Lab meeting
10-03-2020
Lab meeting
10-03-2020

29th - 2nd
3rd
4th - 6th
Training schools
- Early career Invesitigators
- Bioimage Analysts
Taggathon
Satellite
Meeting
Symposium


Centre Broca Nouvelle-Aquitaine
Notes from the conference:
Taggathon



Sattelite
meeting
Sattelite meeting
- Machine learning for Microscopy
- Bioimage Analysis Facility Management


- Uses the Tensorflow Java API
- Easily run pre-trained models on ImageJ
- A community platform for models as if they were plugins
- Macro recordable
- CPU and GPU compatible
- Good way to make your network accessible to biologists
GPU cluster at Pasteur
- Contact: Dmitry Ershov
https://research.pasteur.fr/en/member/dmitry-ershov/ - They use Slurm as workload manager.
https://slurm.schedmd.com/
Maybe it has better GPU support? - Discussion about training in loop to find the best network architechture/parameters
Ilastik
https://www.ilastik.org/

- Pixel classifier with friendly interface
- Easy input from users
-
Supports
-
Pixel classification
-
Object classification
-
Carving (segmentation in 3D)
-
Boundary based segmentation (Multicut)
-
Tracking
-
Now supports DL models
Pre-trained networks with fine-tuning from user input
- 3 facilities joined the discussion
Porto, Nantes and Zurich - 3 different approaches:
Free, Almost free, Expensive - Good discussion, it's important feedback for our Image Facility
Bioimage analysis Facility management

Symposium
QuPath
-
For histopathology data, huge images (60Gb) that are represented in different resolutions.
(google maps style)
-
Cell detection and feature measurement.
Has a classifier that can learn from user input annotation.
- From the public feedback, it’s widely used.
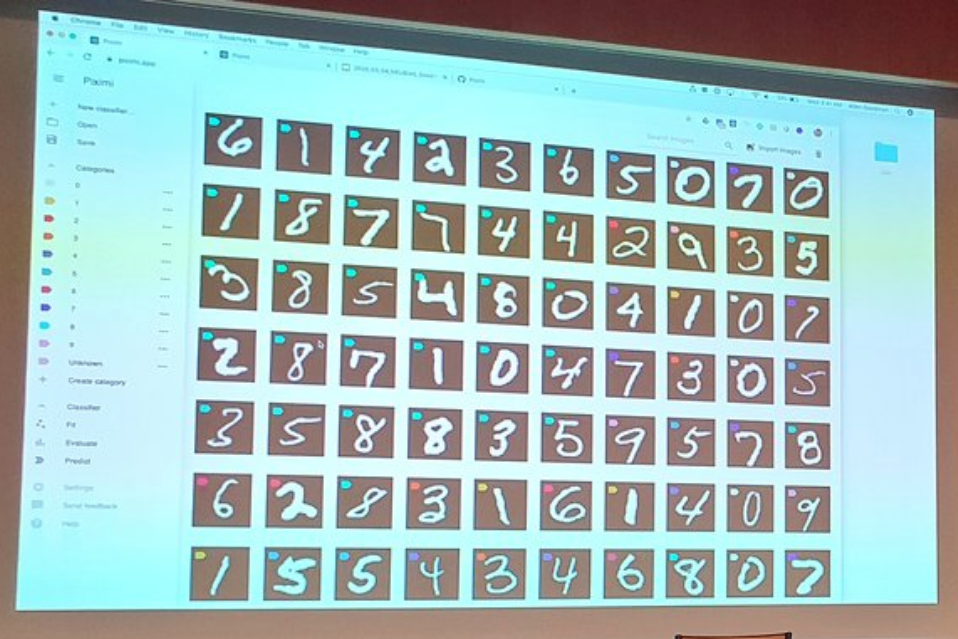
Piximi
-
Open source app for object recognition, from the Broad institute and Carpenter lab
-
It’s a deep learning cell classifier on a web application (piximi.app)
-
The idea is to replace CellProfiler
- Really nice interface


