MaxIV & QIM workshop

Felipe Delestro
Senior Research Software Specialist @ QIM Center
Danmarks Tekniske Universitet (DTU)
fima@dtu.dk



QIM Center
Center for Quantification of Imaging Data from MAX IV





supported by

Anders Bjorholm Dahl
Head of the QIM Center

Jon
Sporring
Deputy Head of the QIM Center

Stephen
Hall
Professor at Lund university

Rebecca
Engberg
Center Manager















What can be achieved in collaboration with the QIM center?
Data acquisition at MaxIV
Start jupyter server
Create project space
Set up permissions
Upload data to DTU servers
Convert data to OME-Zarr
Mount data locally
Data visualization
Export ROI
Load data
Run analysis
Data acquisition at MaxIV
Start jupyter server
Create project space
Set up permissions
Upload data to DTU servers
Convert data to OME-Zarr
Mount data locally
Data visualization
Export ROI
Load data
Run analysis


Projects at the QIM center
- A project is a space in our servers with attached metadata.
- Users can store their raw data and results from analysis.
- Access to the directory is restricted to users who are part of the project.
- Some management aspects are still done by contacting us.

- A centralized space for image analysis at the DTU Computing Center (DCC)
- Easier access to High Performance Computing (HPC) resources
- Projects and resources management
- https://platform.qim.dk/
SFTP data transfer

Any SFTP client can be used, but FileZilla is a good and reliable option
transfer.gbar.dtu.dk
port 22
Globus and large data transfer

open-source Python library, that focuses on 3D volumetric data

Getting started is easy:
pip install qim3d


Documentation at platform.qim.dk/qim3d/
Data handling

Volumetric data can be stored in a variety of file formats.
This can make the very first step of importing the data more complex than it should be
load and save
Data handling

Data handling

Volumetric data can be stored in a variety of file formats.
This can make the very first step of importing the data more complex than it should be
load and save
qim3d.io.load and qim3d.io.save can handle the most common data formats:
-
Tiff(including file stacks) HDF5-
TXRM/TXM/XRM NIfTI-
PIL(including file stacks) -
VOL/VGI DICOM
import qim3d
vol = qim3d.io.load("path/to/image.tif", virtual_stack=True)Data handling

We make a distinction between load/save and import/export when the operation includes transformations to the data.
import and export
Currently we're implementing a 3D import/export following the OME-Zarr standard, which
uses chunked data with a
ulti-scales scheme.

Visualization

Visualization

Easily exploring volumetric data may not be so straightforward. The library provides helper functions this task simple and easy, even within Jupyter Notebooks
import qim3d
vol = qim3d.examples.bone_128x128x128
qim3d.viz.slicer(vol)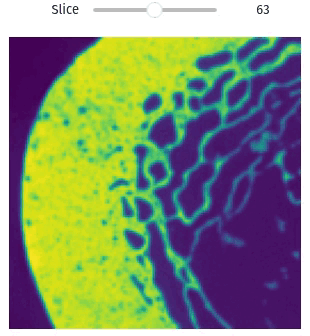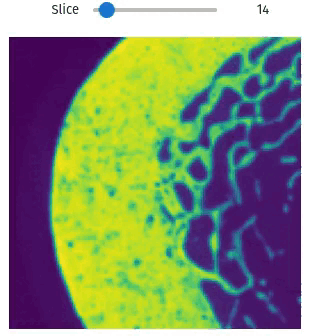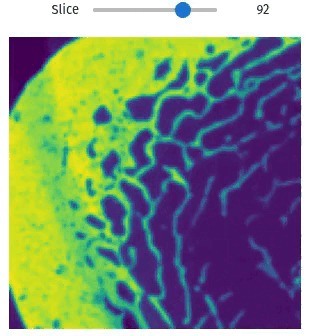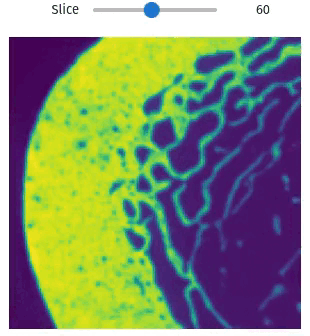
Visualization

