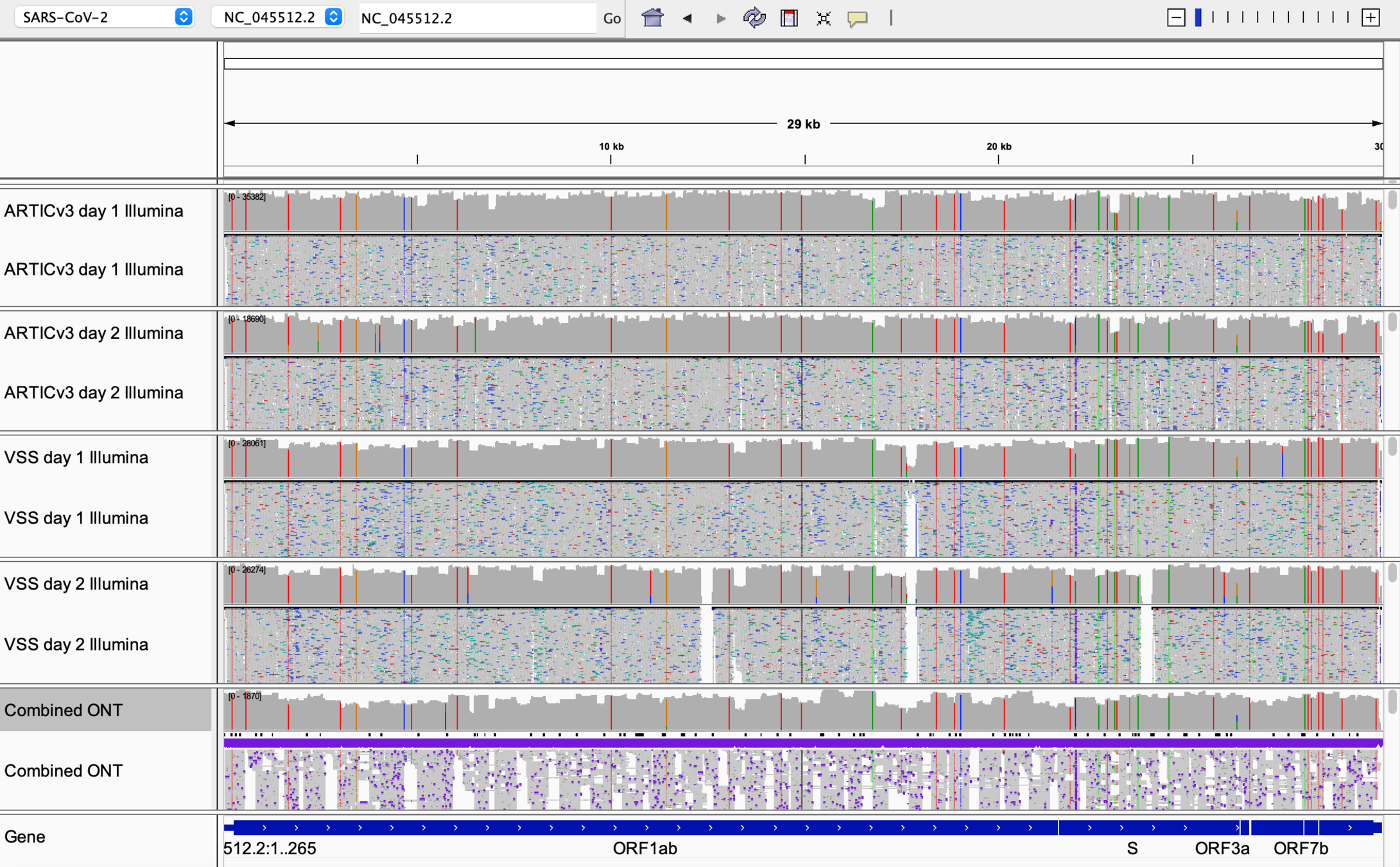
VarSkip Clinical Results
Various Lineages
VarSkip B.1.621
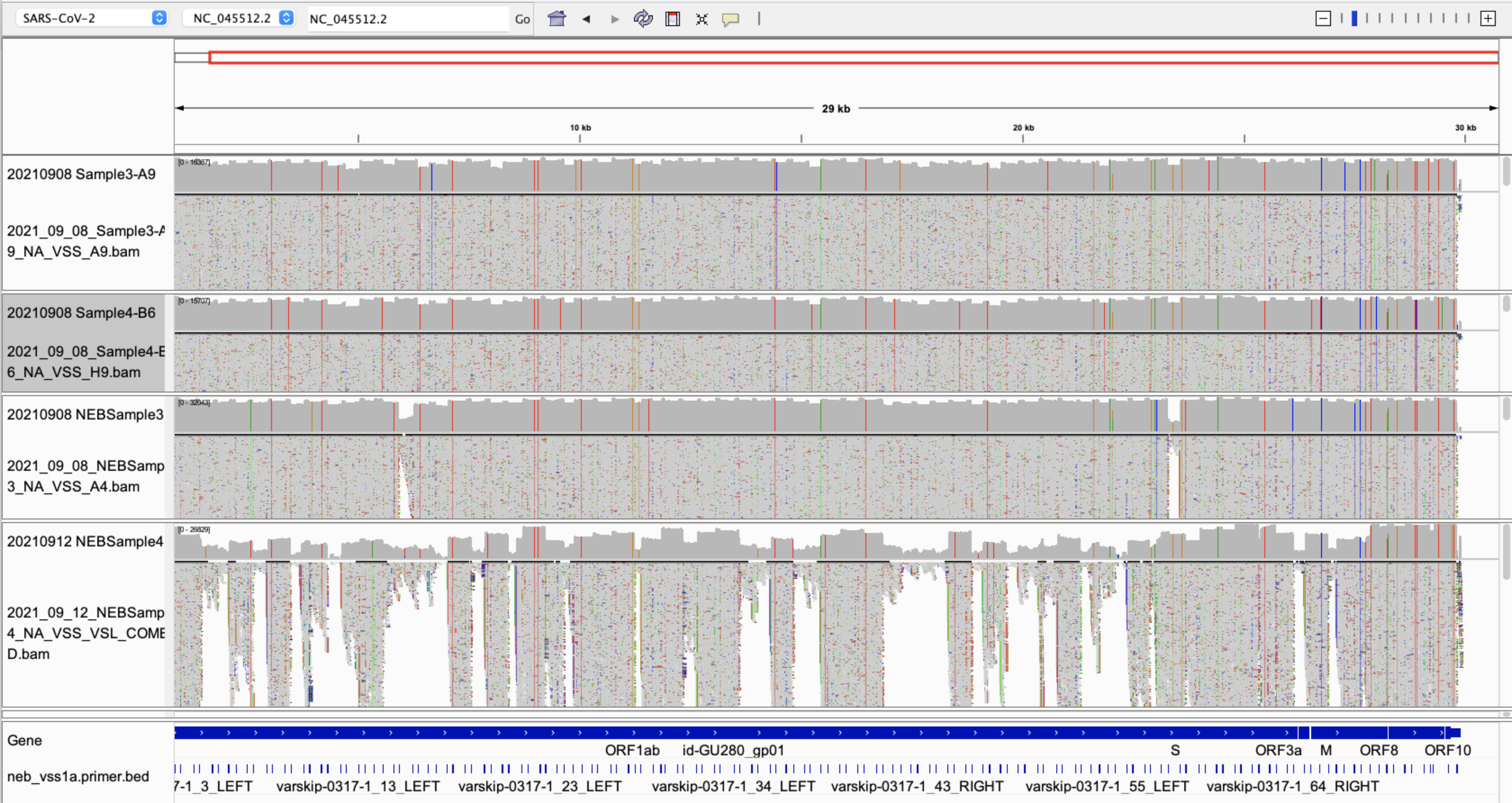
VarSkip Delta 1

VarSkip Delta 2

VarSkip Delta 3
Thanks!
Matt Campbell
NEB Leadership
Eileen Dimalanta
NEBNext Team
NEB Early Access testers
Lynne Apone, Kayli Pinet, Luo Sun
Nicole Nichols
Chris Mason
NEB LAMP, qPCR Teams
primer-monitor.neb.com
VarSkip Sequencing