Open Development
The journey from developing a deliverable to developing communities and back

| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
Donostia, 22.11.23

Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, The Netherlands; physiopy (https://github.com/physiopy)


Open Development
The journey from developing a deliverable to developing communities and back
Donostia, 22.11.23
Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, The Netherlands; physiopy (https://github.com/physiopy)


Disclaimers
1. I have a bias towards the core tenets of Open Science as better scientific practices.
2. I am also fairly biased by my own experience - what I am saying as a whole might not hold for everyone, but hopefully you can get some inspiration from it.
3. In fact, let me come clean now: not everything I'm about to tell you worked for me.


0. Rules & Materials


You're asking questions,
I'm doing that too!
This is a new chapter
Take home #0
This is a take home message
1. Terminology


Replicable, Robust, Reproducible, Generalisable

The Turing Way Community, & Scriberia, 2022 (Zenodo). Illustrations from The Turing Way (CC-BY 4.0)
Guaranteeing reproducibility is important for "reusable, transparent" research.
1
2
3
4


Open Development
Open (Source Scientific Deliverable) Development: the idea of developing a scientific deliverable:
- in an open and public way
- sharing it from the beginning of the development
- fostering a democratic community of contributors in support of the project
- acknowledging all contributions
- using version control and (any sort of) testing when necessary.
Two main elements: the deliverable itself and the community around it.
The deliverable is not necessarily code based!


2. Starting the process


What is the first step in project development?
How do we do Open Development?
- Identify the "optimal form" of your deliverable.
- Find open development projects that get you the closest to your "optimal deliverable", contribute if needed/possible, check licences.
- Decide what (permissive) licence the deliverable and any other artefacts you might create during the development will be released under.
- Decide and publish contribution rules.
- Start open development (share it!): use VCS, test, document the process, write SoPs.
- Standardise the development environment: adopt styles and style-checks, add metadata, create containers.
- Automatise whatever you can automatise: set up CI/CD workflows
- Create releases and assign DOIs to them.
- Foster a community around the deliverable.
- Publish open access.
- Remove embargoes.


The first step in (open) development






Pay attention to the contribution files
Independently from its kind, projects can accept different types of contributions.
Different communities may have different entry requirements, contribution recognitions, or follow different contribution workflows.
Look for a contributors' guidelines
(and a code of conduct).



The "other first step"


| Minimum Viable Product | Unique features |
|---|---|
|
The necessary features |
The feature nothing else has |
| Synergies | Competitors |
|---|---|
|
Projects or deliverables that match (part of) the must have features - How can I collaborate with them? |
Projects that offer the same but do not accept collaboration - Why? |
The "other first step"


| Minimum Viable Product | Unique features |
|---|---|
| Description of practical operations Cover respiratory data Cover cardiac data Bad data examples |
Community driven Version controlled Yearly reviewed |
| Synergies | Competitors |
|---|---|
| neurokit ICP Network physIO toolbox Pinto 2018 paper |
... |
Take home #1
Don't reinvent the wheel:
look for what you need, it might be out already in some other form!
Contribute to development or join a community: it might seem harder at first, but it will provide better artefacts (and improve your network!)


Disclaimer:
I am not a legal expert.
If you ever have any doubts, contact the Technology Transfer Office
of your University.


License your work
A work that is not licensed is not public (paradox!)
There are n+1 (open source) licences to pick up from.
www.choosealicense.org
The licence should be the first commit you make in a project.
Personal picks for science:
Apache 2.0 and CC-BY-ND-4.0
(consider L-GPLv3.0, and CC-BY-4.0 too)


Understand licensing and ownership
- Check the licence of code, data, and libraries you are "borrowing".
- Pay attention to single vs double licensing (e.g. academic vs commercial).
- Check licence compatibility.
- Remember that institutions might have rights to what their employees do:
- However, they can also help you with licensing and license enforcement.
EPFL is the owner of its employees’ inventions and software. Inventors or authors in case of software have the right to one-third of net revenue resulting from the commercialization of their inventions with some exceptions according to directives.


License your work in the right way
- Put a copy of the licence or a link to it as close as possible to "borrowed" material, if not in it.
- If any license requires its adoption for derivatives (e.g. GPL), you must licence your work with the same licence.
- You can ask the original authors to change their licence (e.g. GPL to L-GPL) or give you special permissions.
- Remember to add licences disclaimers in all of your files.
[...]
if __name__ == "__main__":
_main(sys.argv[1:])
"""
Copyright 2022, Stefano Moia & EPFL.
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
"""