import qim3d
vol = qim3d.examples.shell_225x128x128
qim3d.viz.slices_grid(vol, num_slices=15)
Visualization

import qim3d
# Generate synthetic collection of objects
num_objects = 15
vol, labels = qim3d.generate.volume_collection(num_objects = num_objects)# Visualize synthetic collection
qim3d.viz.volumetric(vol)Visualization

import qim3d
vol = qim3d.generate.volume(base_shape=(128,128,128),
final_shape=(128,128,128),
noise_scale=0.03,
order=1,
gamma=1,
max_value=255,
threshold=0.5,
dtype='uint8'
)
mesh = qim3d.mesh.from_volume(vol, step_size=3)
qim3d.viz.mesh(mesh.vertices, mesh.faces)Visualization

import qim3d
vol = qim3d.examples.cement_128x128x128
binary = qim3d.filters.gaussian(vol, sigma = 2) < 60
labeled_volume, num_labels = qim3d.segmentation.watershed(binary)
color_map = qim3d.viz.colormaps.segmentation(num_labels, style = 'bright')
qim3d.viz.slicer(labeled_volume, slice_axis = 1, color_map=color_map)qim3d.viz.colormaps.segmentation creates an colormap designes specifically for segmentation tasks
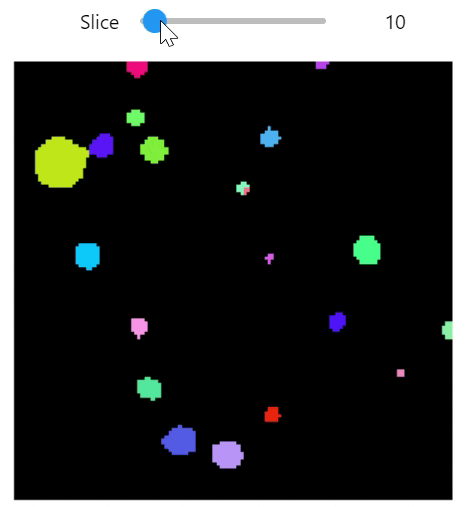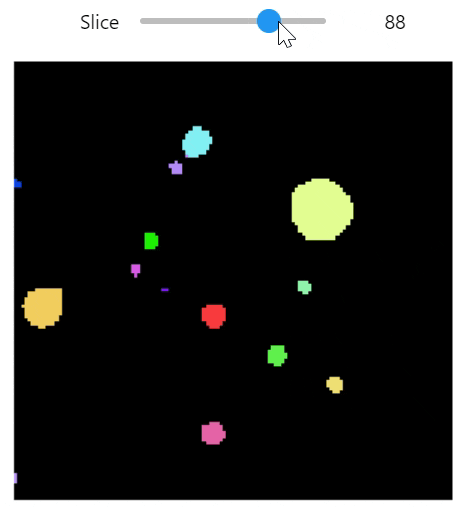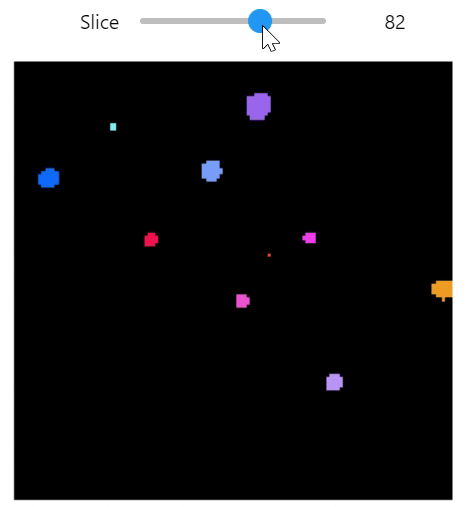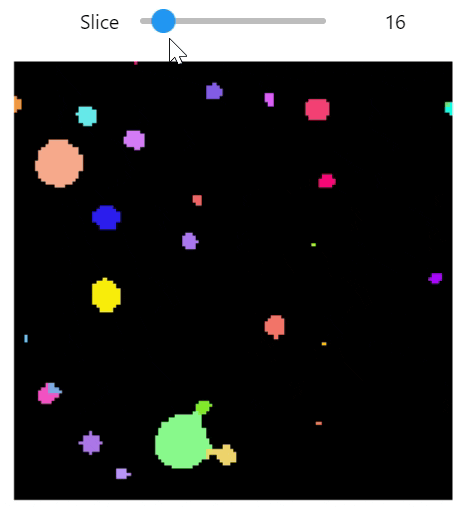
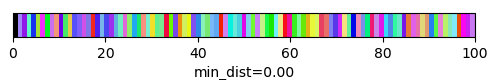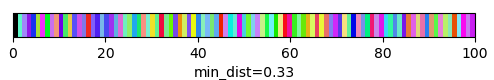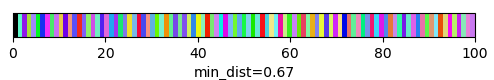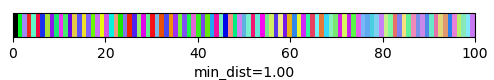
The parameter min_dist allows us to control how different neighbour colors are
Image analysis

