your code (execution)
Stefano Moia
École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland; physiopy (https://github.com/physiopy)
14.02.2023


apptainer exec docker://ghcr.io/apptainer/lolcow cowsay "Hello $USER, and happy B-day Carlo!"Try it now on stiitsrv21/22/23!
Follow from your laptop:
https://slides.com/d/7mDp4d4/live

| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
Where are we in Open Science/Development?
To be Open, Science should be publicly available, reusable,
and transparent¹. It should aim at reaching:

1. The Turing Way Community, 2019 (Zenodo). https://the-turing-way.netlify.app (CC-BY 4.0)
- Open Data
- Open Source Software
- Open Hardware
- Open Access (outreach)
- Reproducible results
- Usable artefacts, independently of setting.
- Complete and clear process documentation
How do we do Open Science?
- Read literature, formulate hypotheses, decide methods, submit a Registered Report (~ Introduction, hypotheses, methods), get through first peer review round.
- Find open development projects, contribute if needed, check licences.
- Decide a (permissive) licence for the artefacts, start open developing code (share it!), use VCS, test and document code, create releases.
- Create containers.
- Collect data, curate them using community schemas (e.g. BIDS), upload them on public databases with embargo ("private time").
- Run analyses in containers, interpret results, write the second part of your Registered Report (~ Results, a posteriori analyses, discussion, conclusions)
- Publish open access.
- Remove embargoes.
- Rinse and repeat.

Replicable, Robust, Reproducible, Generalisable

The Turing Way Community, & Scriberia, 2022 (Zenodo). Illustrations from The Turing Way (CC-BY 4.0)
Guaranteeing reproducibility is important for "reusable, transparent" research.

