jax-cosmo: Finally an Automatically Differentiable Cosmology Library!
2021 July DESC Collaboration Meeting
Francois Lanusse (@EiffL), for the jax-cosmo contributors

And more (not yet in the picture because I'm lagging behind on PR merging)
- Jean-Eric Campagne
- Joe Zuntz
- Tilman Troester
- Ben Horowitz
Why
do you want
any of this?
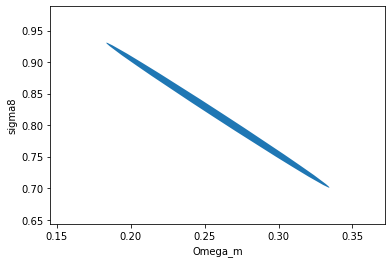
Fisher Forecasts become exact and instantaneous
import jax
import jax.numpy as np
import jax_cosmo as jc
# .... define probes, and load a data vector
def gaussian_likelihood( theta ):
# Build the cosmology for given parameters
cosmo = jc.Planck15(Omega_c=theta[0], sigma8=theta[1])
# Compute mean and covariance
mu, cov = jc.angular_cl.gaussian_cl_covariance_and_mean(cosmo,
ell, probes)
# returns likelihood of data under model
return jc.likelihood.gaussian_likelihood(data, mu, cov)