Image analysis

qim3d covers from basic functionalities to more complex methodologies.
this will allows the library to be a central point for building volumetric data pipelines.
Image analysis

qim3d.filters contains implementations for common filter operations, including the option to run them on chunked data using Dask
qim3d.filters.Pipeline allows the creation of classes that act as filter pipelines
import qim3d
from qim3d.filters import Pipeline, Median, Gaussian, Maximum, Minimum
# Get data
vol = qim3d.examples.fly_150x256x256
# Show original
fig1 = qim3d.viz.slices_grid(vol, num_slices=5, display_figure=True)
# Create filter pipeline
pipeline = Pipeline(
Median(size=5),
Gaussian(sigma=3, dask = True)
)
# Append a third filter to the pipeline
pipeline.append(Maximum(size=3))
# Apply filter pipeline
vol_filtered = pipeline(vol)
# Show filtered
fig2 = qim3d.viz.slices_grid(vol_filtered, num_slices=5, display_figure=True)
Image analysis

The modules qim3d.detection and qim3d.segmentation contain wrappers for common methods that are useful when building larger pipelines
import qim3d
# Get data
vol = qim3d.examples.cement_128x128x128
vol_blurred = qim3d.filters.gaussian(vol, sigma=2)
# Detect blobs, and get binary_volume
blobs, binary_volume = qim3d.detection.blobs(
vol_blurred,
min_sigma=1,
max_sigma=8,
threshold=0.001,
overlap=0.1,
background="bright"
)
# Visualize detected blobs
qim3d.viz.circles(blobs, vol, alpha=0.8, color='blue')
Image analysis

The modules qim3d.detection and qim3d.segmentation contain wrappers for common methods that are useful when building larger pipelines
import qim3d
vol = qim3d.examples.cement_128x128x128
bin_vol = qim3d.filters.gaussian(vol, sigma = 2)<60
fig1 = qim3d.viz.slices_grid(bin_vol, slice_axis=1, display_figure=True)labeled_volume, num_labels = qim3d.segmentation.watershed(bin_vol)
cmap = qim3d.viz.colormaps.segmentation(num_labels)
fig2 = qim3d.viz.slices_grid(labeled_volume, slice_axis=1, color_map=cmap, display_figure=True)

Image analysis

qim3d.processing is meant for more complex methods, that perform quantification tasks that are specific but common for volumetric data
The current implementations are:
- qim3d.processing.structure_tensor
- qim3d.processing.local_thickness
Image analysis