Licence compatibility
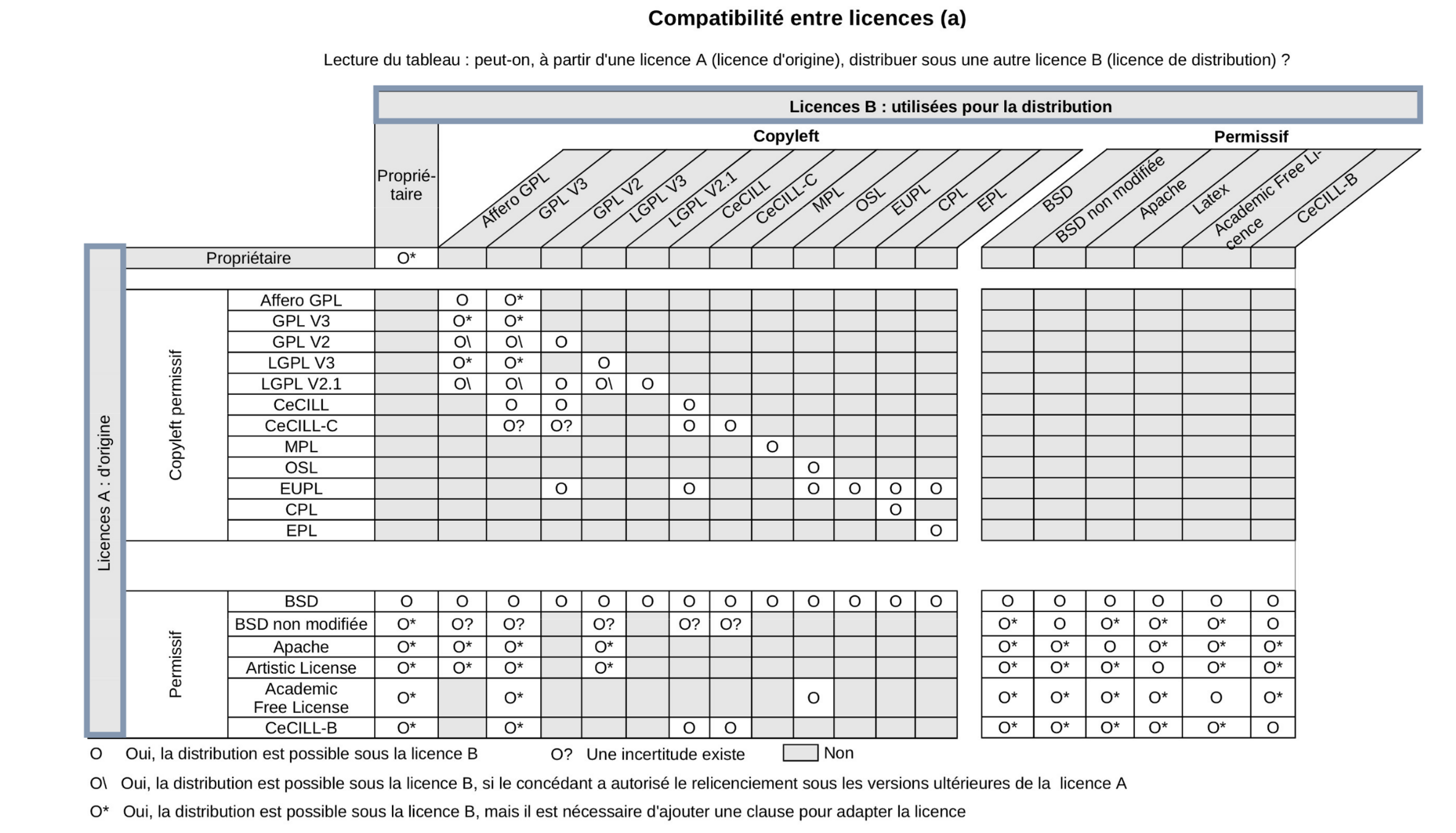
© Sebastien Adams, I WANT TO DISTRIBUTE MY SOFTWARE DEVELOPMENTS. HOW TO DEFINE AN OPEN LICENSING STRATEGY?
© Benjamin Jean (2011), Option libre. Du bon usage des licences libres.


License your work in the right way
MATLAB users:
- If you include external functions/scripts/libraries, your work is considered a derivative. Report licence, authors, and origin of the code inside them and respect their licence.
- Alternatively, don't include anything but state requirements / create install scripts.
- If you are releasing a build, the build is considered a derivative.
Python users:
- If you copy-paste code, your work is a derivative.
- Imports are trickier:
- Technically, GPL or © licences triggers on import.
- Practically, it's a really grey area. Make those imports optional, and specify their licences as clearly as possible.


License your work in the right way

[...]


Take home #2
Licensing is as complicated as it is important.
Double check licenses of borrowed material, report them in your own work
for licence tracking.
Licence your work
and implement the licence properly.


3. Your future self best friend


Does any of these situations look familiar?
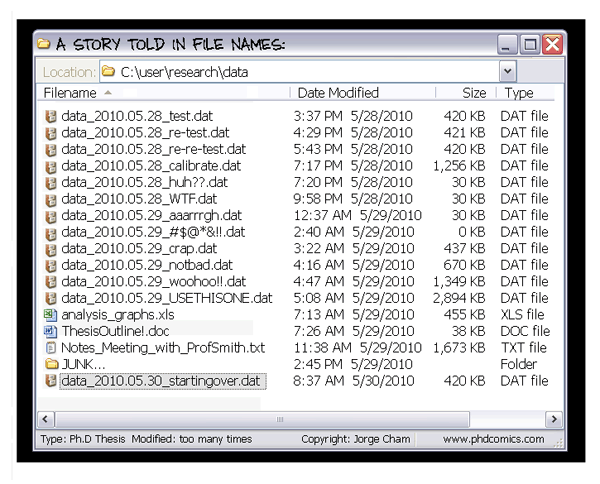
I can't work on that project now because my colleague/friend/dog is working on [a different part than what I'd modify of] it at the moment...


Version Control Systems
Version Control Systems
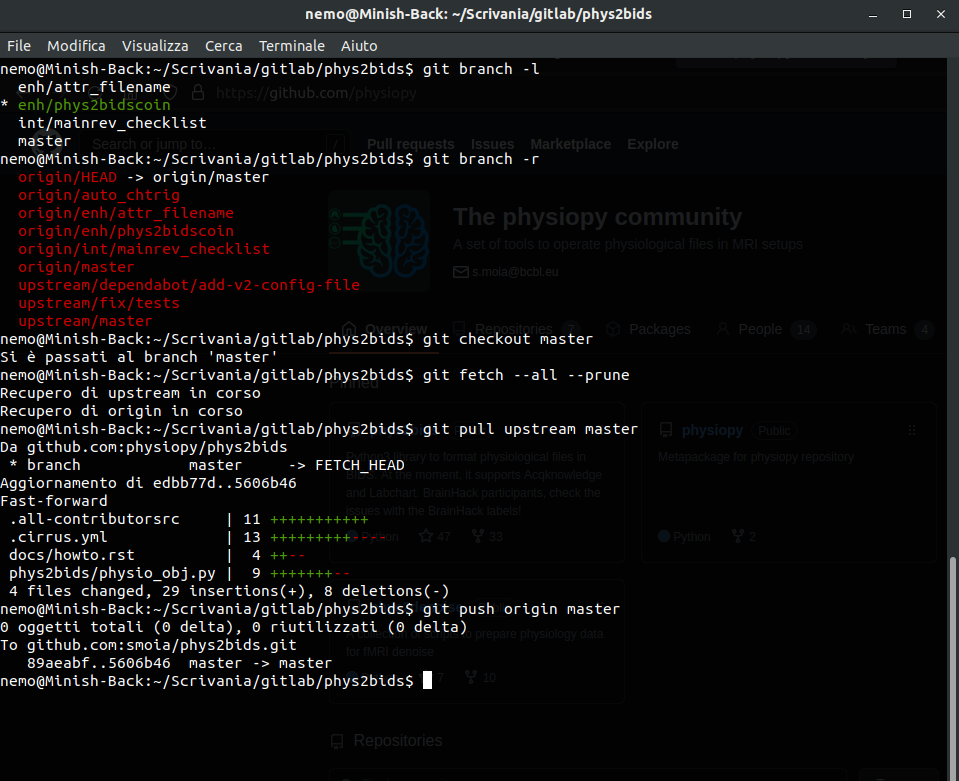



