The gains and hurdles of making science a community effort
The case of Physiopy

| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
MRITogether 08.12.2023

Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, The Netherlands; physiopy (https://github.com/physiopy)



Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, The Netherlands; physiopy (https://github.com/physiopy)


The gains and hurdles of making science a community effort
The case of Physiopy
MRITogether 08.12.2023
Disclaimers
1. I am a member of the physiopy community
2. I have a bias towards the core tenets of Open Science as better scientific practices.
3. I am also fairly biased by my own experience - what I am saying as a whole might not hold for everyone, but hopefully you can get some inspiration from it.


0. Rules & Materials


This is a new chapter


Take home #0
This is a take home message


1. physiopy


Why physiopy
In neuroimaging, integration of physiological measures to data collection and analyses are still a niche topic. By raising awareness, we can inspire researchers and clinicians become interested in the topic.
*Open Source Software Development is the idea of developing a software publicly, sharing it from the beginning of the development, fostering a democratic community of contributors in support of the project, using version control and software testing.
physiopy adopts a Community driven, BIDS-based, Open Development* approach.
Sharing physiological data, toolboxes, and documentation following the concepts of Open Science could improve the exposition to this topic.


Community practices meetings, community consensus, and community guidelines.
Spark interest!
The more we share, the better it becomes
This is (not)
the way!
Of the People, by the People, for the People
physiopy's aims


The main goal of physiopy is to help collect, analyze and share physiological data by:
- Writing packages to make user-friendly pipelines to work with physiological data.
- Specializing in physiological data use in neuroimaging (i.e. MRI) data analysis.
- Providing documentation containing tips and strategies on how to collect such data and use our packages.
- Helping set a standard for these data, albeit without forcing users to use it.
Core components
A set of easily adoptable toolboxes
Community of users, developers, and researchers interested in physiology
Clear and approachable documentation


Community practices based on consensus
Core components


Core components


physiopy's libraries
Raw data
BIDSification
physiological data preprocessing
phys. denoising
phys. imaging
Data acquisition
Process description


QA/QC
physiopy's libraries
Raw data
phys2bids
peakdet
phys2denoise
phys. imaging
Data acquisition
physiopy's documentation
physioQC
Coordinated testing suite


BIDS Extension Proposal
physiopy's status


- All volunteers are... volunteering (so far)
- The project has virtually no funds (so far)
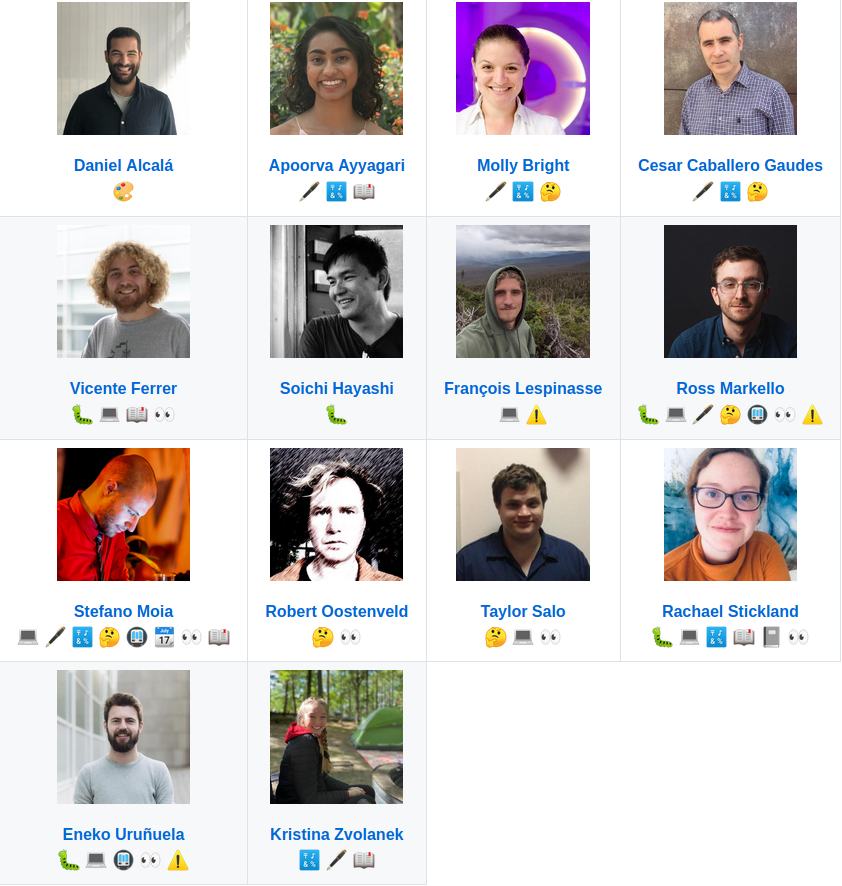
- Founded in 2019 by 9 people in 3 labs
- In 2023 it counts over 30 contributors from over 24 institutions
2. Communities