Structure tensor
import qim3d
vol = qim3d.examples.NT_128x128x128
val, vec = qim3d.processing.structure_tensor(vol, visualize=True, axis=2)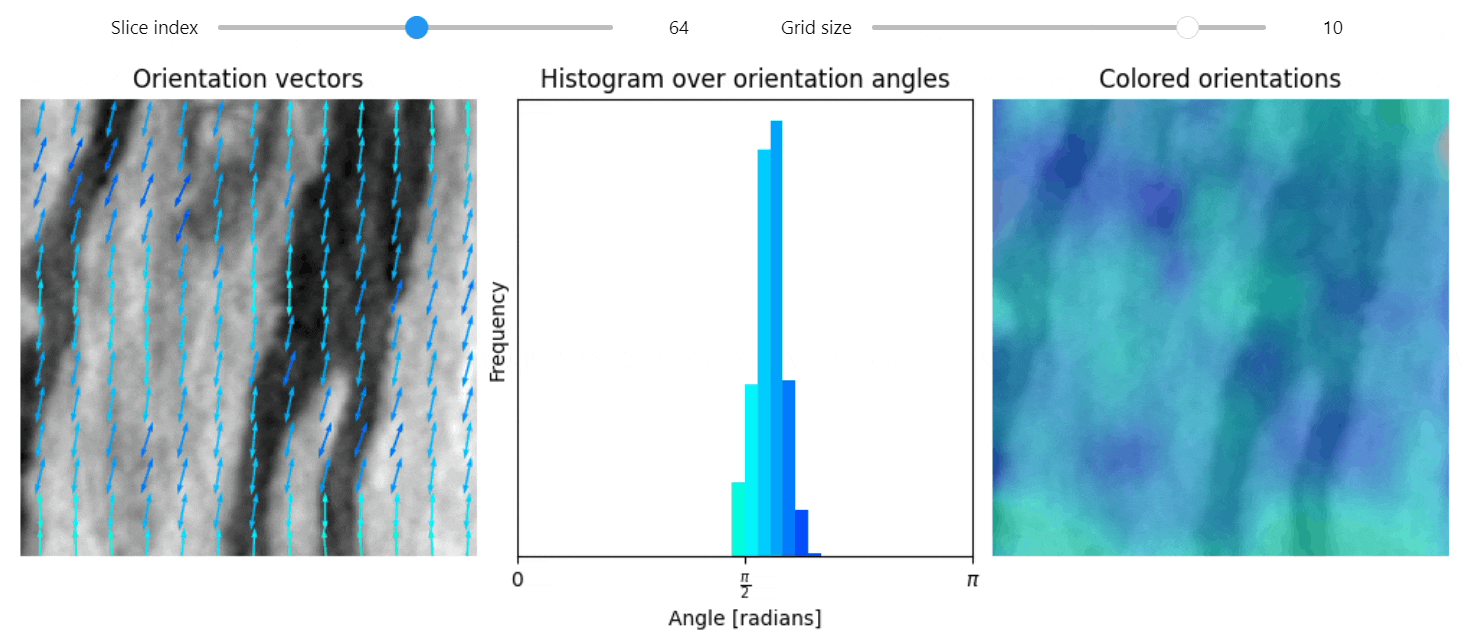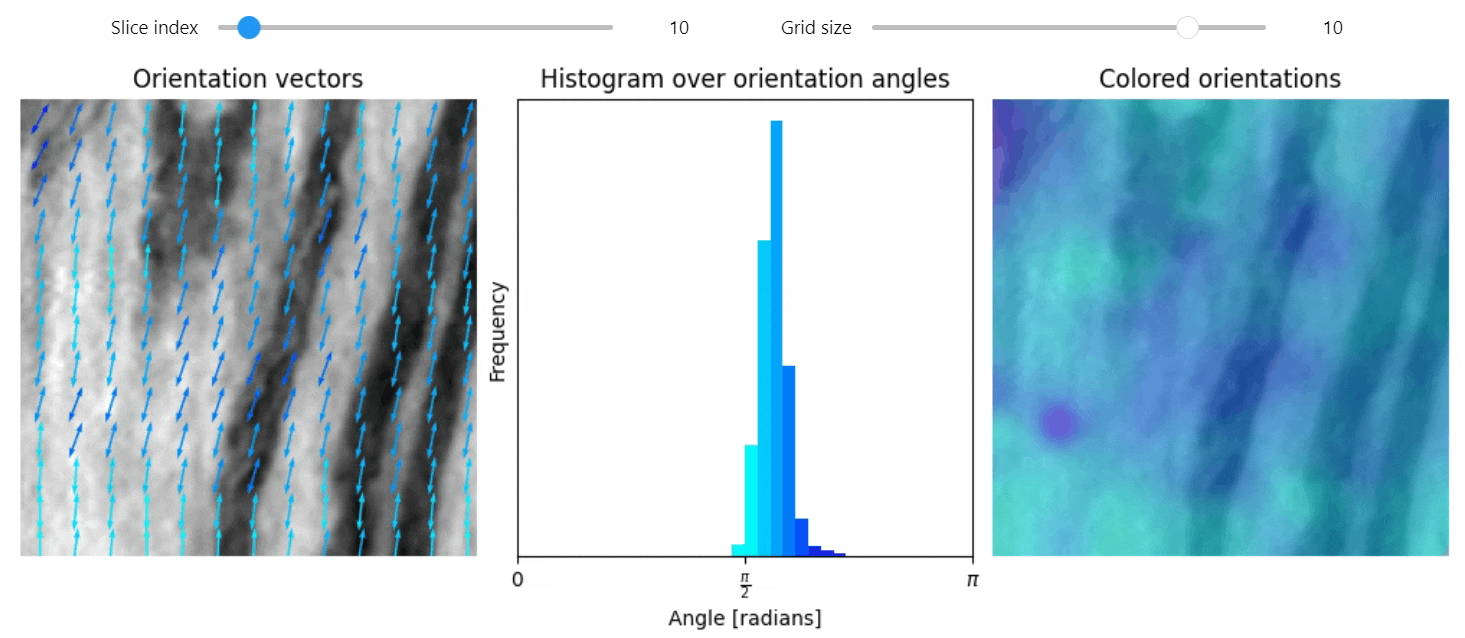