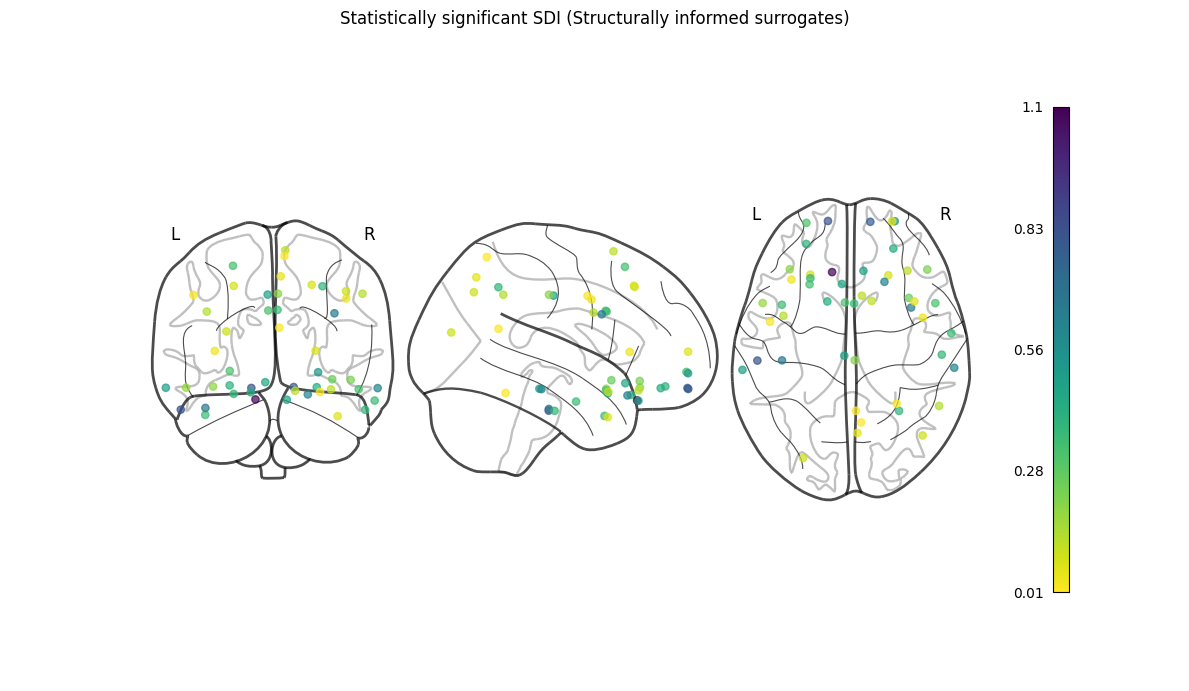
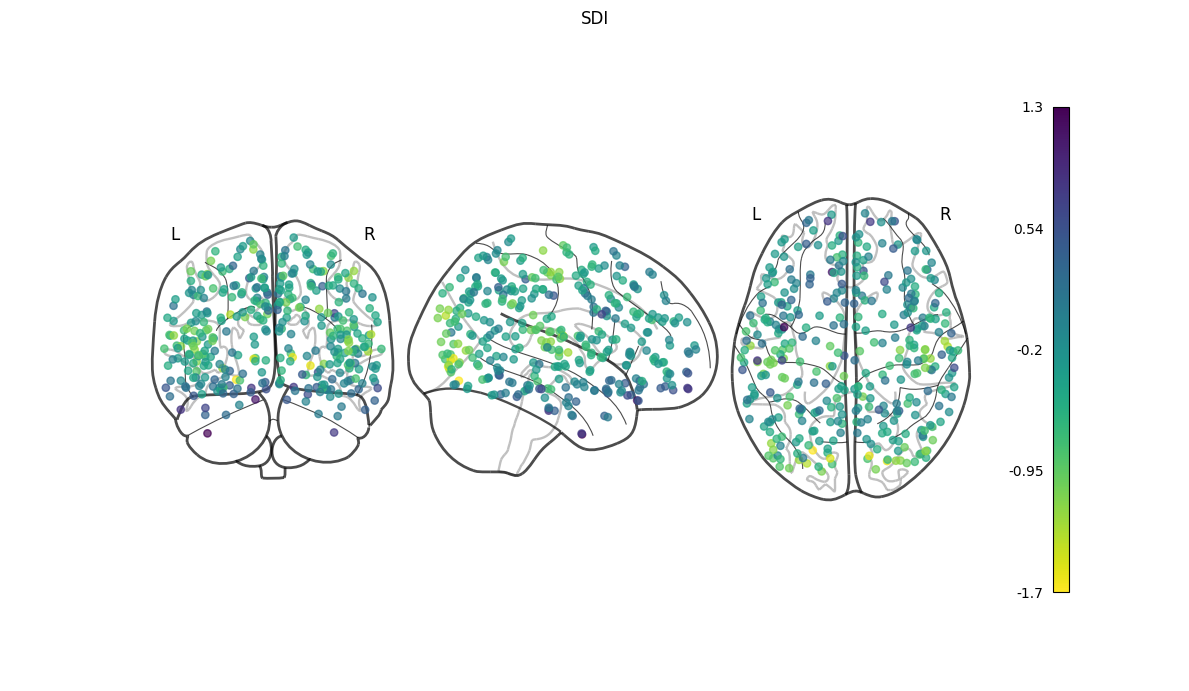
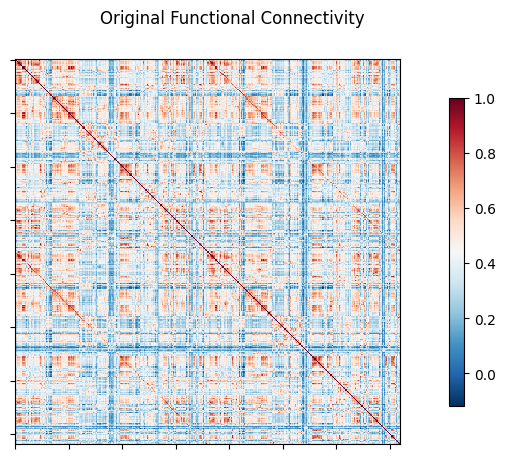
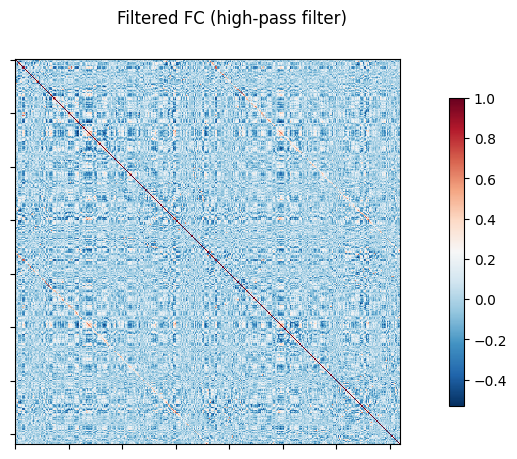