Version Control
Version control systems are a way to manage and track changes to files.







Content
Aggregation/delivery


VCS for data
File history & parallel development
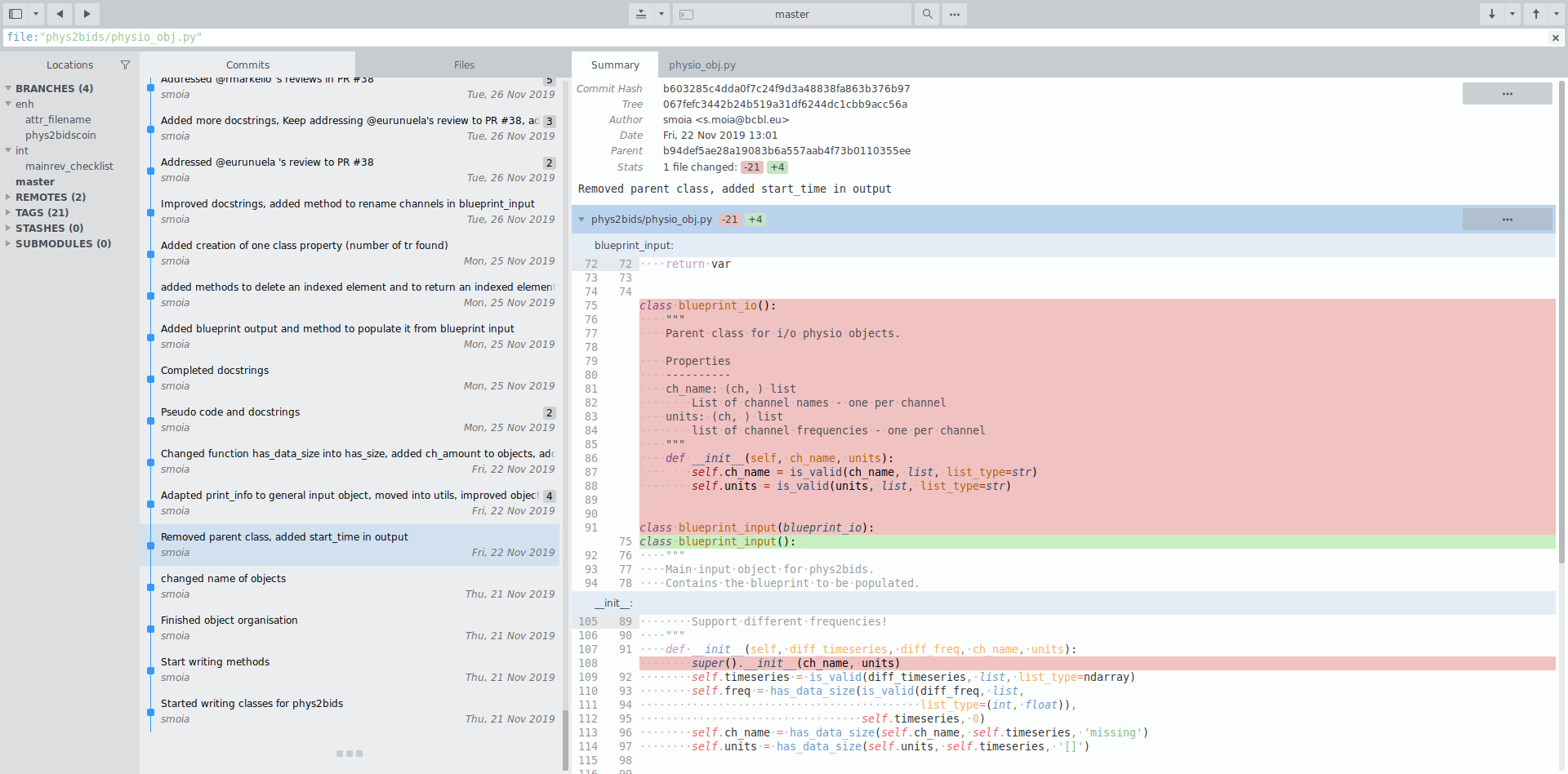
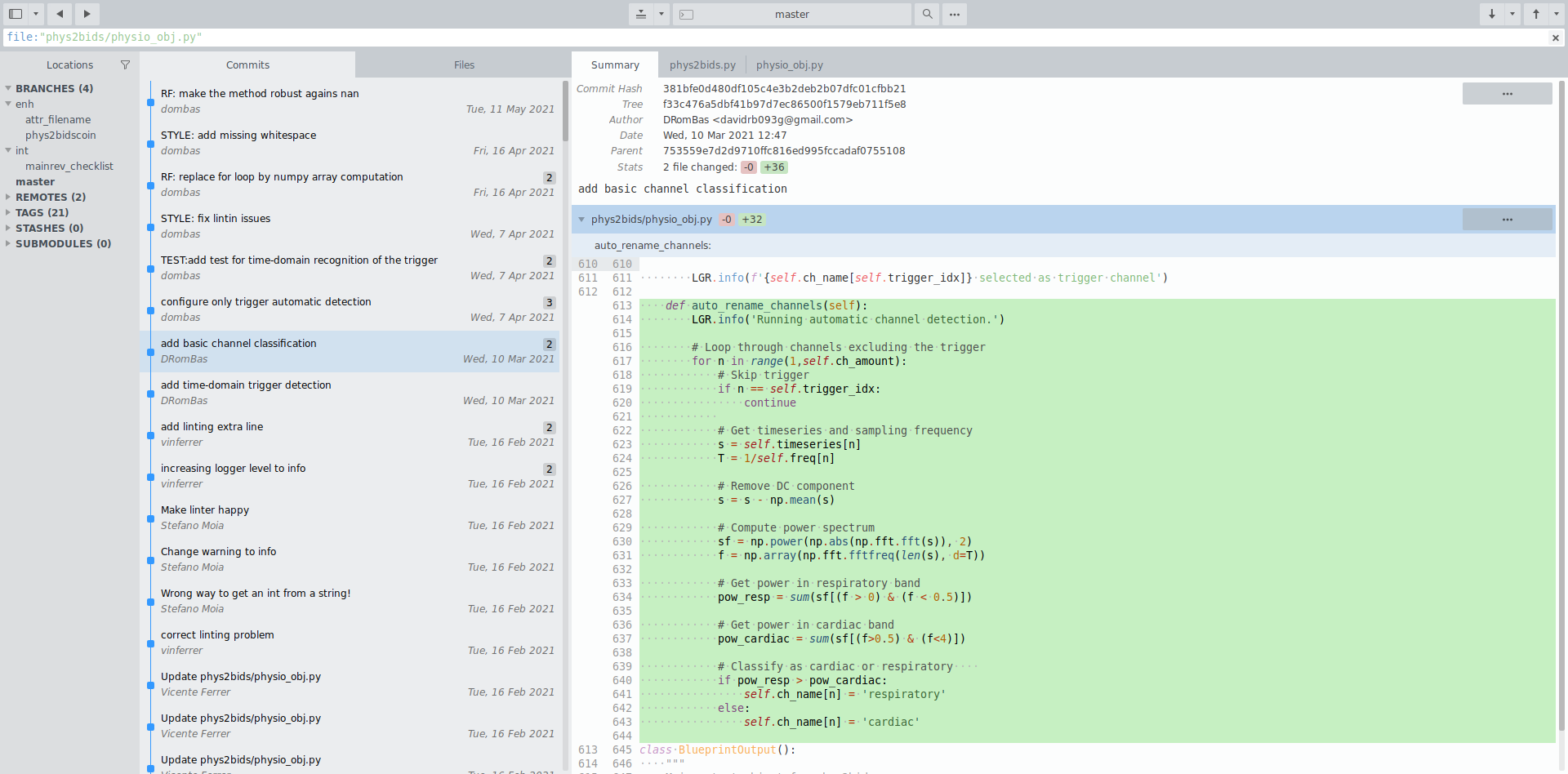
Attribution
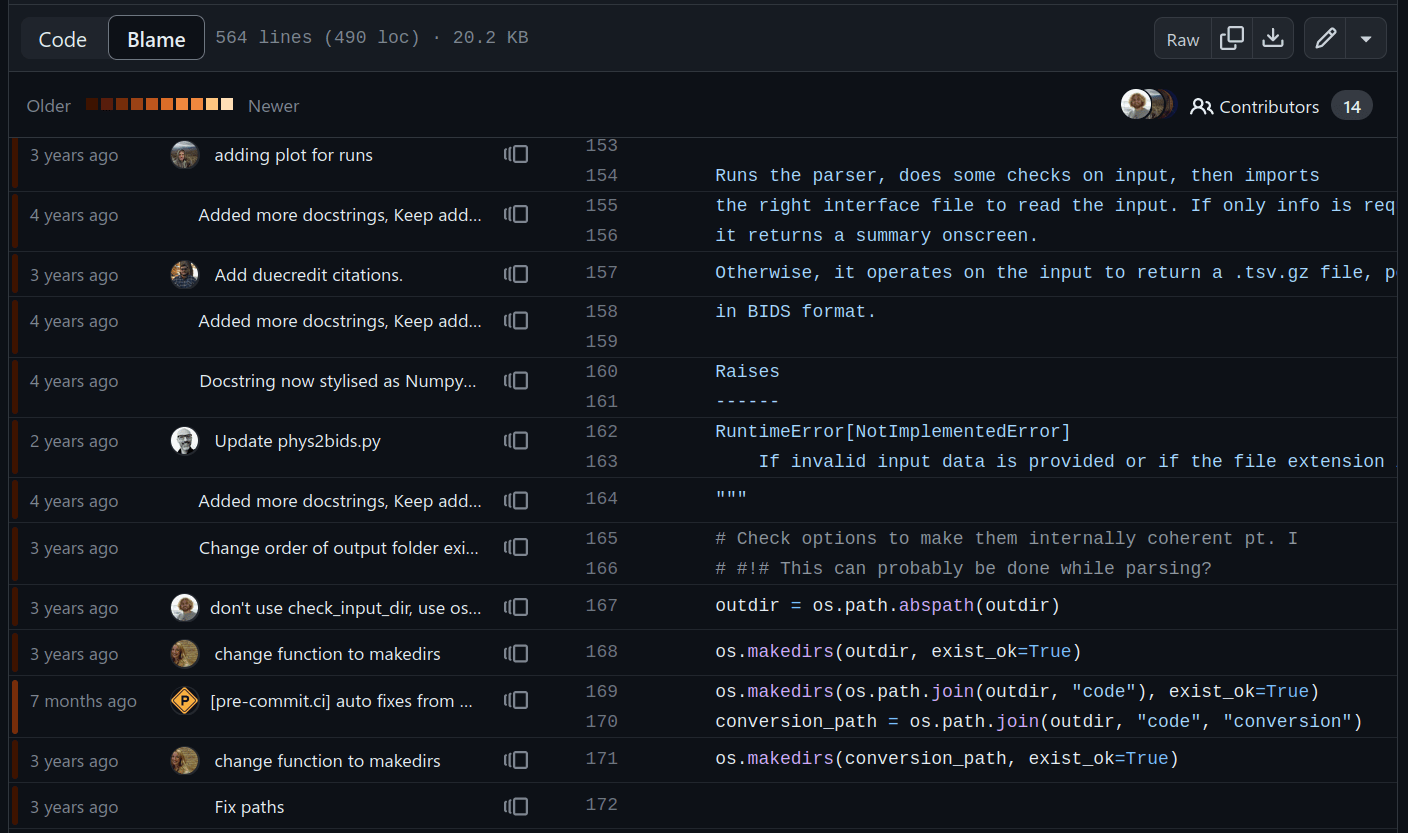