Really Replicable?
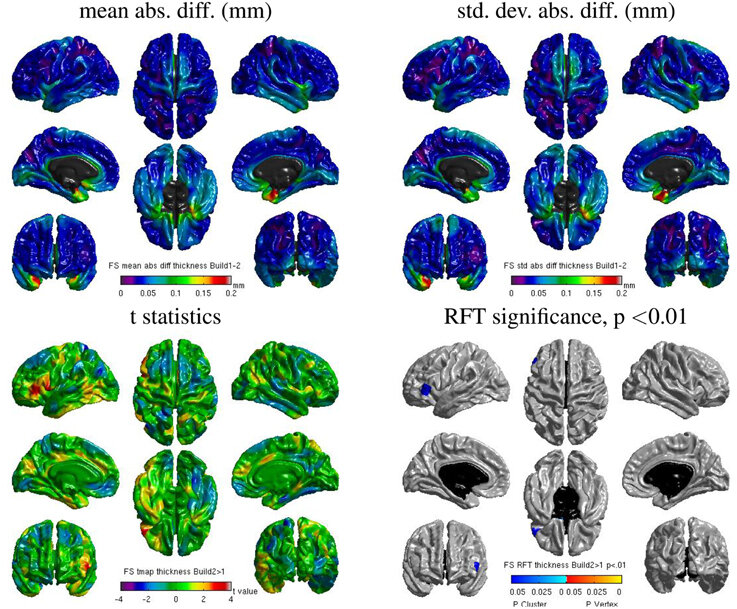
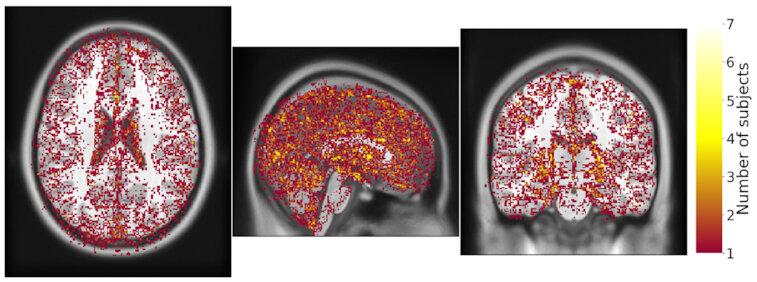
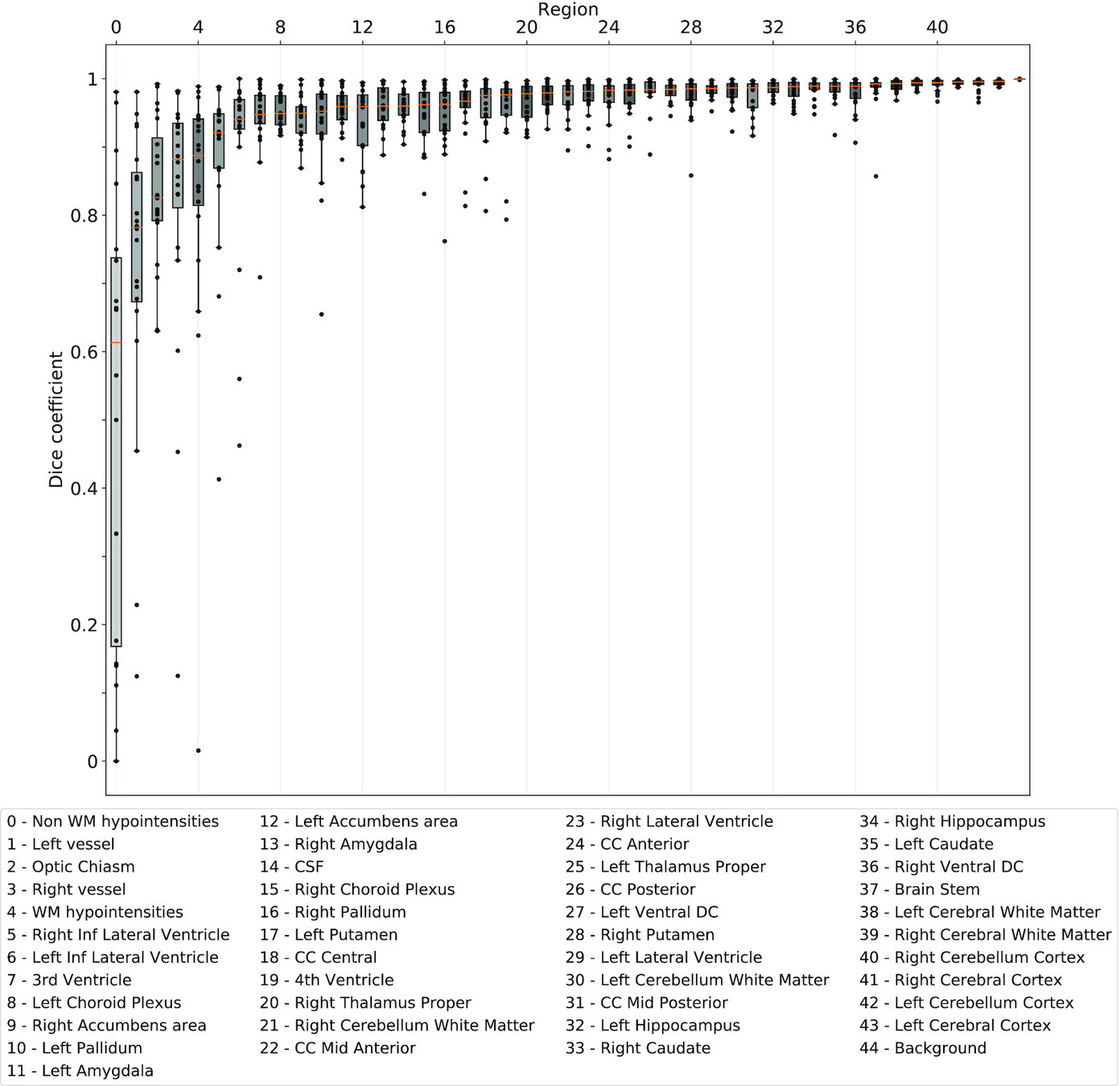
Same hardware, two Freesurfer builds (different glibc version)
Difference in estimated cortical tickness.¹
Same hardware, same FSL version, two glibc versions
Difference in estimated tissue segmentation.²
Same hardware, two Freesurfer builds (two glibc versions)
Difference in estimated parcellation.²
1. Glatard, et al., 2015 (Front. Neuroinform.) 2. Ali, et al., 2021 (Gigascience)