Adapt & juggle
- Stir the contributors, but follow them
- Be ready to switch focus when needed


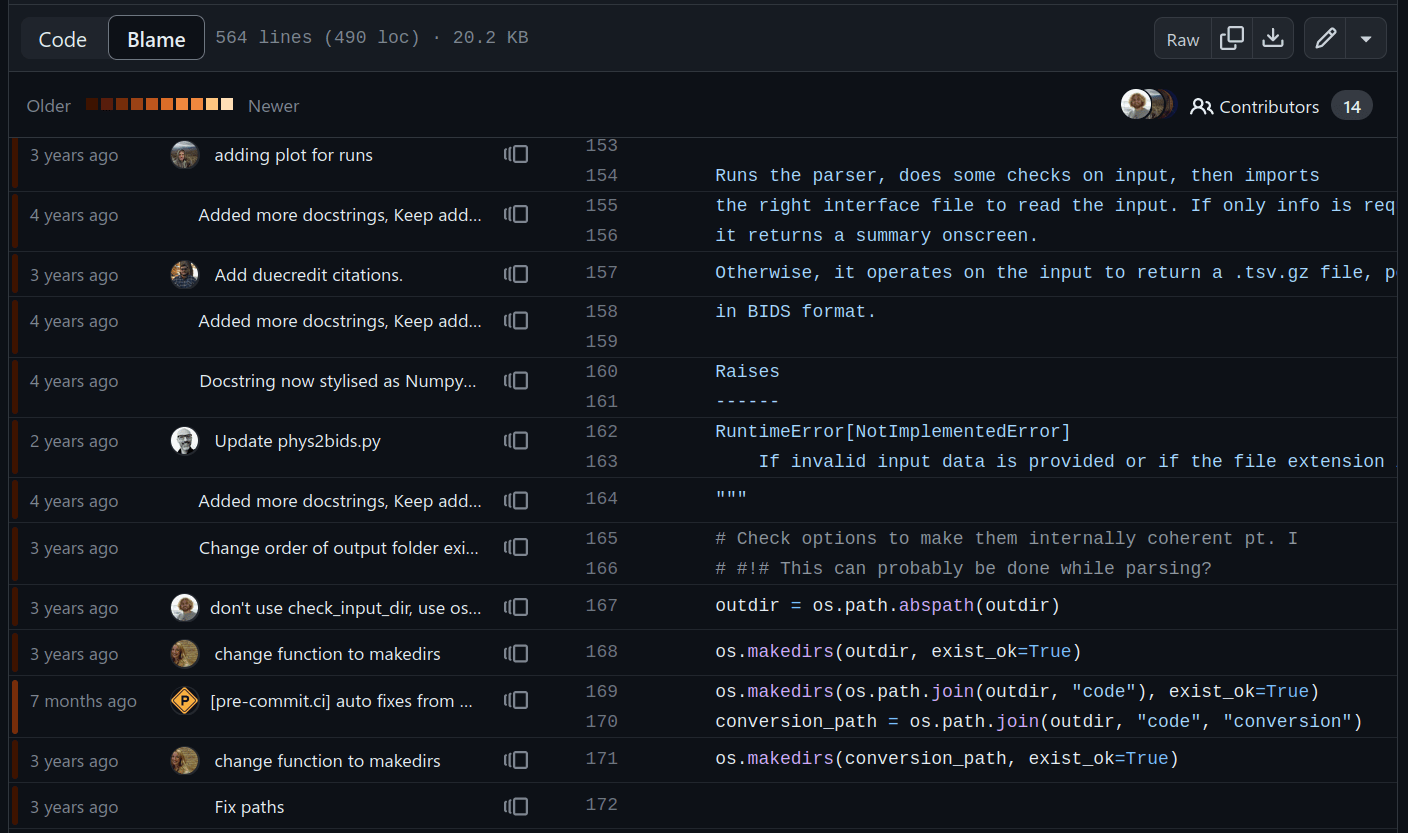
Keep track of everything
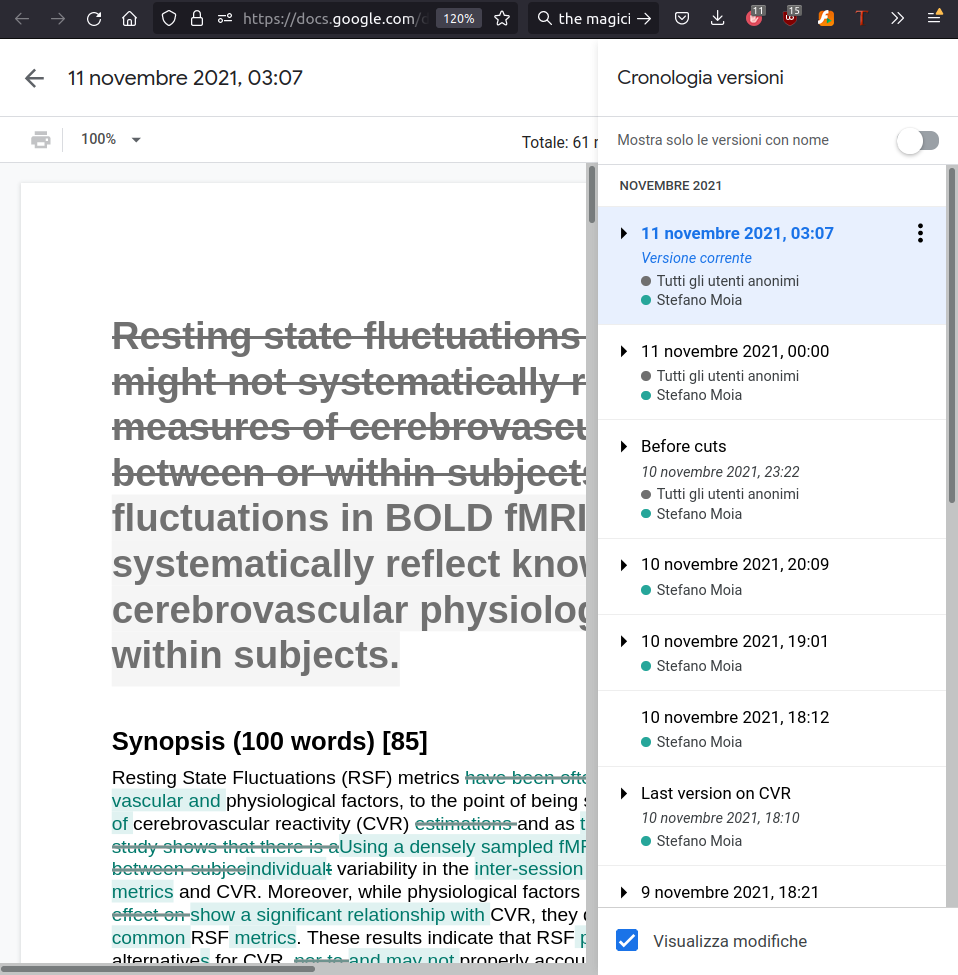