Parallel working


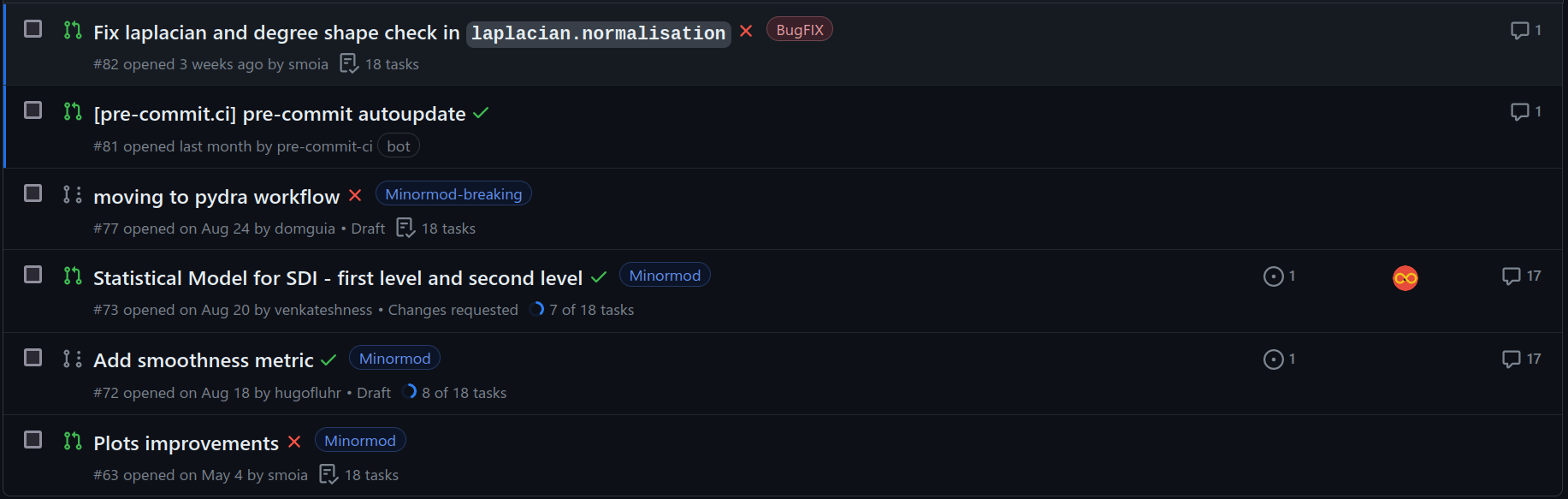
Automation pt. 1: git hooks
pip install pre-commit # Install via pip, or
# Comes installed with development extras
pip install -e /path/to/phys2cvr[dev]
cd /path/to/phys2cvr
pre-commit init
pre-commit run

(Local and remote) simple automations, e.g:
- Code style (black, isort, ...)
- File checks (empty lines, indent, executables)
- Language and typos (!!!)
Take home #3
Working with VCS allows you to:
-
track changes in time
-
access to automations
-
work in parallel on new features without disrupting the "main" version of your project
Bonus: it can force a team to double check projects!


4. Going public the right way


Test your deliverable
Testing a project is as important as developing it.
Arguably, it's even more important, so spend time on it!
There are multiple types of tests:
- User tests: a person (user) uses the tool and make a report on it.
- Automated tests: developers write tests to be run after each change.
- Unit tests: they test (new) parts of the project on their own.
- Integration/End-to-end tests: they test the project as a whole.