# Fisher matrix in just one line:
F = - jax.hessian(gaussian_likelihood)(theta)- No derivatives were harmed by finite differences in the computation of this Fisher!
- You can scale to thousands of input parameters, no issues of numerical precision.
Inference becomes fast and scalable
- Gradients of the log posterior are required for modern efficient and scalable inference techniques:
- Variational Inference
- Hamiltonian Monte-Carlo
- With gradients, you can scale to thousands of parameters
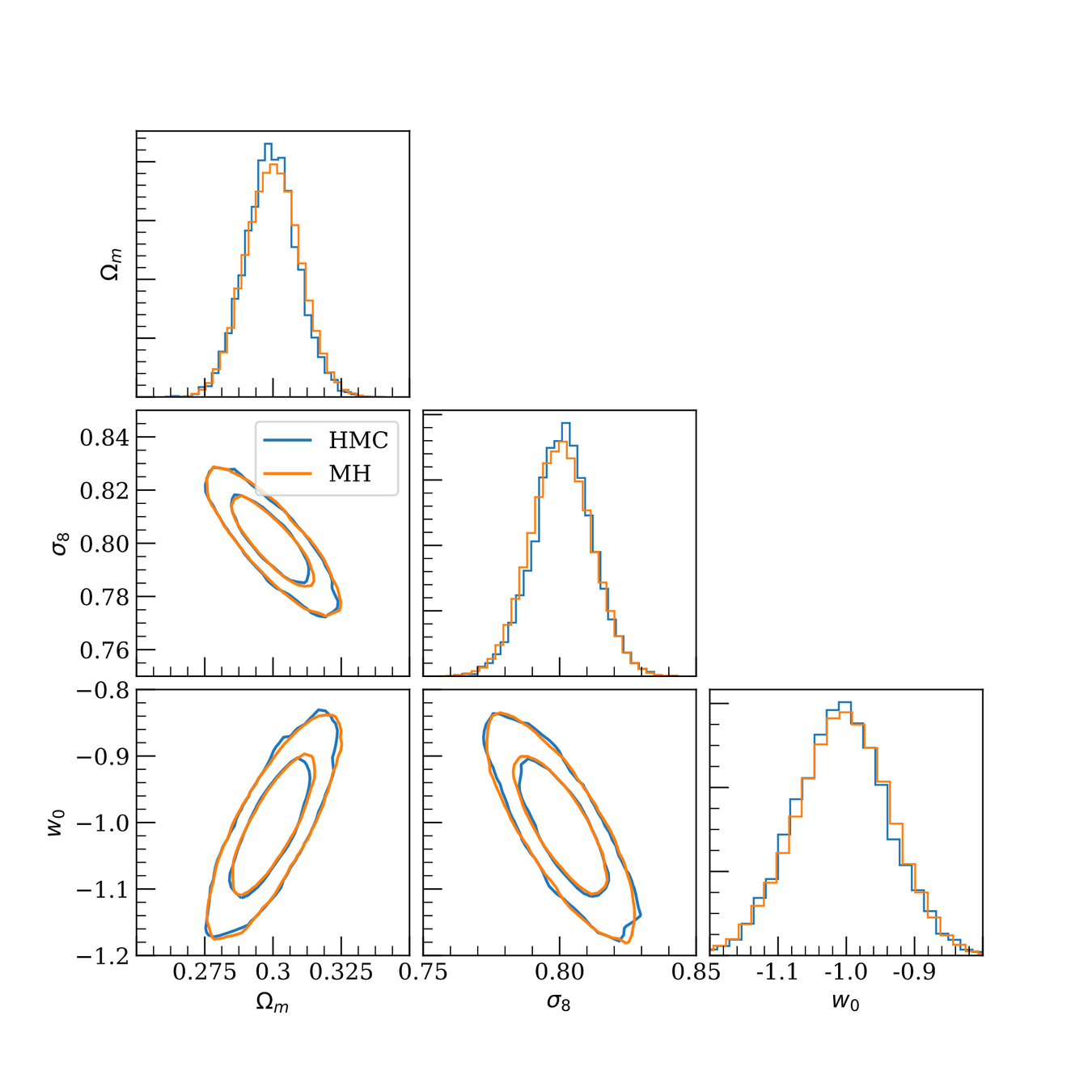
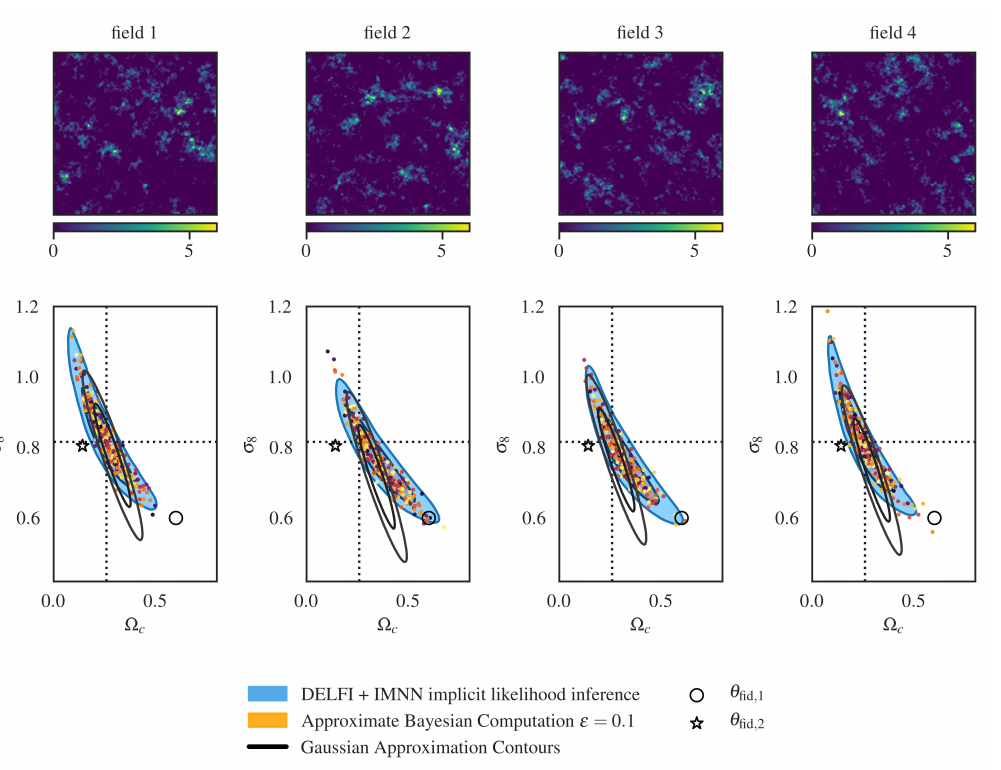
DES Y1 posterior, jax-cosmo HMC vs Cobaya MH
(credit: Joe Zuntz)
def log_posterior( theta ):
return gaussian_likelihood( theta ) + log_prior(theta)
score = jax.grad(log_posterior)(theta)You can differentiate through Cosmology
Given (g)riz photometry, find a tomographic bin assignment method that optimizes a 3x2pt analysis.