Containerisation (vs VM)



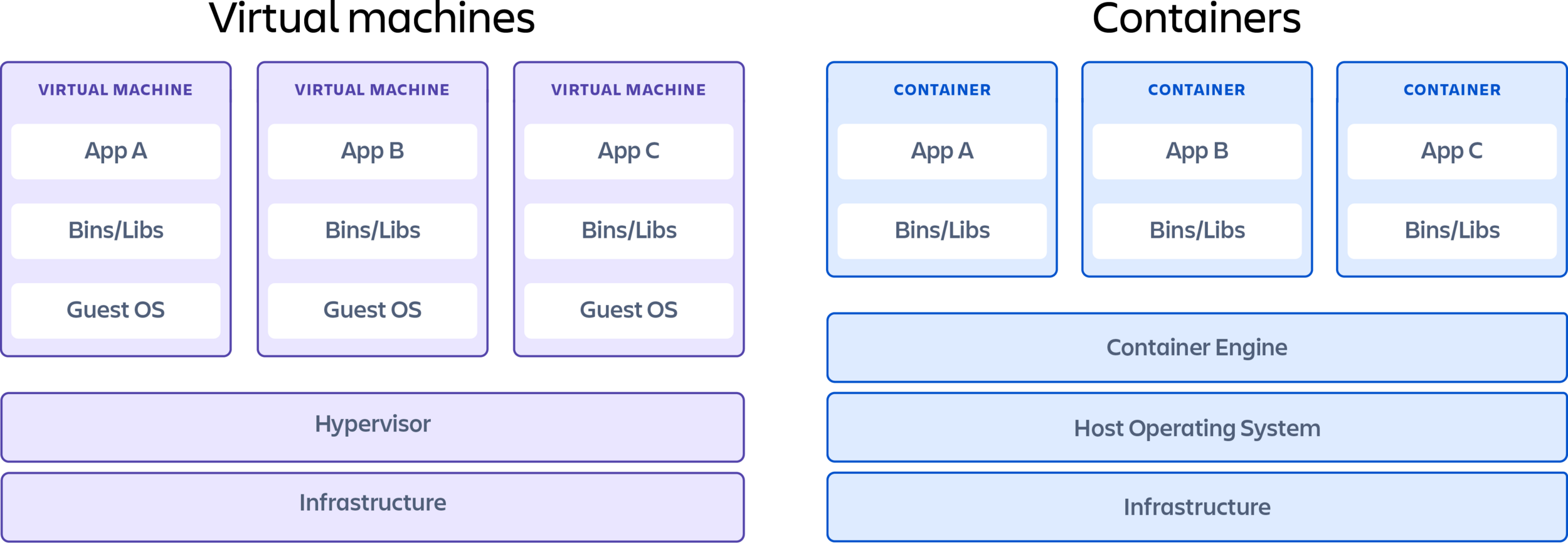
Containers vs VM:
Pros:
- Cost (less CPU, less memory)
- Faster runtime = faster runs
- Smaller sizes
- More portable*
Cons:
- Security (can be exploited)*
- Graphic User Interface is secondary*
Docker vs Apptainer

Docker:
- Targeting Laptops: better OS support (yes, you, mac/win peeps)
- Hosts public hub to share built containers
- Works with layer images to build containers
- Docker images can be used as bases for Apptainer recipes

Apptainer:
- Built for HPCs (Unix only), maintained by the Linux Foundation
- Easier "recipe" syntax
- Containers are a full root system folder that can be built as sandboxes
- Supports Docker images as bases
- It is safer than docker