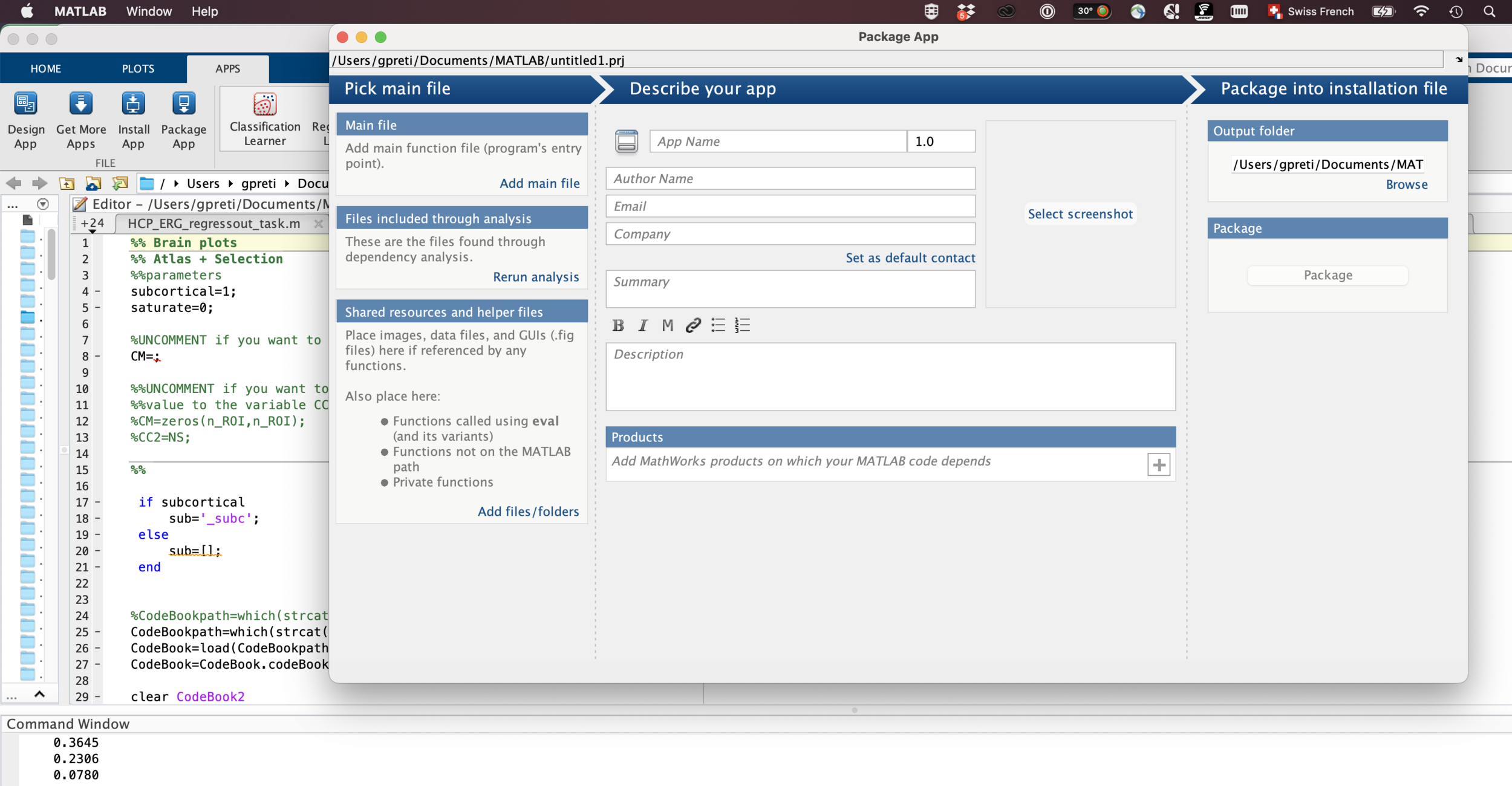
Make releases
Releases make your work easier to retrieve (and cite).
Imagine them as hard links to a certain moment in time.
(e.g. paper #1 vs paper #2).
You can create, package, and distribute releases, all automagically through automatic workflows.


MATLAB packaging
*feat Giulia's laptop


Make your project identifiable (and citable)
{
"license": "Apache-2.0",
"title": "physiopy/phys2bids: BIDS formatting of physiological recordings",
"upload_type": "software",
"creators": [
[...]
{
"orcid": "0000-0002-7796-8795",
"affiliation": "Florida International University",
"name": "Katie Bottenhorn"
},
[...]
],
"access_right": "open"
}

Publish!
If your project is software related, think about publishing it.
While there are various journals that can be targeted for a software publication, JOSS is free and completely integrated in GitHub.


If your deliverable is data, SOPs,
or documentation:
publish those as well!
Take home #2
Go public the right way:
-
make a release of your deliverable,
-
assign a DOI to it,
-
publish!


5. Automation


Let bots do the hard work
Automated workflows are your friend.
Everything can be automated, from testing to releasing (packaging end etc.).
Workflows can require a bit more work to be set up, but they can save a lot of time and energy in the long run!







Automation at work
- Continuous Integration: frequently integrating new changes into the main branch of a tool. Normally, workflows run automatic steps at each integration, e.g. automatic testing.
- Continous Deployment: frequently deploying (releasing) new versions of a tool using automated workflows (e.g. right after integration).





Automation at work



- Pre-commit [local, remote]: Automate code checks and styling on git commit
- Pytest, pytest-cov [local, remote]: (automated) testing and coverage
- Codecov [remote]: automated code coverage change check
- Auto [remote]: automated version update, tag, release, and changelog
- Zenodo, PyPI [remote]: automated DOI and package publishing
- Readthedocs [remote]: (automated) documentation publishing

pip install pre-commit # Install via pip, or
# Comes installed with development extras
pip install -e /path/to/phys2cvr[dev]
cd /path/to/phys2cvr
pre-commit init
pre-commit run


Let's not
reinvent the wheel
Take advantage of the marketplace: there is a very high probability that what you are looking for is already available.


Take home #4
Set up automated workflows
to manage your project development.
They will not only help you, but also increase the stability and reliability
of your outcome.


6. Communities


What is the first step in project development?
Contributions and communities
- The development can be very driven and focused on key points.
- Decision making is quick.
- Users might not be engaged enough to value your project.
- One developer = more time needed for new features, less reviewing.
- Less stability in the group = more time training.
- Smaller user base = deliverable is less tested and consensus is not guaranteed.
- The development can become more based on the help of the volunteers.
- Decision making is slower.
- Sense of involvement and responsibility might increase recognition!
- Many developers = less time needed for new features and better quality!
- More involved people = more mentors.
- Bigger user base = better tested deliverable and widespread consensus.


Recognise your contributors
Depending on the community and the governance scheme, contributions might be recognised differently.
Be clear about how you will recognise contributions.
One way of recognising contributors is the all-contributors specification.


Aim for readability
Improve the readability of your deliverable
a, b = rui()
c = s(a, b)
p(c)a, b = read_user_input()
c = sum_two_numbers(a, b)
print(c)def very_important_function(template: str, *variables, file: os.PathLike, engine: str, header: bool = True, debug: bool = False):
"""Applies `variables` to the `template` and writes to `file`."""
with open(file, 'w') as f:
...
def very_important_function(
template: str,
*variables,
file: os.PathLike,
engine: str,
header: bool = True,
debug: bool = False,
):
"""Applies `variables` to the `template` and writes to `file`."""
with open(file, "w") as f:
...