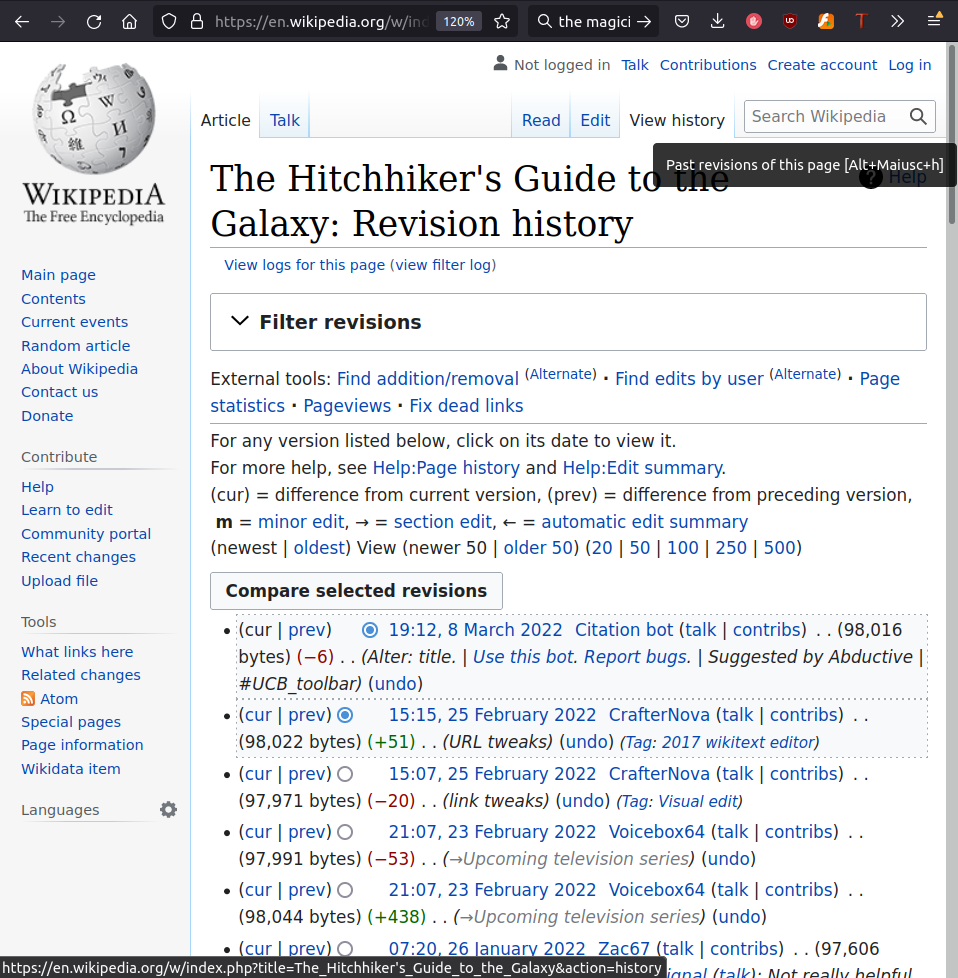
Keep track of everything
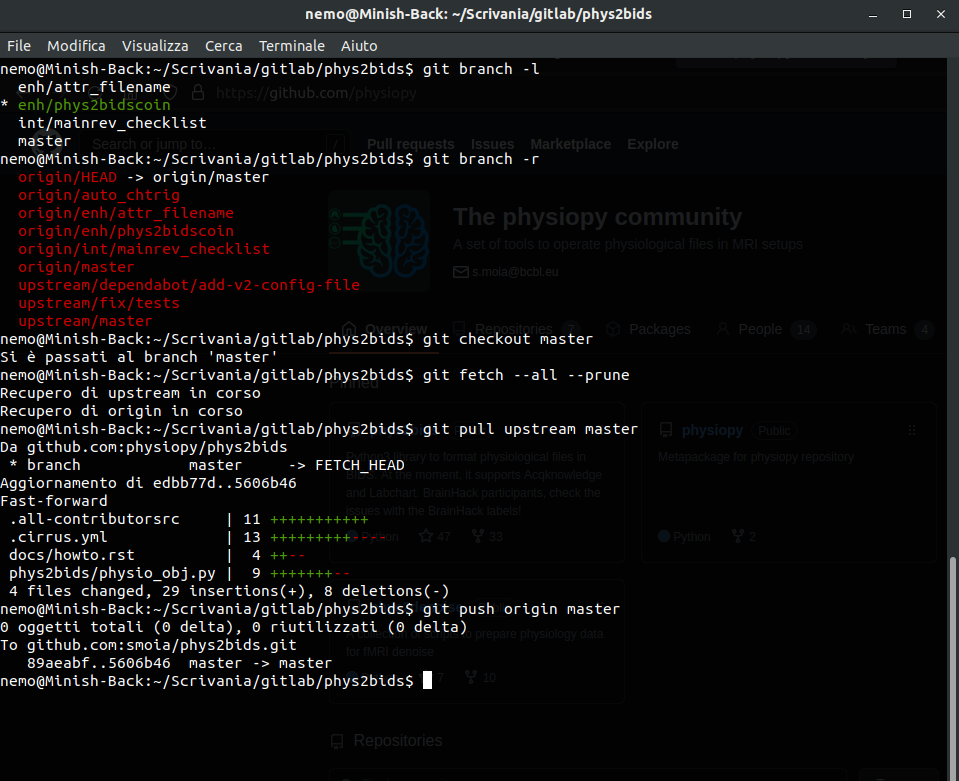



