Bayesian model reduction for nonlinear regression
Dimitrije Marković
DySCO meeting 18.01.2023

Outline
- Bayesian deep learning
- Structured shrinkage priors
- Bayesian model reduction
- Regression
- Non-linear regression
Outline
- Bayesian deep learning
- Structured shrinkage priors
- Bayesian model reduction
- Regression
- Non-linear regression
Deep learning
Optimization
Bayesian deep learning
Inference
Advantages
- More robust, accurate and calibrated predictions
- Learning from small datasets
- Continuous learning (inference)
- Distributed or federated learning (inference)
- Marginalization
Few references
Murphy, Kevin P. Probabilistic machine learning: an introduction. MIT press, 2022.
Wilson, Andrew Gordon. "The case for Bayesian deep learning." arXiv preprint arXiv:2001.10995 (2020).
Bui, Thang D., et al. "Partitioned variational inference: A unified framework encompassing federated and continual learning." arXiv preprint arXiv:1811.11206 (2018).
Murphy, Kevin P. Probabilistic machine learning: Advanced Topics. MIT Press 2023
Outline
- Bayesian deep learning
- Structured shrinkage priors
- Bayesian model reduction
- Regression
- Non-linear regression
Deep learning
Optimization
Structured shrinkage priors
Nalisnick, Eric, José Miguel Hernández-Lobato, and Padhraic Smyth. "Dropout as a structured shrinkage prior." International Conference on Machine Learning. PMLR, 2019.
Ghosh, Soumya, Jiayu Yao, and Finale Doshi-Velez. "Structured variational learning of Bayesian neural networks with horseshoe priors." International Conference on Machine Learning. PMLR, 2018.
Dropout as a spike-and-slab prior
Better shrinkage priors
Regularized horseshoe prior
Piironen, Juho, and Aki Vehtari. "Sparsity information and regularization in the horseshoe and other shrinkage priors." Electronic Journal of Statistics 11.2 (2017): 5018-5051.