Panel discussion
F1000 Research
-
Immediate publication (after editorial review) and actual peer-review post-publication
-
There’s a clear label on the paper during this waiting peer review period
-
Lots of different categories to publish
-
-
Open peer review (reviewers always named)
-
Report of reviewers is accessible
-
-
Peer review for the technical aspect, not the impact of research (PLoS like)
-
All versions of the paper are accessible and citable
-
Interactive figures with Plotly or Shiny apps
-
Readers get notified if a new version of the paper they downloaded is available

| F1000 | Nature Methods | Nature Comm |
Scientific Reports | PLoS One |
PLoS Comp Bio |
|---|---|---|---|---|---|
| 2.64 | 23.03 | 11.80 | 4.12 | 2.87 | 4.38 |
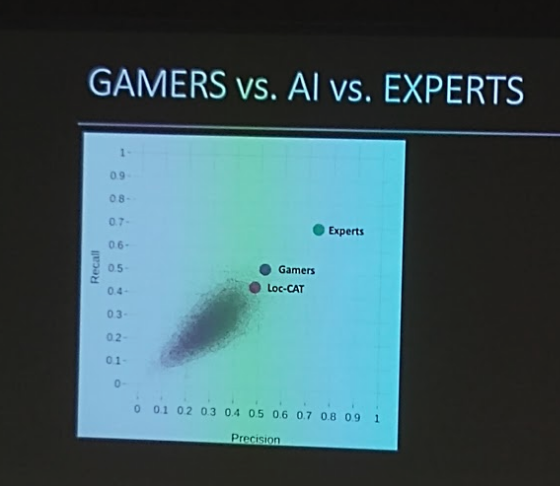
Emma Lundberg
One of the responsibles for The human protein atlas and also the organization of the Kaggle competition for cell classification, that's finished and published now.
How to get ground truth for millions of images?