Keep track of everything
Keep track of everything
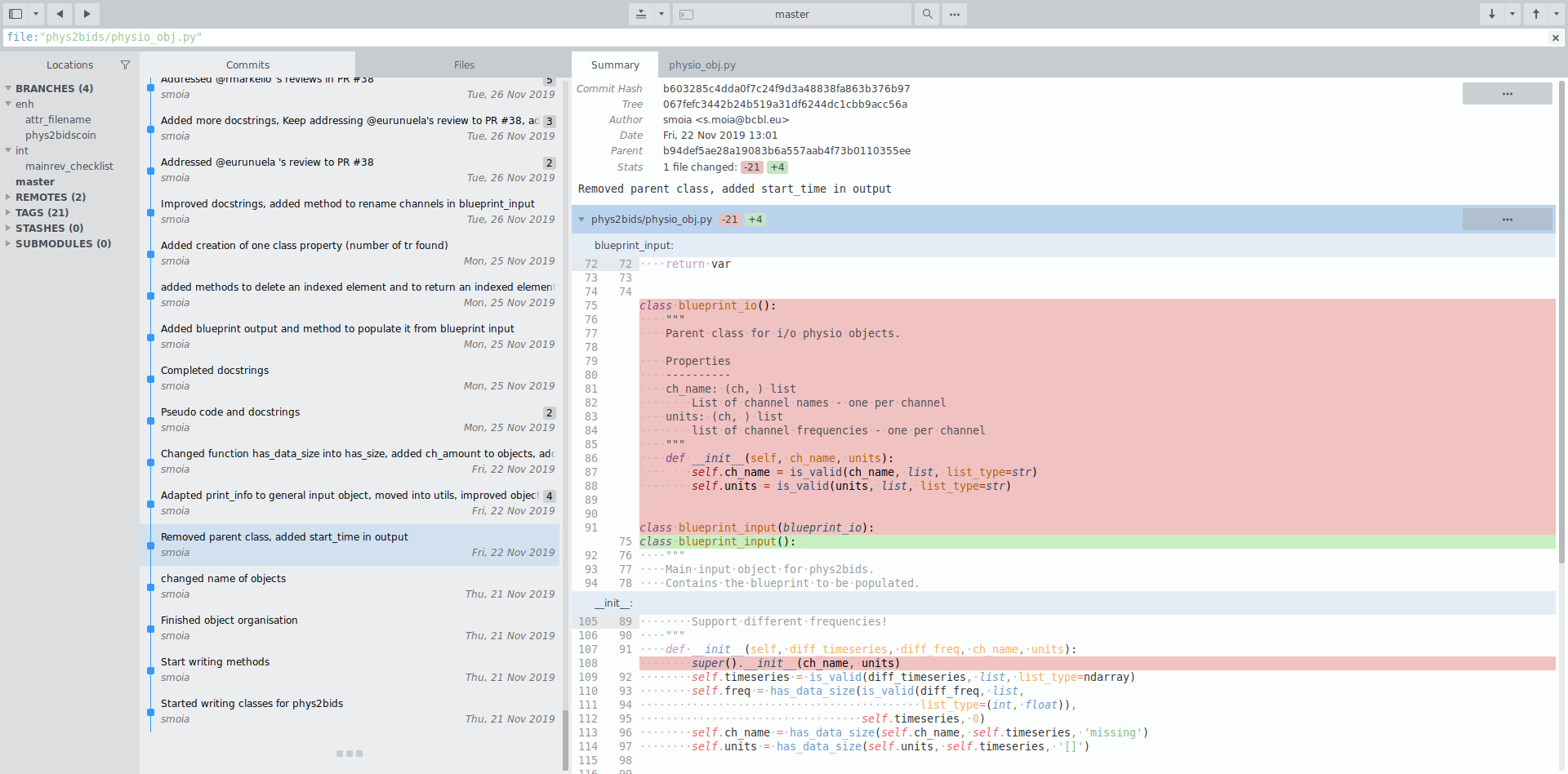
Attribution