- Strategy with differentiable physics:
- Introduce a parametric "bin assignment function"
- Optimize this function to maximize DETF FoM
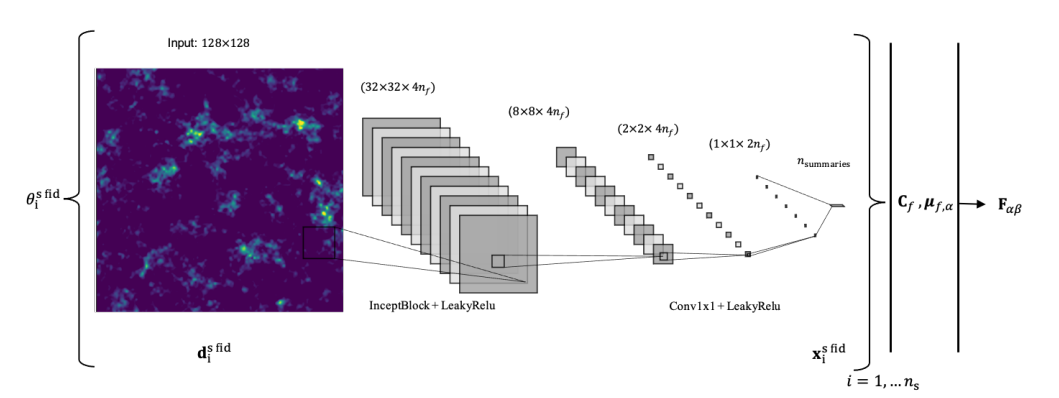
- Differentiable Physics for Score Compression
Makinen et al. 2021, arXiv:2107.07405
What Framework Should you Adopt?



Some top-level considerations
-
[Low Barrier to Entry] You don't want contributors to have to learn an entirely new language to start contributing (I'm looking at you Julia!).
-
[Maintainability] You want readable code, without several layers of implementation (I'm looking at you GalSim and CCL!)
-
[Trivially installable] This should always work:
- [Performance] Duh! But more precisely, you want to be able to take advantage of next gen platforms like Perlmutter. Code should work 100% the same on CPU or GPU for the user.
$ pip install mylibrary=> a JAX-based library will meet all of these criteria
What
Is
JAX?
JAX: NumPy + Autograd + XLA
- JAX uses the NumPy API
=> You can copy/paste existing code, works pretty much out of the box
- JAX is a successor of autograd
=> You can transform any function to get forward or backward automatic derivatives (grad, jacobians, hessians, etc)
- JAX uses XLA as a backend
=> Same framework as used by TensorFlow (supports CPU, GPU, TPU execution)
import jax.numpy as np
m = np.eye(10) # Some matrix
def my_func(x):
return m.dot(x).sum()
x = np.linspace(0,1,10)
y = my_func(x)from jax import grad
df_dx = grad(my_func)
y = df_dx(x)
-
Pure functions
=> JAX is designed for functional programming: your code should be built around pure functions (no side effects)- Enables caching functions as XLA expressions
- Enables JAX's extremely powerful concept of composable function transformations
-
Just in Time (jit) Compilation
=> jax.jit() transformation will compile an entire function as XLA, which then runs in one go on GPU.