Docker vs Apptainer


Bootstrap: docker
From: python:3.8.13-slim-buster
%files
.. /opt
%environment
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
%post
# Set install variables, create tmp folder
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
# Prepare repos and install dependencies
pip3 install /opt/.[all]
%runscript
nigsp
%labels
org.label-schema.name="NiGSP"
org.label-schema.description="NiGSP: python library for Graph Signal Processing on Neuroimaging data"
org.label-schema.url="https://github.com/miplabch/nigsp"
org.label-schema.vcs-url="https://github.com/miplabch/nigsp"
org.label-schema.schema-version="1.0"FROM python:3.8.13-slim-buster AS nigspdock
WORKDIR /app
# Prepare environment
COPY .. .
RUN pip3 install .[all]
ENV LANG="en_US.UTF-8" \
LC_ALL="en_US.UTF-8"
CMD nigsp
ARG BUILD_DATE
ARG VCS_REF
ARG VERSION
LABEL org.label-schema.build-date=$BUILD_DATE \
org.label-schema.name="NiGSP" \
org.label-schema.description="NiGSP: python library for Graph Signal Processing on Neuroimaging data" \
org.label-schema.url="https://github.com/miplabch/nigsp" \
org.label-schema.vcs-ref=$VCS_REF \
org.label-schema.vcs-url="https://github.com/miplabch/nigsp" \
org.label-schema.version=$VERSION \
org.label-schema.schema-version="1.0"
Docker
Apptainer
Example container: NiGSP


Bootstrap: docker
From: python:3.8.13-slim-buster
%environment
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
%post
# Set install variables, create tmp folder
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
# Prepare repos and install dependencies
pip3 install nigsp[all]
%runscript
nigsp
FROM python:3.8.13-slim-buster
# Prepare environment
ENV DEBIAN_FRONTEND=noninteractive \
TZ=Europe/Brussels
RUN pip3 install nigsp[all]
CMD nigsp
Docker
Apptainer
# Create a sandbox image from a python image on the Docker Hub
apptainer build --fakeroot --sandbox nigsp.img docker://python:3.8.13-slim-buster
# Start an interactive session to modify the sandbox (short and long flags)
apptainer shell -f -e -w --no-home nigsp.img
apptainer shell --fakeroot --cleanenvironment --writable --no-home nigsp.img
# Install nigsp in the container
pip3 install nigsp[all]
pip3 list
# Exit the container
exit
# Create an unmodifiable image from the previous sandbox
apptainer build -f nigsp.sif nigsp.img
# Execute a command in the new image, in this case call the help of nigsp
apptainer exec -e --no-home nigsp.sif nigsp --helpExample container