Outline
- Bayesian deep learning
- Structured shrinkage priors
- Bayesian model reduction
- Regression
- Non-linear regression
Factorization
Approximate posterior
Hierarchical model
Non-centered parameterization
Approximate posterior
Hierarchical model
Variational free energy
Stochastic variational inference
Stochastic gradient
Bayesian model reduction
Two generative processes for the data
flat model
extended model
Friston, Karl, Thomas Parr, and Peter Zeidman. "Bayesian model reduction." arXiv preprint arXiv:1805.07092 (2018).
Bayesian model reduction
flat model
extended model
BMR algorithm
Step 1
BMR algorithm
Step 2
New epoch
\( p(\pmb{z}_i) \propto \exp\left[ \int d \pmb{z}_{i+1} p_{i|i+1}q_{i+1} \right] \)
step 1
\(\vdots\)
step 2
Outline
- Bayesian deep learning
- Structured shrinkage priors
- Bayesian model reduction
- Regression
- Non-linear regression
Regression
Linear (D=(1,100), N=100)
Logistic (D=(1,100), N=200)
Multinomial (D=(10,10), N=400)
Generative model
Regression comparison
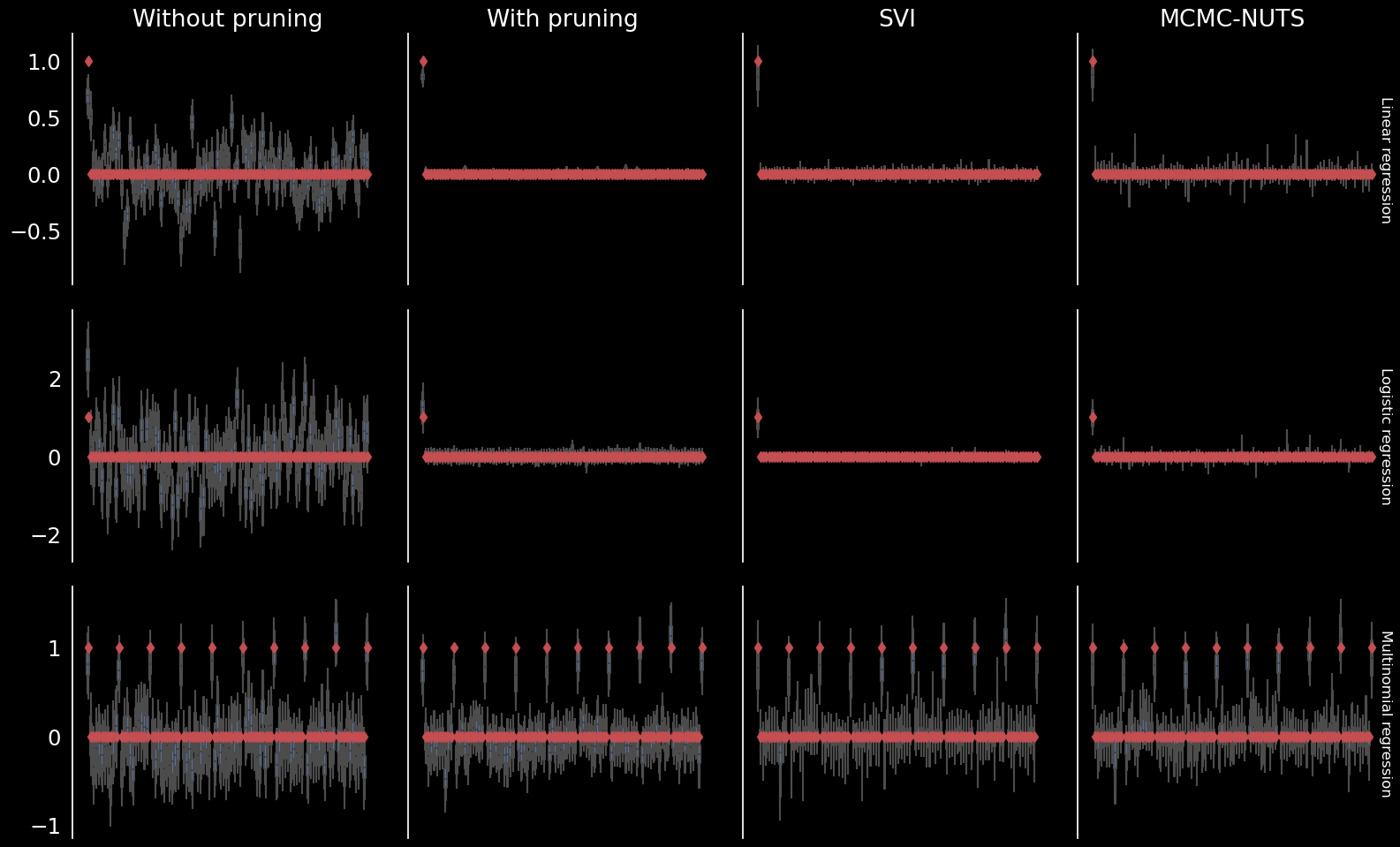
Outline
- Bayesian deep learning
- Structured shrinkage priors
- Bayesian model reduction
- Regression
- Non-linear regression
Simulated data
Normal likelihood
Bernoulli likelihood
Categorical likelihood
Neural network model
Normal and Bernoulli likelihoods
Categorical likelihood
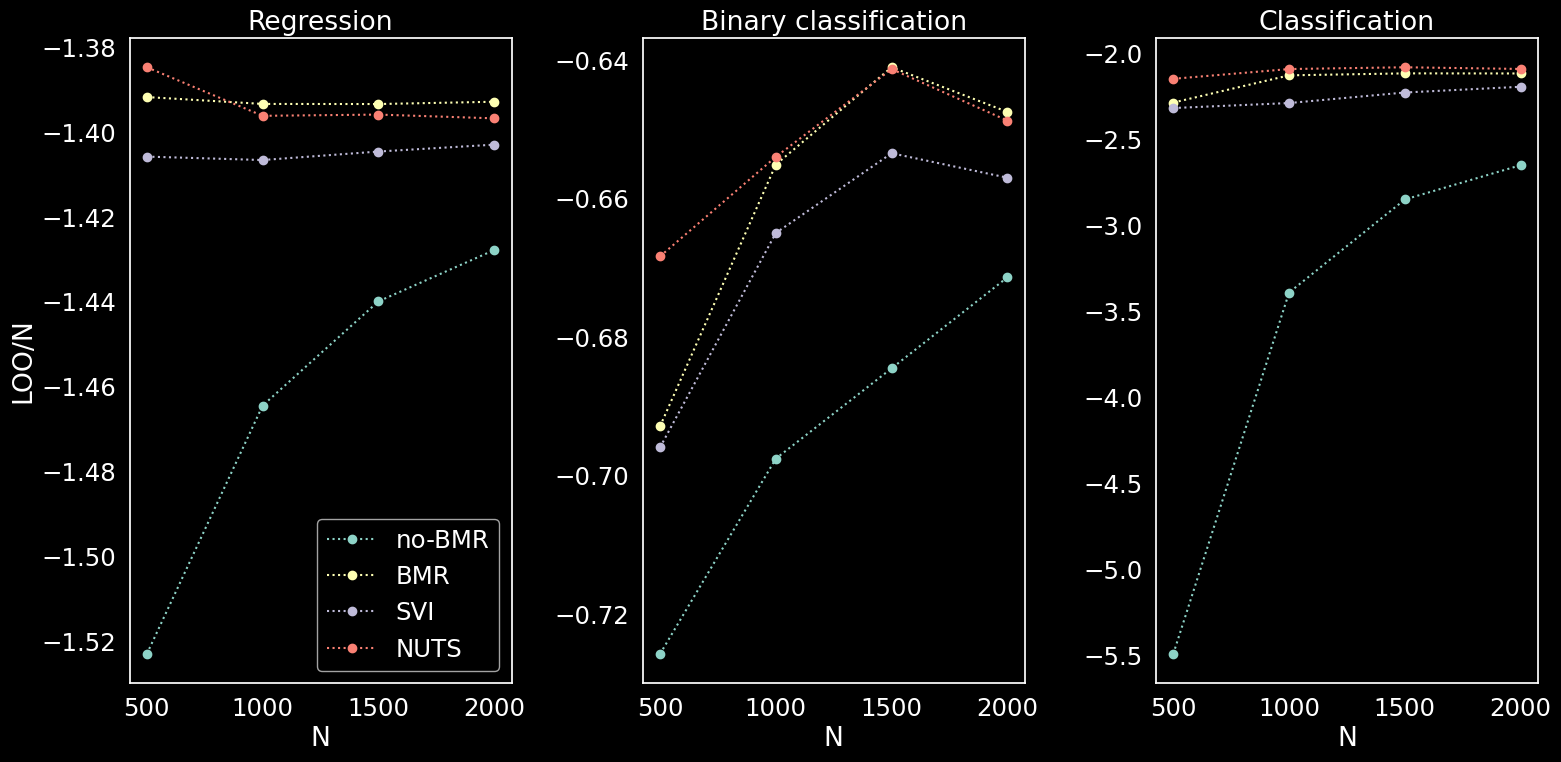
Comparison
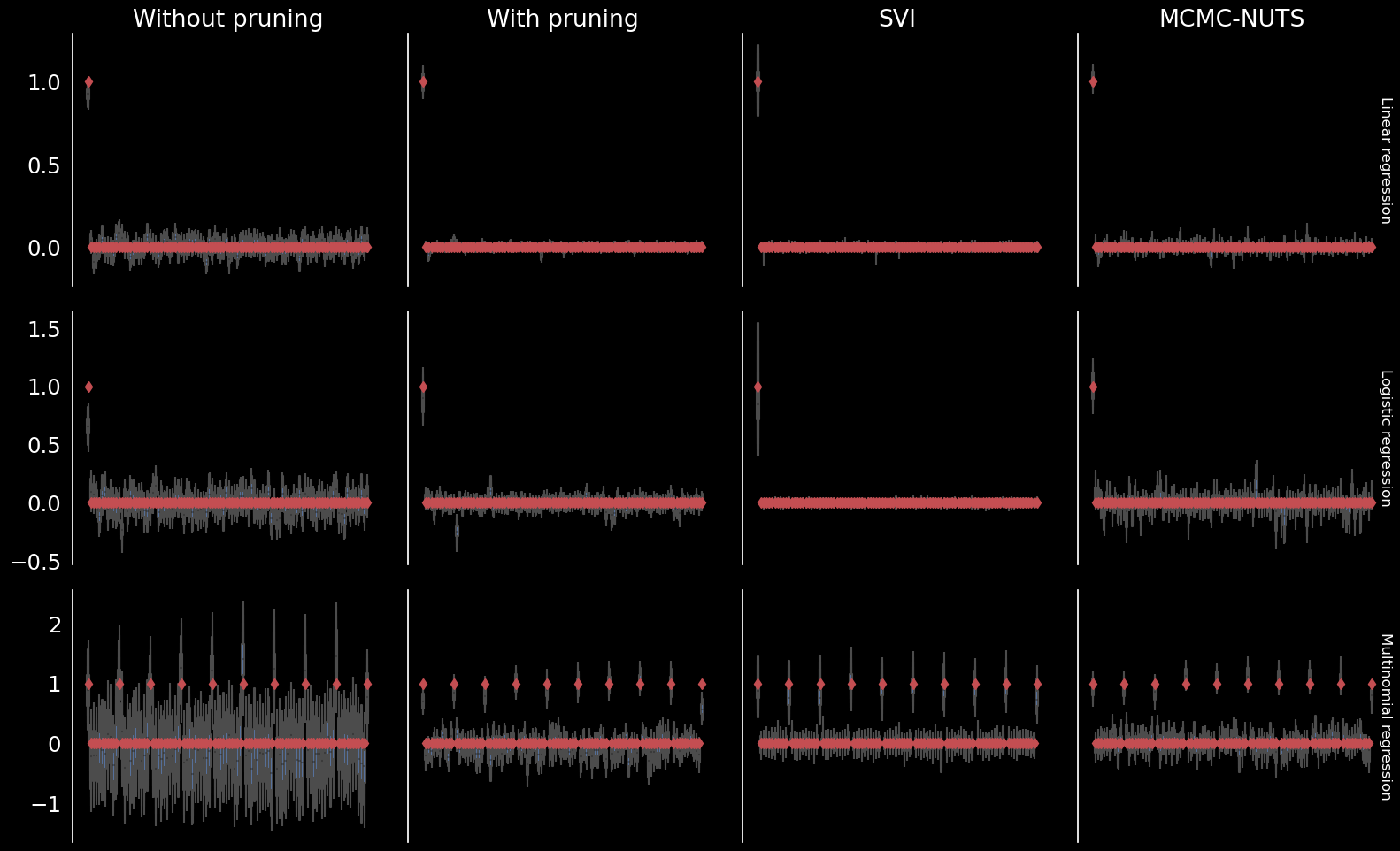
Leave one out cross validation
Real data
TODO: UCI Machine learning repository
| label | N | D |
|---|---|---|
| Yacht | 308 | 6 |
| Boston | 506 | 13 |
| Energy | 768 | 8 |
| Concrete | 1030 | 8 |
| Wine | 1599 | 11 |
| Kin8nm | 8192 | 8 |
| Power Plant | 9568 | 4 |
| Naval | 11,934 | 16 |
| Protein | 45,730 | 9 |
| Year | 515,345 | 90 |
Image classification
TODO:
- Compare BMR with SVI, and MAP/MLE.