-
Arbitrary order forward and backward autodiff
=> jax.grad() transformation will apply the d/dx operator to a function f
-
Auto-vectorization
=> jax.vmap() transformation will add a batch dimension to the input/output of any function
def pure_fun(x):
return 2 * x**2 + 3 * x + 2
def impure_fun_side_effect(x):
print('I am a side effect')
return 2 * x**2 + 3 * x + 2
C = 10. # A global variable
def impure_fun_uses_globals(x):
return 2 * x**2 + 3 * x + C# Decorator for jitting
@jax.jit
def my_fun(W, x):
return W.dot(x)
# or as an explicit transformation
my_fun_jitted = jax.jit(my_fun)def f(x):
return 2 * x**2 + 3 *x + 2
df_dx = jax.grad(f)
jac = jax.jacobian(f)
hess = jax.hessian(f)
# As decorator
@jax.vmap
def f(x):
return 2 * x**2 + 3 *x + 2
# Can be composed
df_dx = jax.jit(jax.vmap(jax.grad(f)))- Cosmology library embracing JAX computing model
=> Autodiff, GPU acceleration, automated batching
- MIT licensed, collaboratively developed
=> Follows the all-contributors guidelines
- Inspired by CCL's tracer mechanism
=> See design document
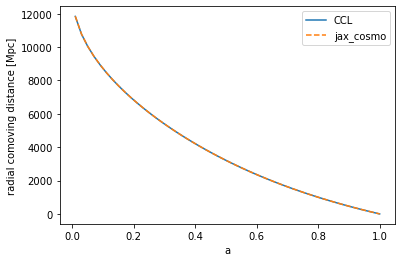
- Unit tested against CCL
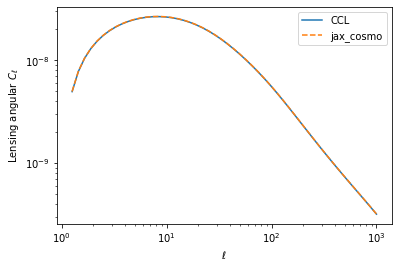
Status of Implementation
- Certainly not feature-complete but approximately DES Y1 3x2pt capable
- Power spectrum:
- Eisenstein-Hu Power
- halofit
- Tracers:
- Lensing (including NLA, multiplicative bias)
- NumberCounts
- 2pt functions
- Limber, harmonic space
- Gaussian covariance tools
- Power spectrum:
- These were the main features needed for the DESC 3x2pt Tomo Challenge
- Took about a week to implement, another week to tidy up a bit
Let's take it for a spin
My
Two
Cents

JAX vs Julia vs TensorFlow vs PyTorch
-
Let's go with JAX:
- It's normal NumPy, don't need to learn anything new
- Under the hood, they will all use XLA, so no difference
- Very powerful function transformation mechanisms
- It's normal NumPy, don't need to learn anything new
Do We Need to Rewrite CCL from Scratch?
-
Yes, if we want differentiability there is no workaround.
Trying to progressively adapt CCL with differentiable components would be a nightmare.
- But:
- When you already have a validated numerical scheme and implementation to compare to, there is no "research/uncertainty", it's purely a matter of translating code. I coded jax-cosmo in about a week.
=> CCL would continue to play a major role as a standard reference.
- It can be an opportunity to bring all of our experience in the design of a new library.
- When you already have a validated numerical scheme and implementation to compare to, there is no "research/uncertainty", it's purely a matter of translating code. I coded jax-cosmo in about a week.
What about CAMB and CLASS?
- Quick and Easy solution: train an emulator
- E.g. CosmoPower from A. Spurio Mancini
- E.g. CosmoPower from A. Spurio Mancini
- Longer term solution:
- Someone (maybe Zack) will code a differentiable version of those
- We could kickstart such a project
Where to host such projects?
- These tools will be useful to all modern cosmological surveys, but they are non trivial to implement, test, and maintain.
- You want to ensure broad user adoption
-
Help build a community to support the maintenance and development of the project
- 2 options:
- A DESC-sponsored project (like CCL), but developed completely openly (think astropy)
- Open project hosted in some sort of external entity, enabling anyone to contribute, across any collaboration.
- That was the idea behind the Differentiable Universe Initiative
- That was the idea behind the Differentiable Universe Initiative
- A DESC-sponsored project (like CCL), but developed completely openly (think astropy)
- With our new DESC Software policy, I think option 1 is now doable.
In Conclusion...
But more seriously, happy to chat anytime if you are interested in contributing, or just want to know more about JAX :-)