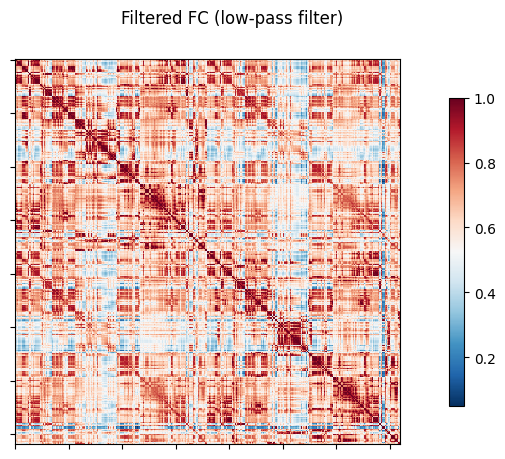
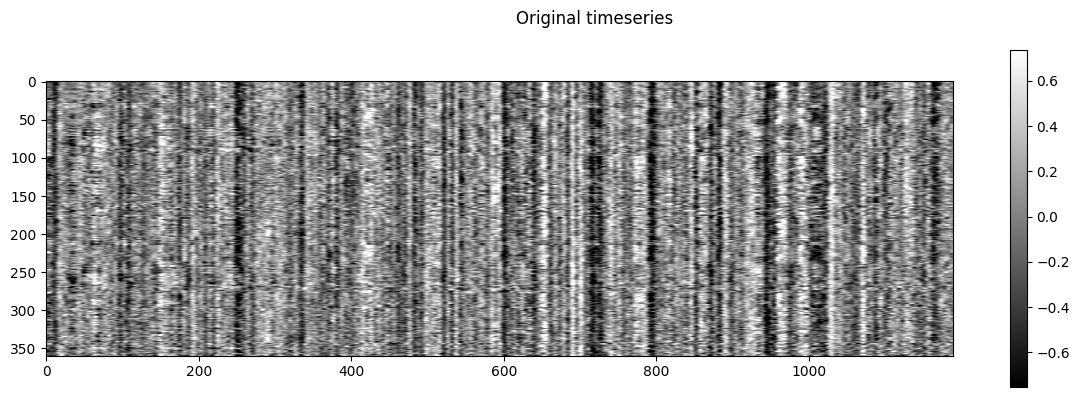
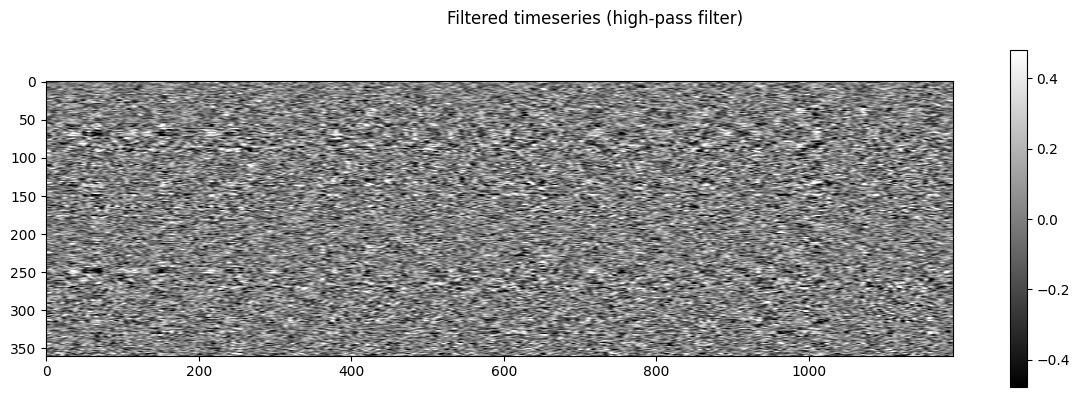
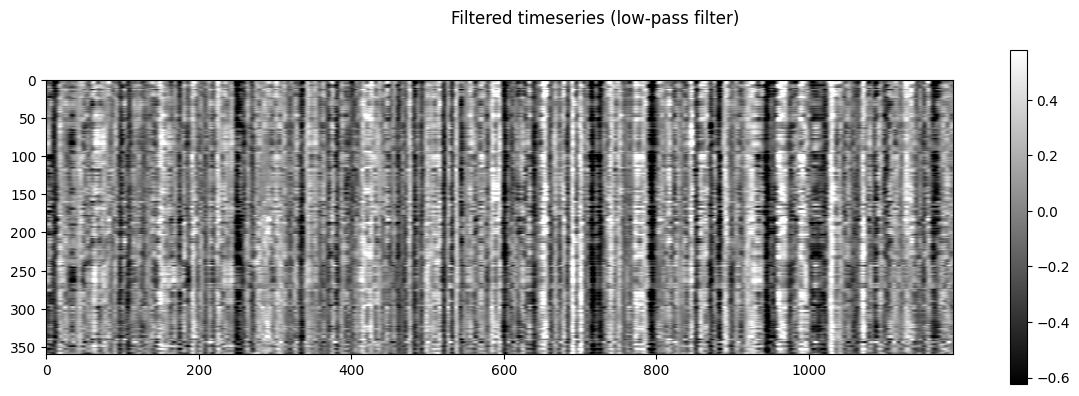
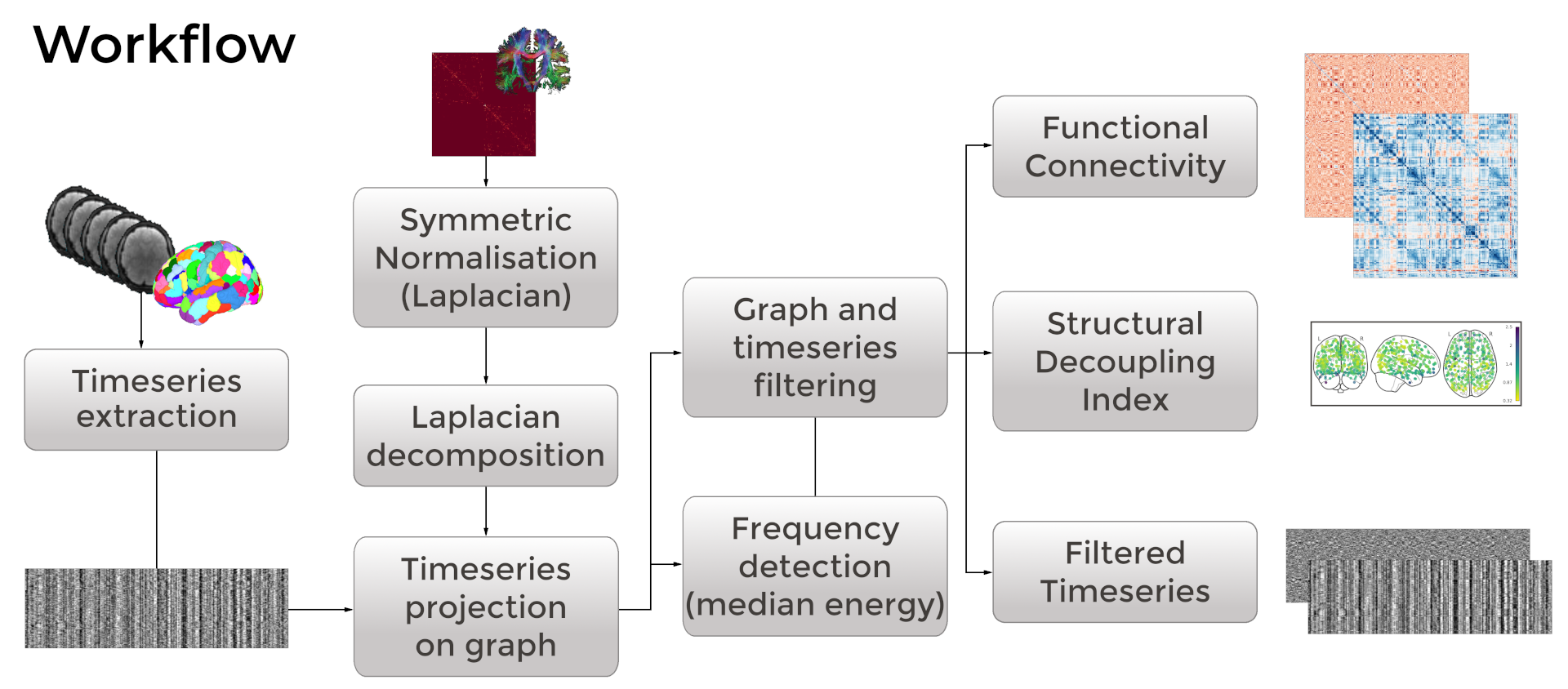