- Vary data set size, network depth.
- Maybe NUTS can handle some cases?
- Compute relevant values on the testing data set.
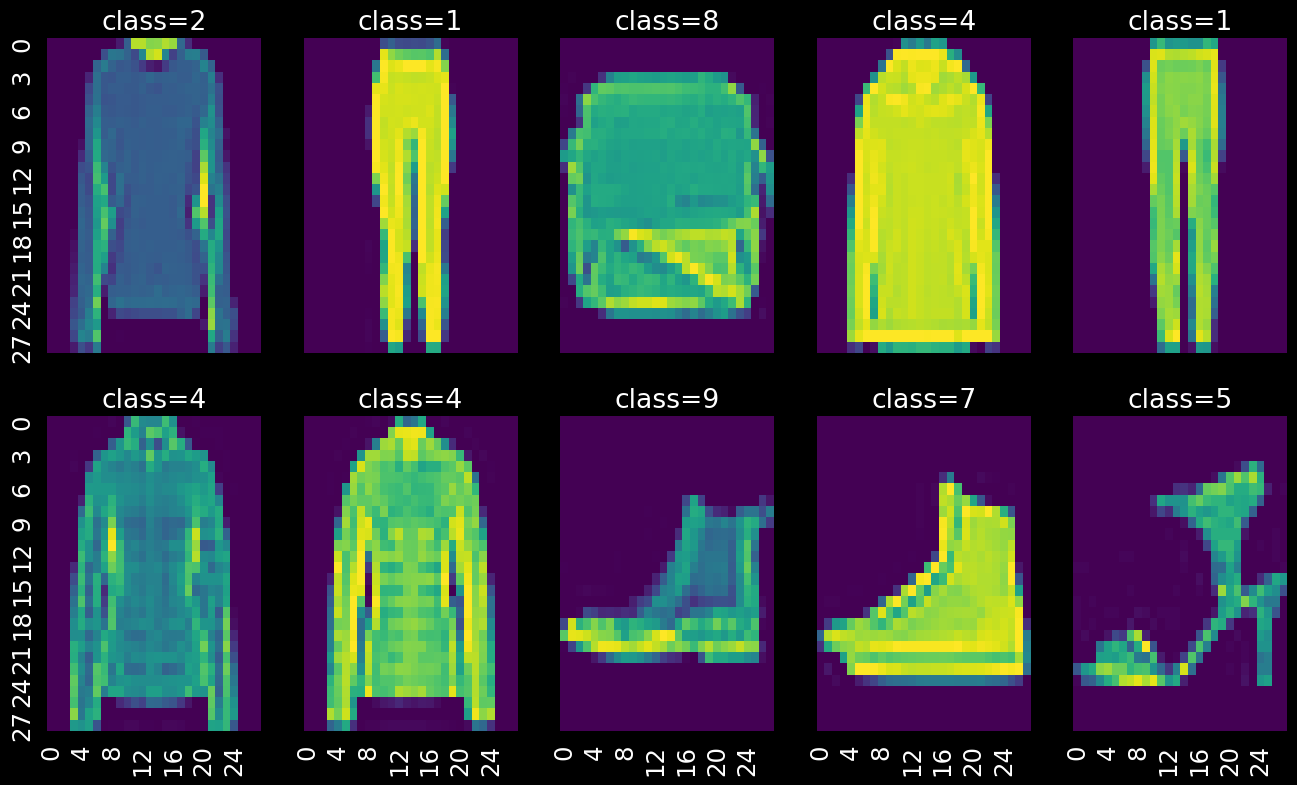
Fashion MNIST
Discussion
BMR seems to work great and has great potential for a range of deep learning applications.
https://github.com/dimarkov/numpc
Might be possible to prune pre-trained models using Laplace approximation.
But ... we can do better and use BMR to formulate Bayesian sparse predictive coding models.
Naturally complements distributed and federated inference problems.