Bootstrap: docker
From: ubuntu:20.04
%environment
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
export FSLDIR="/opt/fsl-6.0.6.2"
source ${FSLDIR}/etc/fslconf/fsl.sh
export FSLOUTPUTTYPE="NIFTI_GZ"
export FSLMULTIFILEQUIT="TRUE"
export FSLTCLSH="$FSLDIR/bin/fsltclsh"
export FSLWISH="$FSLDIR/bin/fslwish"
export FSLLOCKDIR=""
export FSLMACHINELIST=""
export FSLREMOTECALL=""
export FSLGECUDAQ="cuda.q"
export PATH="$FSLDIR/bin:$PATH"
%post
# Set install variables, create tmp folder
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
# Prepare repos and install dependencies
apt update -qq
apt install -y -q --no-install-recommends \
bc \
build-essential \
ca-certificates \
curl \
dc \
dirmngr \
file \
g++ \
gcc \
git \
less \
make \
nano \
sudo \
wget \
python3-distutils \
python3-pip \
python3-rpy2 \
python-is-python3
# Install FSL
mkdir -p ${TMPDIR}/fsl
cd ${TMPDIR}/fsl || exit 1
curl -fL https://fsl.fmrib.ox.ac.uk/fsldownloads/fslinstaller.py --output ./fslinstaller.py
chmod +x fslinstaller.py
python3 fslinstaller.py -d /opt/fsl-6.0.6.2 -V 6.0.6.2
# echo "Installing FSL conda environment ..."
# bash /opt/fsl-6.0.6.2/etc/fslconf/fslpython_install.sh -f /opt/fsl-6.0.6.2
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/fsl
# Install PYTHON things.
pip3 install pip==22.3.1 setuptools==65.5.1 wheel==0.38.4
# Install wxPython in a particular way.
pip3 install --no-cache -f https://extras.wxpython.org/wxPython4/extras/linux/gtk3/ubuntu-20.04 wxpython==4.2.0
# Install datalad, fsleyes, nilearn, peakdet, phys2cvr.
pip3 install \
annexremote==1.6.0 \
datalad==0.17.10 \
fsleyes==1.5.0 \
fsleyes-props==1.8.2 \
fsleyes-widgets==0.12.3 \
fslpy==3.10.0 \
ipython \
h5py==3.7.0 \
matplotlib==3.6.2 \
nibabel==4.0.2 \
nilearn==0.10.0 \
numpy==1.23.5 \
pandas==1.5.2
# Final removal of lists and cleanup
rm -rf /var/lib/apt/lists/*
%runscript
fslmaths
FROM ubuntu:20.04 AS ubuntudock
# Prepare environment
ENV DEBIAN_FRONTEND=noninteractive \
TZ=Europe/Brussels
ENV FSLDIR="/opt/fsl-6.0.5.1" \
PATH="/opt/fsl-6.0.5.1/bin:$PATH" \
FSLOUTPUTTYPE="NIFTI_GZ" \
FSLMULTIFILEQUIT="TRUE" \
FSLTCLSH="/opt/fsl-6.0.5.1/bin/fsltclsh" \
FSLWISH="/opt/fsl-6.0.5.1/bin/fslwish" \
FSLLOCKDIR="" \
FSLMACHINELIST="" \
FSLREMOTECALL="" \
FSLGECUDAQ="cuda.q"
RUN apt-get update -qq \
&& apt-get install -y -q --no-install-recommends \
bc \
build-essential \
ca-certificates \
curl \
dc \
dirmngr \
file \
g++ \
gcc \
git \
gnupg \
libfontconfig1 \
libfreetype6 \
libgl1-mesa-dev \
libgl1-mesa-dri \
libglu1-mesa-dev \
libgomp1 \
libice6 \
libopenblas-base \
libxcursor1 \
libxft2 \
libxinerama1 \
libxrandr2 \
libxrender1 \
libxt6 \
less \
make \
nano \
sudo \
wget \
python3-distutils \
python3-pip \
python3-rpy2 \
python-is-python3 \
&& rm -rf /var/lib/apt/lists/* \
&& echo "Downloading FSL ..." \
&& mkdir -p /opt/fsl-6.0.5.1 \
&& curl -fL https://fsl.fmrib.ox.ac.uk/fsldownloads/fsl-6.0.5.1-centos7_64.tar.gz \
| tar -xz -C /opt/fsl-6.0.5.1 --strip-components 1 \
&& echo "Installing FSL conda environment ..." \
&& bash /opt/fsl-6.0.5.1/etc/fslconf/fslpython_install.sh -f /opt/fsl-6.0.5.1
RUN pip3 install pip==22.3.1 setuptools==65.5.1 wheel==0.38.4
RUN pip3 install --no-cache -f https://extras.wxpython.org/wxPython4/extras/linux/gtk3/ubuntu-20.04 wxpython==4.2.0
RUN pip3 install annexremote==1.6.0 \
datalad==0.17.10 \
fsleyes==1.5.0 \
fsleyes-props==1.8.2 \
fsleyes-widgets==0.12.3 \
fslpy==3.10.0 \
ipython \
h5py==3.7.0 \
matplotlib==3.6.2 \
nibabel==4.0.2 \
nilearn==0.10.0 \
numpy==1.23.5 \
pandas==1.5.2
RUN ln -s /opt/fsl-6.0.5.1/bin/eddy_cuda9.1 /opt/fsl-6.0.5.1/bin/eddy_cuda
ENV FSLOUTPUTTYPE="NIFTI_GZ"
RUN apt-get update -qq \
&& apt-get install -y -q --no-install-recommends \
locales \
&& rm -rf /var/lib/apt/lists/*
RUN sed -i -e 's/# en_US.UTF-8 UTF-8/en_US.UTF-8 UTF-8/' /etc/locale.gen && locale-gen
CMD source ${FSLDIR}/etc/fslconf/fsl.sh; fslmaths
Docker
Apptainer
Apptainer example: complete data analysis.