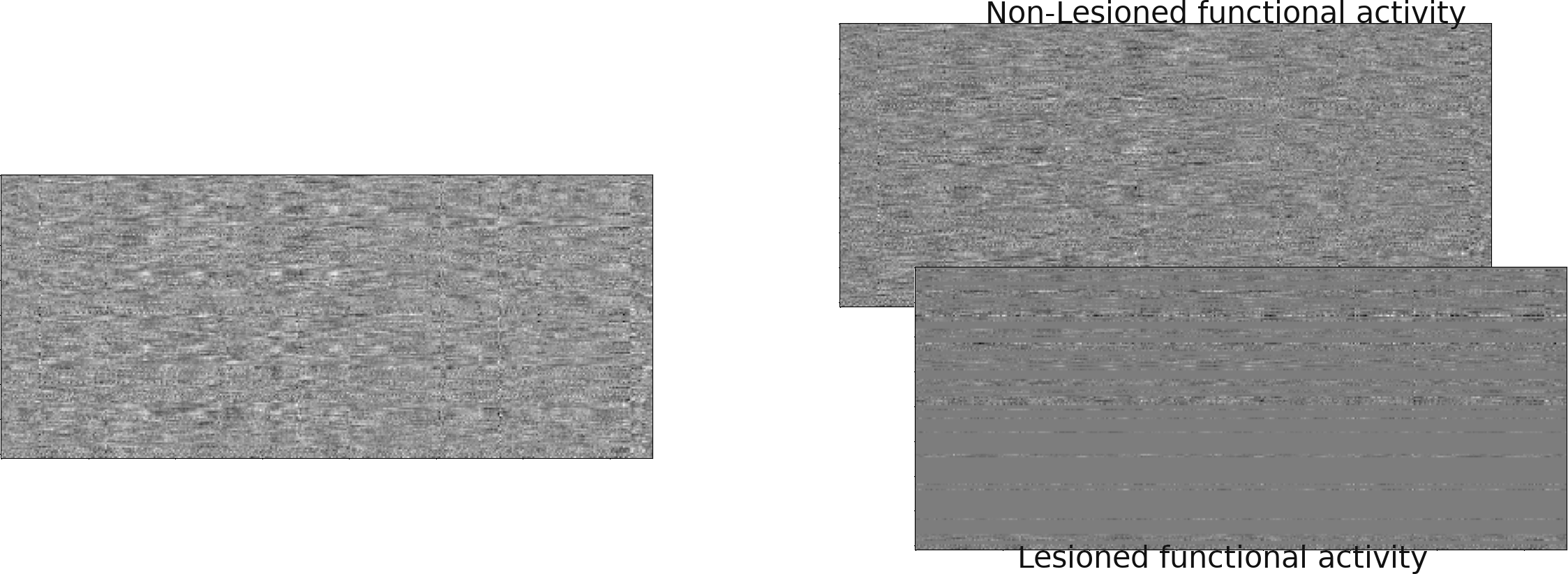
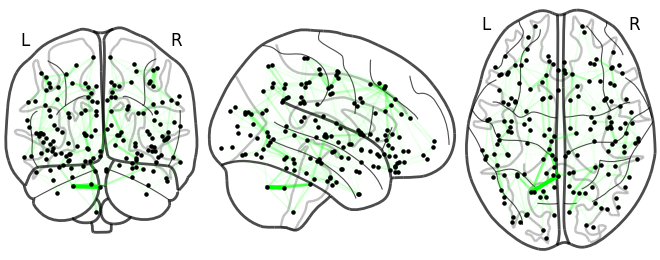
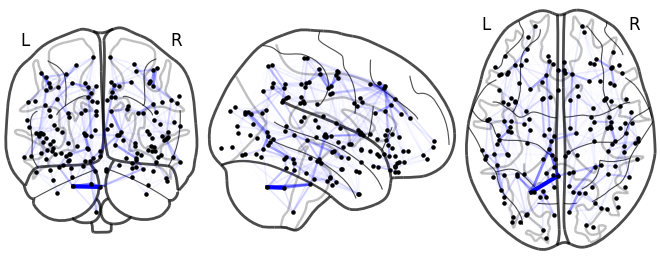
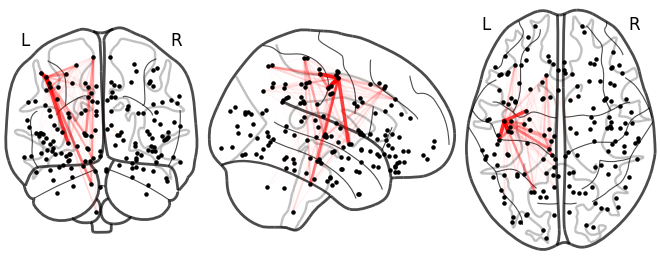