Engage (others) & disengage (yourself)
- Keep meeting regularly, even if nothing is happening
- Give space to other contributors by distancing yourself
- Eventually, discuss a governance


Contributions and communities
- Development is a straight(er) line
- Time effective (on the short run)
- Low(er) engagement
- Less mentors
- Small(er) user base
- Less tests
- Consensus not guaranteed
- Clear attribution
- Development takes its own path
- (More) Time consuming
- High(er) engagement
- More mentors
- Big(ger) user base
- More tests
- Consensus based
- Fuzzier attribution


Take home #1
There are advantages and disadvantages in working within a community - choose what better fit your aims and interest.


3. (Some) Do's


Discuss good practices and/or LEARN


Recognise the contributors
Depending on the community and the governance scheme, contributions might be recognised differently.
Be clear about how you will recognise contributions.
One way of recognising contributors is the all-contributors specification.
(authorship is more complicated)


Create meaningful deliverables
when you can
(even multiple times)
Aim for readability
a, b = rui()
c = s(a, b)
p(c)a, b = read_user_input()
c = sum_two_numbers(a, b)
print(c)def very_important_function(template: str, *variables, file: os.PathLike, engine: str, header: bool = True, debug: bool = False):
"""Applies `variables` to the `template` and writes to `file`."""
with open(file, 'w') as f:
...
def very_important_function(
template: str,
*variables,
file: os.PathLike,
engine: str,
header: bool = True,
debug: bool = False,
):
"""Applies `variables` to the `template` and writes to `file`."""
with open(file, "w") as f:
...



Automate as much as you can



- Pre-commit [local, remote]: Automate code checks and styling on git commit
- Pytest, pytest-cov [local, remote]: (automated) testing and coverage
- Codecov [remote]: automated code coverage change check
- Auto [remote]: automated version update, tag, release, and changelog
- Zenodo, PyPI [remote]: automated DOI and package publishing
- Readthedocs [remote]: (automated) documentation publishing





Take home #2
Engage your community
to learn more/better.
Especially, learn how
to automate your work!


Thanks to...
...you for the (sustained) attention!
That's all folks!
...the organisers, for having me here
...the Physiopy contributors


| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |

| github.com/physiopy | |
| physiopy.github.io | |
| physiopy.community@gmail.com |
Open meetings every third Thursday of the month at 16h00 UTC