Bootstrap: docker
From: ubuntu:20.04
%environment
# export templateloc=/usr/share/afni/atlases
export AFNIPATH=/opt/afni-AFNI_22.3.07
export AFNI_PLUGINPATH="$AFNIPATH"
export templateloc=/usr/share/afni/atlases
export AFNI_AUTOGZIP=YES
export AFNI_COMPRESSOR=GZIP
export ANTSPATH="/opt/ants-2.4.2/bin"
export ANTSSCRIPTS="/opt/ants-2.4.2/Scripts"
export C3DPATH="/opt/convert3d-1.0.0"
export FSLDIR="/opt/fsl-6.0.6.2"
source ${FSLDIR}/etc/fslconf/fsl.sh
export FSLOUTPUTTYPE="NIFTI_GZ"
export FSLMULTIFILEQUIT="TRUE"
export FSLTCLSH="$FSLDIR/bin/fsltclsh"
export FSLWISH="$FSLDIR/bin/fslwish"
export FSLLOCKDIR=""
export FSLMACHINELIST=""
export FSLREMOTECALL=""
export FSLGECUDAQ="cuda.q"
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
export R_LIBS="/usr/lib/R"
export LD_LIBRARY_PATH="/opt/ants-2.4.2/lib:$LD_LIBRARY_PATH"
export PREPROCPATH="/opt/preprocessing"
export PATH="$AFNIPATH:$ANTSPATH:$ANTSSCRIPTS:$C3DPATH/bin:$FSLDIR/bin:$PREPROCPATH:$PREPROCPATH/00.pipelines:$PATH"
%post
# Set install variables, create tmp folder
export TMPDIR="/tmp/general_preproc_build_$( date -u +"%F_%H-%M-%S" )"
[[ -d ${TMPDIR} ]] && rm -rf ${TMPDIR}
mkdir -p ${TMPDIR}
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
apt update -qq
apt install -y -q --no-install-recommends ca-certificates dirmngr gnupg
# Prepare repos and install dependencies
apt-key adv --keyserver keyserver.ubuntu.com --recv-keys C9A7585B49D51698710F3A115E25F516B04C661B
apt-key adv --keyserver keyserver.ubuntu.com --recv-keys 6E12762B81063D17BDDD3142F142A4D99F16EB04
echo "deb https://ppa.launchpadcontent.net/marutter/rrutter4.0/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb-src https://ppa.launchpadcontent.net/marutter/rrutter4.0/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb https://ppa.launchpadcontent.net/c2d4u.team/c2d4u4.0+/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb-src https://ppa.launchpadcontent.net/c2d4u.team/c2d4u4.0+/ubuntu focal main" | tee -a /etc/apt/sources.list
apt update -qq
apt install -y -q --no-install-recommends \
bc \
build-essential \
bzip2 \
cmake \
curl \
dc \
file \
freeglut3-dev \
g++ \
gcc \
git \
less \
libcurl4-openssl-dev \
libeigen3-dev \
libexpat1-dev \
libf2c2-dev \
libfftw3-3 \
libfftw3-dev \
libgdal-dev \
libgfortran4 \
libgfortran-8-dev \
libglew-dev \
libgl1-mesa-dev \
libgl1-mesa-dri \
libgl1-mesa-glx \
libglib2.0-dev \
libglu1-mesa-dev \
libglw1-mesa \
libgomp1 \
libgsl-dev \
libgts-dev \
libjpeg8-dev \
liblapack3 \
libopenblas-dev \
libmotif-dev \
libnetpbm10-dev \
libnode-dev \
libpng16-16 \
libpng-dev \
libquadmath0 \
libtiff5 \
libtiff5-dev \
libudunits2-dev \
libxext-dev \
libxi-dev \
libxm4 \
libxmhtml-dev \
libxml2-dev \
libxmu-dev \
libxmu-headers \
libxpm-dev \
libxt-dev \
m4 \
make \
mesa-common-dev \
nano \
r-base-dev \
rsync \
tcsh \
python3-distutils \
python3-pip \
python3-rpy2 \
python-is-python3 \
qhull-bin \
xvfb \
zlib1g-dev
# Install AFNI
mkdir -p ${TMPDIR}/afni
cd ${TMPDIR}/afni || exit 1
ln -s /usr/lib/x86_64-linux-gnu/libgsl.so.23 /usr/lib/x86_64-linux-gnu/libgsl.so.19
ln -s /usr/lib/x86_64-linux-gnu/libXp.so.6 /usr/lib/x86_64-linux-gnu/libXp.so
git clone https://github.com/afni/afni.git source
cd source || exit 1
git fetch --tags
git -c advice.detachedHead=false checkout AFNI_22.3.07