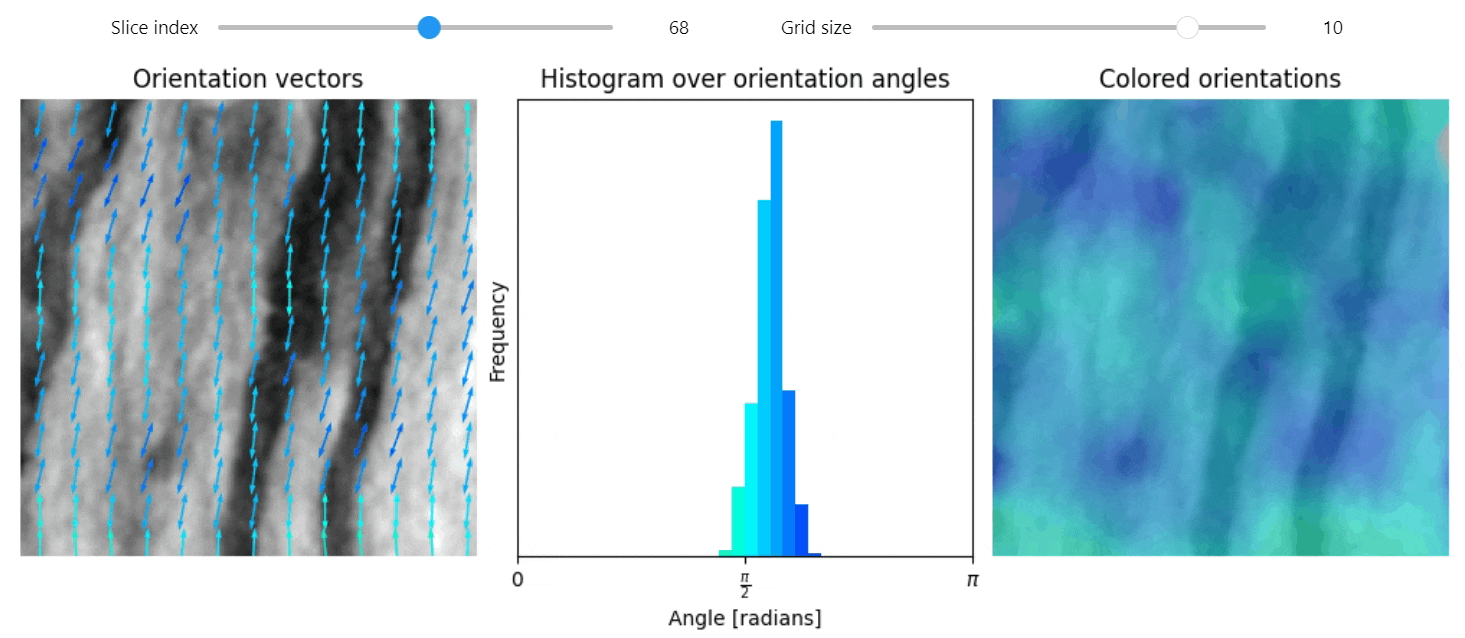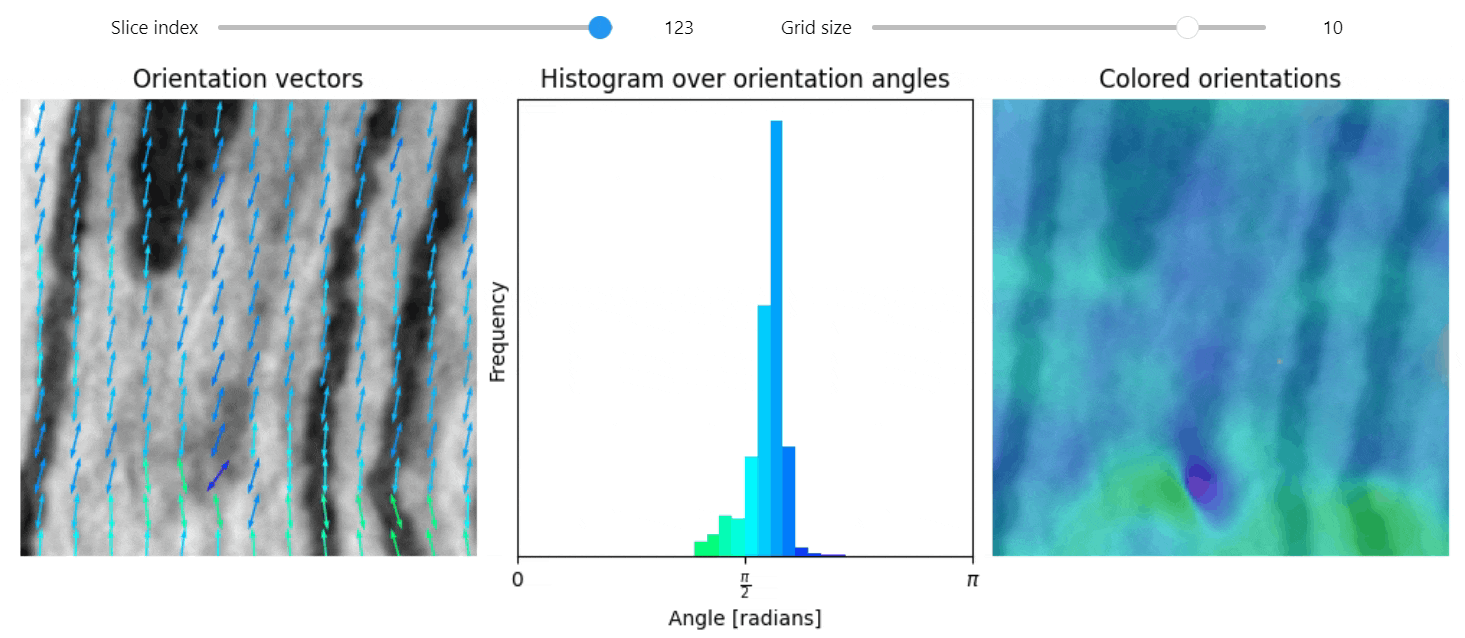
Image analysis