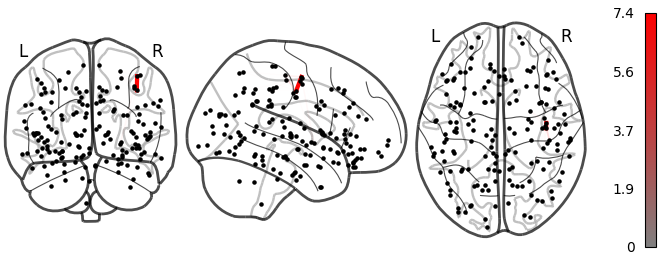
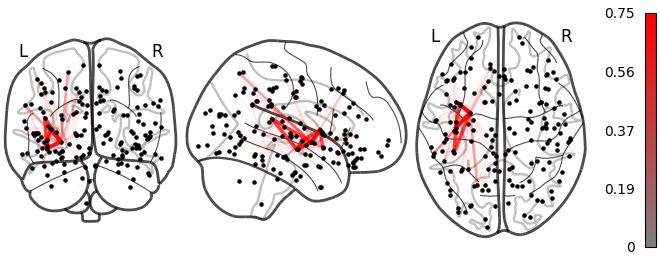
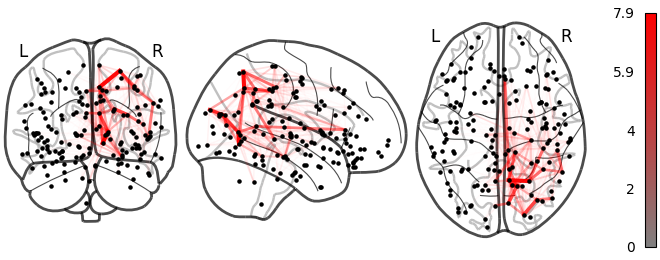
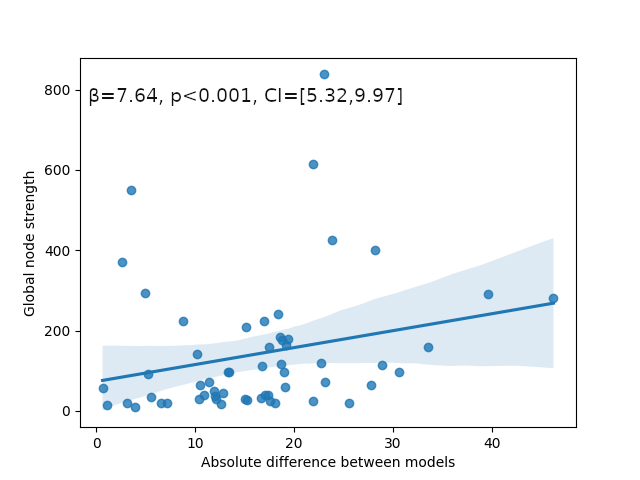
NiGPS: Graph Signal Processing on Multimodal MRI data (ISMRM 2023)
By Stefano Moia
NiGPS: Graph Signal Processing on Multimodal MRI data (ISMRM 2023)
CC-BY 4.0 Stefano Moia, 2023. Images are property of the original authors and should be shared following their respective licences. This presentation is otherwise licensed under CC BY 4.0. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/