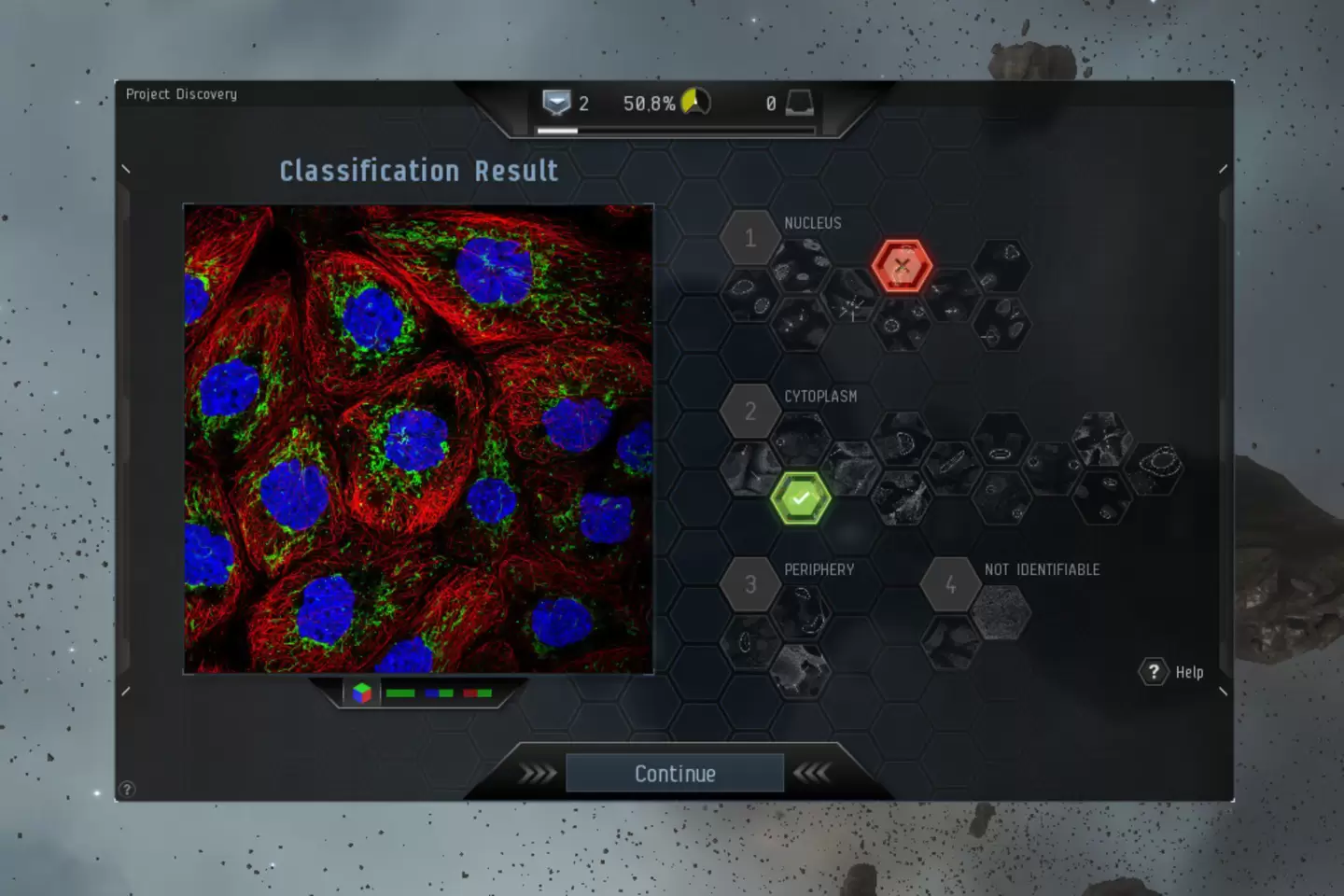

32 million classifications
70 working years
CLIJ - GPU support for Fiji
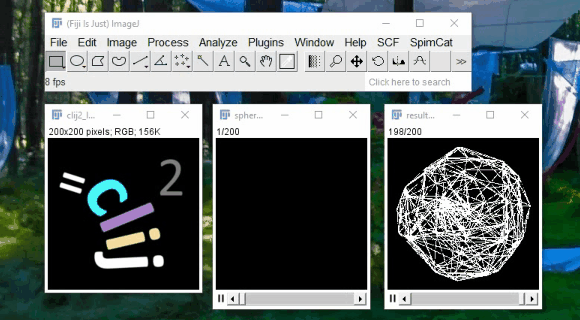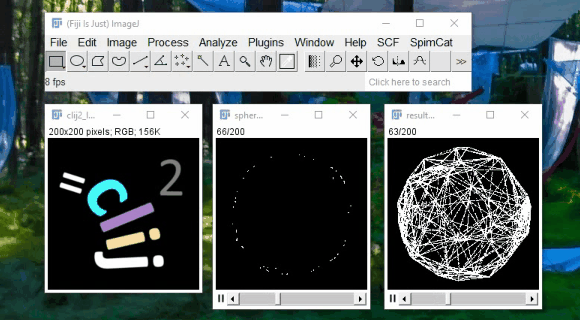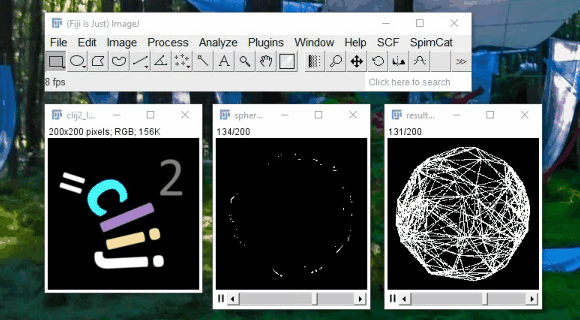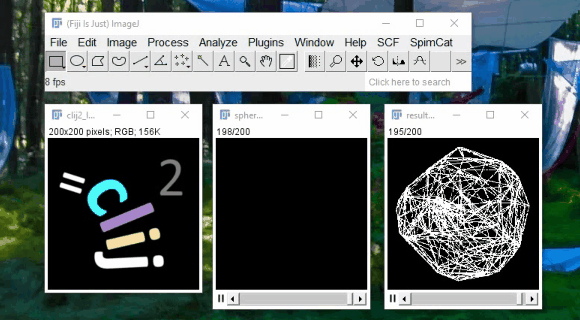
Spot detection example:
2h40 hours on CPU
11min on GPU
Kristin Branson
- Deep learning model for tracking flies in videos
- Automated system analysed 400 000 flies
- All the acquired data allowed to create another DL model that simulates the social behavior fo synthetic flies
Conclusions
- Everything is Deep learning now. More than 90% of the image analysis conference was about DL.
- People talked a lot about a "Model Zoo". A single place where DL trained models can be downloaded. Tensor flow is, without doubt, the standard.
-
image.sc is building strength, lots of developers are talking about it.
- Artificial Labeling is getting mainstream, especially with the industry software (Nikon, Zeiss, etc)
thanks!