cd src || exit 1
cp other_builds/Makefile.linux_ubuntu_16_64_glw_local_shared Makefile
make itall
mv linux_ubuntu_16_64_glw_local_shared /opt/afni-AFNI_22.3.07
export PATH="/opt/afni-AFNI_22.3.07:$PATH"
export R_LIBS="/usr/lib/R"
rPkgsInstall -pkgs ALL
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/afni
# Install ANTs
mkdir -p ${TMPDIR}/ants/build
git clone https://github.com/ANTsX/ANTs.git ${TMPDIR}/ants/source
cd ${TMPDIR}/ants/source || exit 1
git fetch --tags
git -c advice.detachedHead=false checkout v2.4.2
cd ${TMPDIR}/ants/build || exit 1
cmake -DCMAKE_INSTALL_PREFIX=/opt/ants-2.4.2 -DBUILD_SHARED_LIBS=ON -DBUILD_TESTING=OFF ${TMPDIR}/ants/source
make -j 10
mkdir -p /opt/ants-2.4.2
cd ANTS-build || exit 1
make install
mv ../../source/Scripts/ /opt/ants-2.4.2
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/ants
# Install C3D
echo "Downloading Convert3D ..."
mkdir -p /opt/convert3d-1.0.0
curl -fsSL https://sourceforge.net/projects/c3d/files/c3d/1.0.0/c3d-1.0.0-Linux-x86_64.tar.gz/download \
| tar -xz -C /opt/convert3d-1.0.0 --strip-components 1
# Install FSL
mkdir -p ${TMPDIR}/fsl
cd ${TMPDIR}/fsl || exit 1
curl -fL https://fsl.fmrib.ox.ac.uk/fsldownloads/fslinstaller.py --output ./fslinstaller.py
chmod +x fslinstaller.py
python3 fslinstaller.py -d /opt/fsl-6.0.6.2 -V 6.0.6.2
# echo "Installing FSL conda environment ..."
# bash /opt/fsl-6.0.6.2/etc/fslconf/fslpython_install.sh -f /opt/fsl-6.0.6.2
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/fsl
# Clone EuskalIBUR preprocessing.
git clone https://github.com/smoia/EuskalIBUR_preproc.git /opt/preprocessing
apt install -y -q csvtool
# Install PYTHON things.
pip3 install pip==22.3.1 setuptools==65.5.1 wheel==0.38.4
# Install wxPython in a particular way.
pip3 install --no-cache -f https://extras.wxpython.org/wxPython4/extras/linux/gtk3/ubuntu-20.04 wxpython==4.2.0
# Install datalad, fsleyes, nilearn, peakdet, phys2cvr.
pip3 install \
annexremote==1.6.0 \
boto==2.49.0 \
certifi==2022.12.7 \
cffi==1.15.1 \
chardet==4.0.0 \
charset-normalizer==2.1.1 \
contourpy==1.0.6 \
cryptography==38.0.4 \
cycler==0.11.0 \
datalad==0.17.10 \
dill==0.3.6 \
distro==1.8.0 \
fasteners==0.18 \
fonttools==4.38.0 \
fsleyes==1.5.0 \
fsleyes-props==1.8.2 \
fsleyes-widgets==0.12.3 \
fslpy==3.10.0 \
h5py==3.7.0 \
humanize==4.4.0 \
idna==3.4 \
importlib-metadata==5.1.0 \
iso8601==1.1.0 \
jaraco.classes==3.2.3 \
jeepney==0.8.0 \
Jinja2==3.1.2 \
joblib==1.2.0 \
keyring==23.11.0 \
keyrings.alt==4.2.0 \
kiwisolver==1.4.4 \
lxml==4.9.2 \
MarkupSafe==2.1.1 \
matplotlib==3.6.2 \
more-itertools==9.0.0 \
msgpack==1.0.4 \
nibabel==4.0.2 \
nilearn==0.9.2 \
numpy==1.23.5 \
packaging==22.0 \
pandas==1.5.2 \
patool==1.12 \
peakdet==0.2.0rc1 \
phys2cvr==0.16.0 \
Pillow==9.3.0 \
platformdirs==2.6.0 \
pycparser==2.21 \
PyOpenGL==3.1.6 \
pyparsing==2.4.7 \
python-dateutil==2.8.2 \
python-gitlab==3.12.0 \
pytz==2022.6 \
requests==2.28.1 \
requests-toolbelt==0.10.1 \
scikit-learn==1.2.0 \
scipy==1.9.3 \
SecretStorage==3.3.3 \
simplejson==3.18.0 \
six==1.16.0 \
threadpoolctl==3.1.0 \
tqdm==4.64.1 \
urllib3==1.26.13 \
Whoosh==2.7.4 \
zipp==3.11.0
# Final removal of lists and cleanup
cd /tmp || exit 1
rm -rf ${TMPDIR}
rm -rf /var/lib/apt/lists/*Containers in action (resources limitation)