Any question [/opinions/objections/...]?
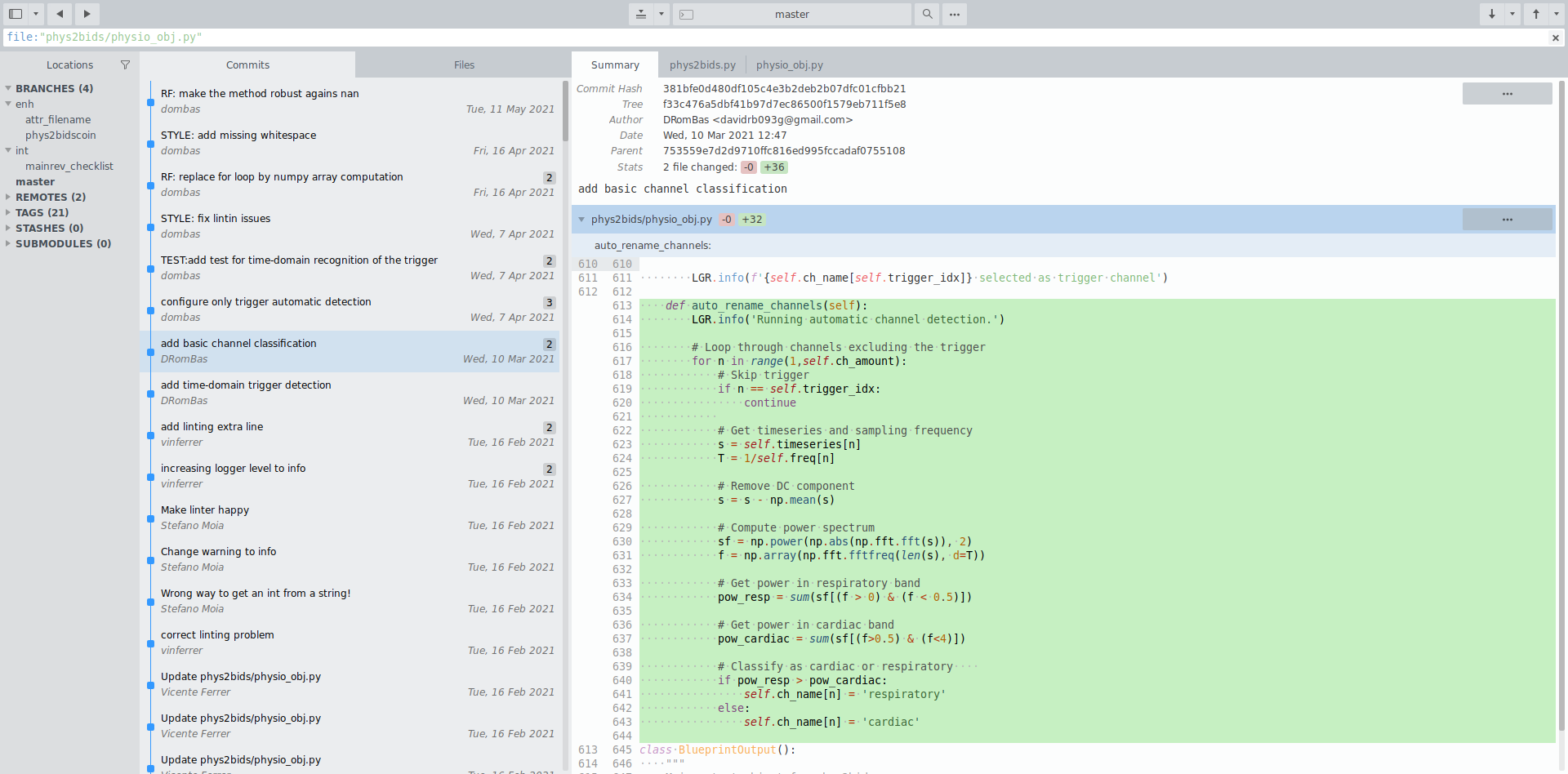
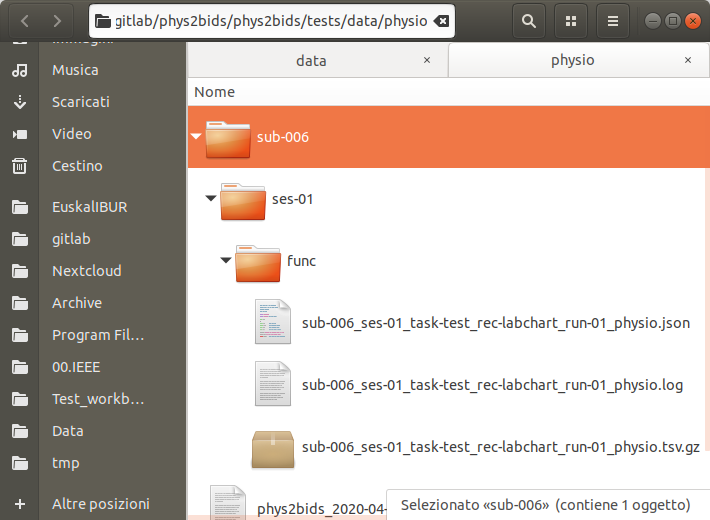
BIDSifying physiological data: phys2bids
physiopy/phys2bids is our flagship repository, introduced in December 2019.
Its aim is reorganising physiological recordings into BIDS format, and it currently supports AcqKnowledge (BIOPAC), Labchart (ADInstruments), Spike2, and GE files.
BIDScoin¹ integrates phys2bids as a plugin, granting it a GUI and facilitating its use.
1. Zwiers, Moia, & Oostenveld, 2022 (Front Neuroinform)


Preprocess physio data: peakdet
physiopy/peakdet was "donated" by its maintaner, Ross Markello, and is currently maintained by physiopy.
Its aim is denoising physiological data and detect peaks in the signal.
It supports automatic peak detection and manual result correction.
Denoising fMRI data: phys2denoise
physiopy/phys2denoise is our second repository, introduced in May 2020.
Its aim is creating denoising regressors for fMRI from physiological recordings.
It is in alpha stage (partially tested code), and it supports common denoising methods based (at the moment) on cardiac and respiratory data.


physiopy's documentation
physiopy's documentation is an important pillar of physiopy's aim, as it is meant to guide new users in their physiological data approach.
We are compiling best practices based on bimonthly discussions within the community.


The gains and hurdles of making science a community effort: the case of Physiopy (MRI Togetherr 2023)
By Stefano Moia
The gains and hurdles of making science a community effort: the case of Physiopy (MRI Togetherr 2023)
CC-BY 4.0 Stefano Moia, 2023. Images are property of the original authors and should be shared following their respective licences. This presentation is otherwise licensed under CC BY 4.0. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/
- 79