Local thickness
import qim3d
vol = qim3d.examples.fly_150x256x256
lt_vol = qim3d.processing.local_thickness(vol, visualize=True, axis=0)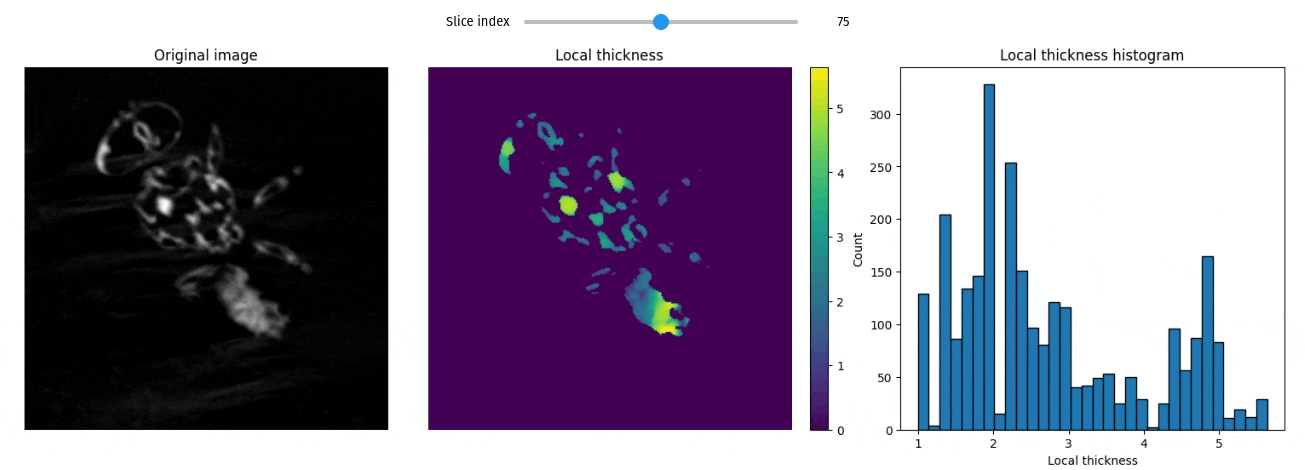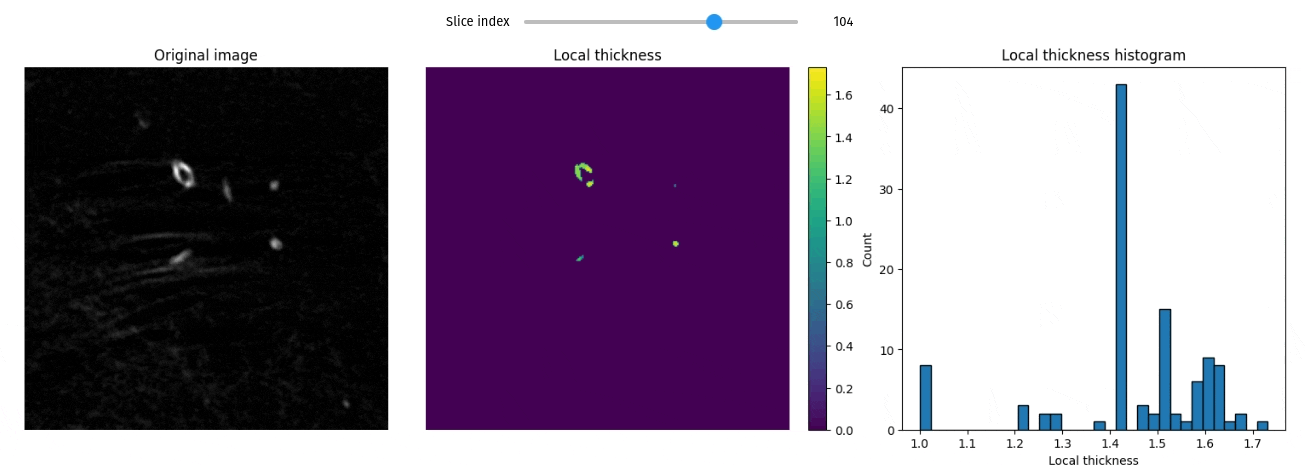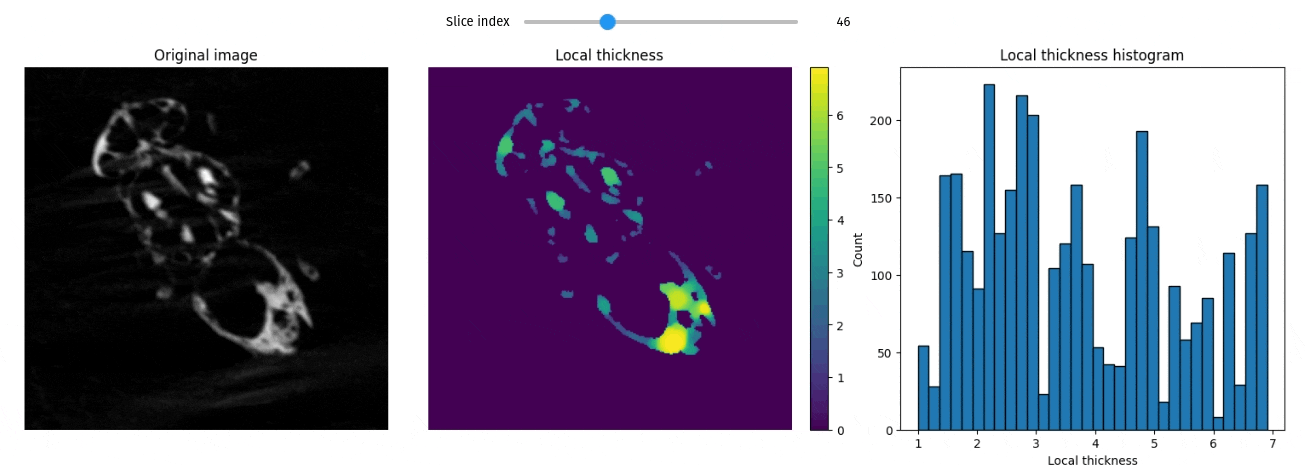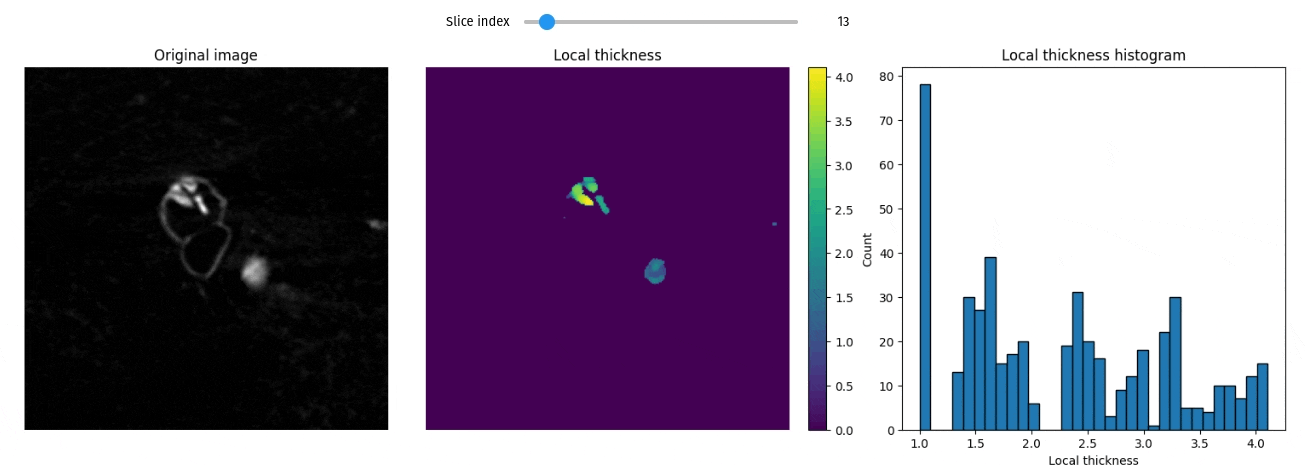
The library is an open-source project, and you're all welcome to contribute!

documentation and
installation instructions:
☕
🧑💻 Practical session
- Upload a file to your home directory at DTU
https://platform.qim.dk/qim-public/examples/bone.tif
- Launch a Jupyter server using the QIM platform
- Follow along with some examples on how to use qim3d
Final thoughts
- The platform and the qim3d library are constantly being developed, so your feedback is essential!
- Always feel free to reach out with any questions
- Felipe Delestro ⇨
fima@dtu.dk - Rebecca Engberg ⇨
reen@dtu.dk
- Felipe Delestro ⇨