apptainer run --cpus 1.3 --memory-reservation 10G nigsp.sif nigsp --help
mkdir -p mipcontainers/tmp && cd mipcontainers
cp -r /media/miplab-nas2/Data3/mipcontainers/phenotype .
git clone https://github.com/smoia/fuzzy-invention.git
chmod +x fuzzy-invention/*.py
apptainer build -f sing2_apptainer.sif fuzzy-invention/sing2.def
cp /media/miplab-nas2/Data3/mipcontainers/sing2_apptainer.sif .
apptainer exec -e --no-home -B ./phenotype:/data -B ./fuzzy-invention:/scripts -B ./tmp:/tmp sing2_apptainer.sif \
/scripts/prepare_questionnaire.py /data/derivatives /data
BIDSapps: containers for BIDS pipelines

Easily build a neuroimaging container

Easily build a neuroimaging container

MATLAB container

That's all folks!
Thank you!
Any question [/opinions/objections/...]?
Love your code execution (MIP:Lab meeting 14.02.2023)
By Stefano Moia
Love your code execution (MIP:Lab meeting 14.02.2023)
CC-BY 4.0 Stefano Moia, 2023. Images are property of the original authors and should be shared following their respective licences. This presentation is otherwise licensed under CC BY 4.0. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/
- 146