Discuss good practices


Take home #5
There are advantages and disadvantages in any form of governance - choose yours from the start and state it clearly.
In any case, don't forget to recognise other people's work and take time to develop the community!


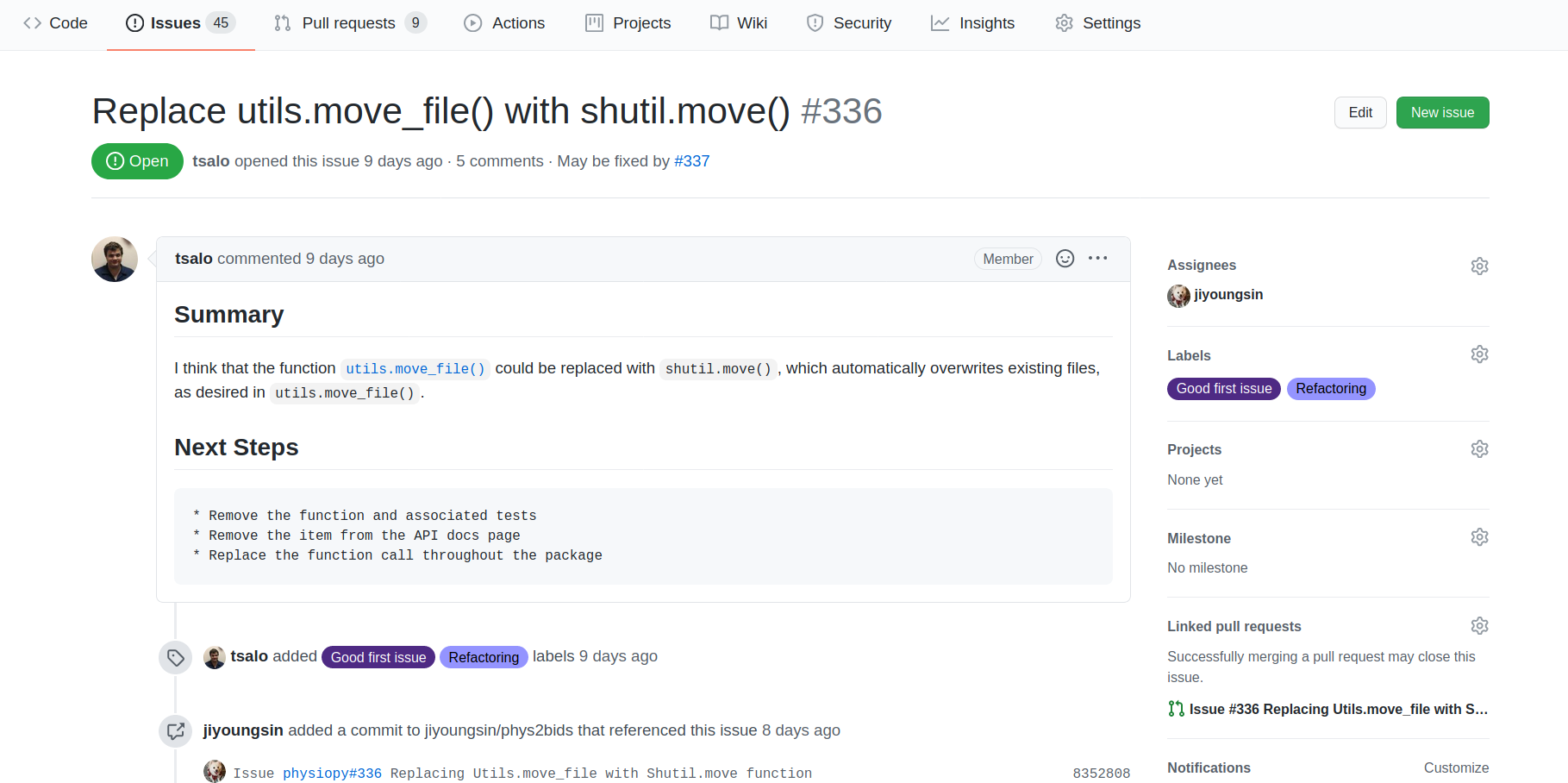
Guide the development: issues
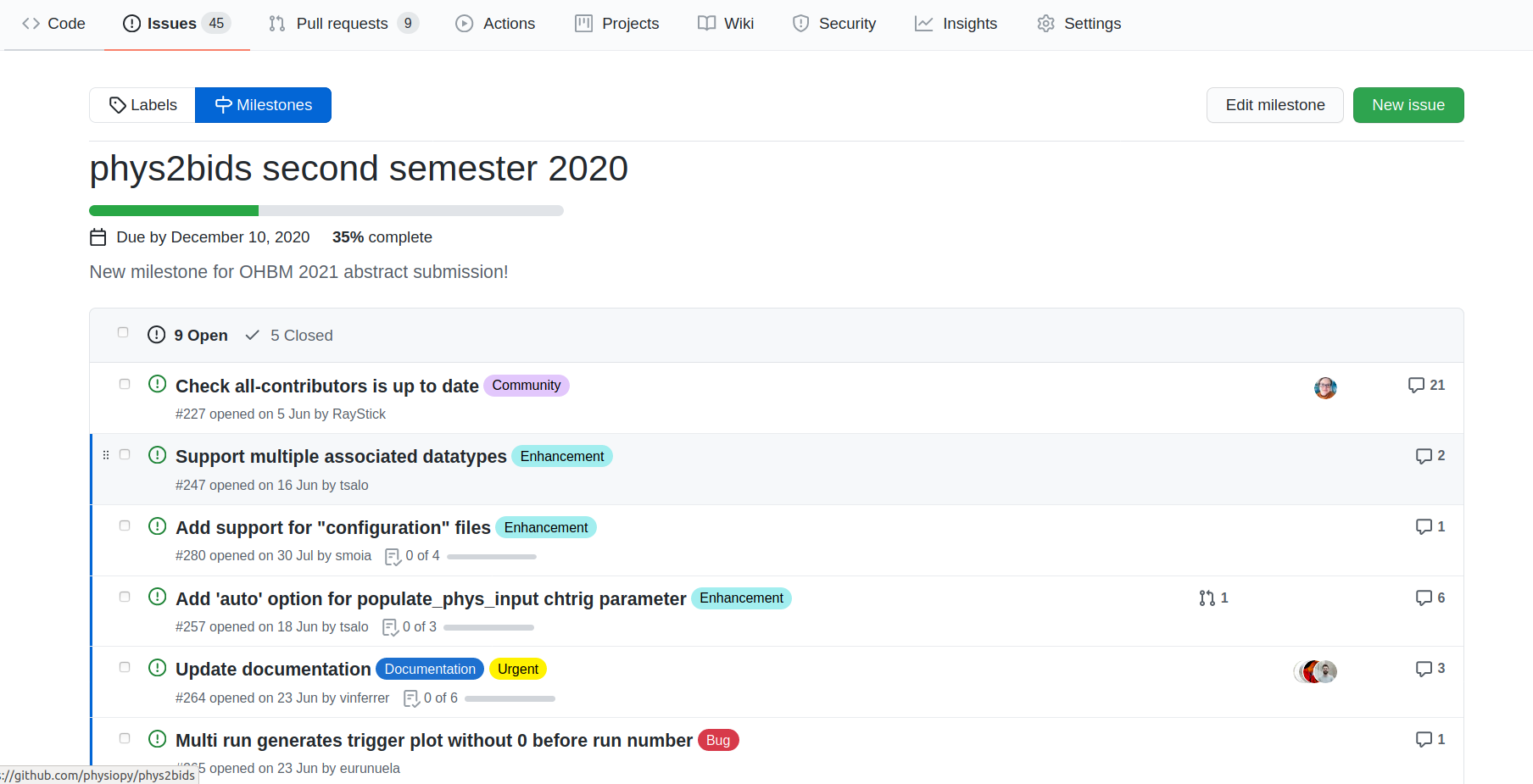
Guide the development: milestones
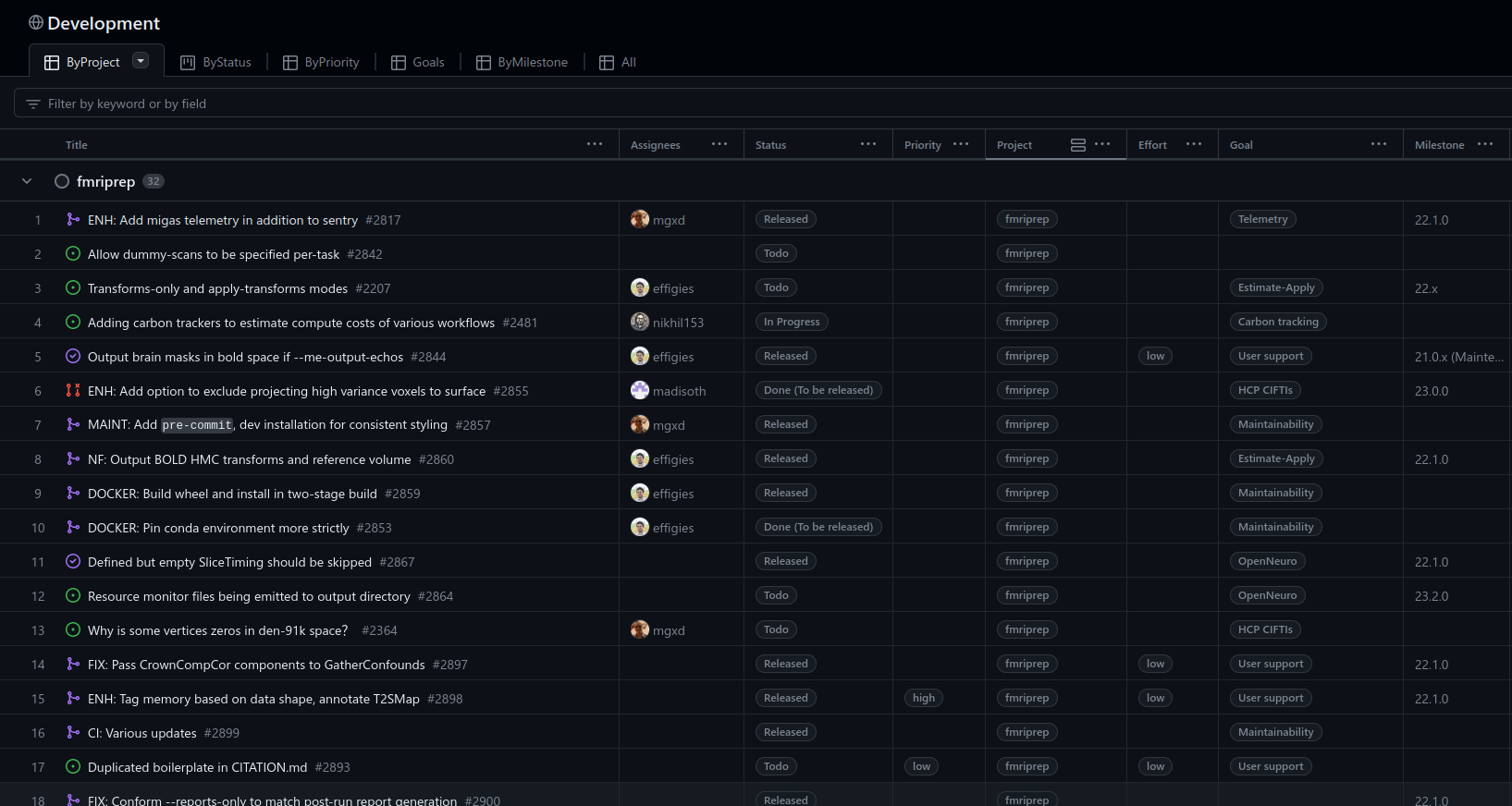
Guide the development: projects
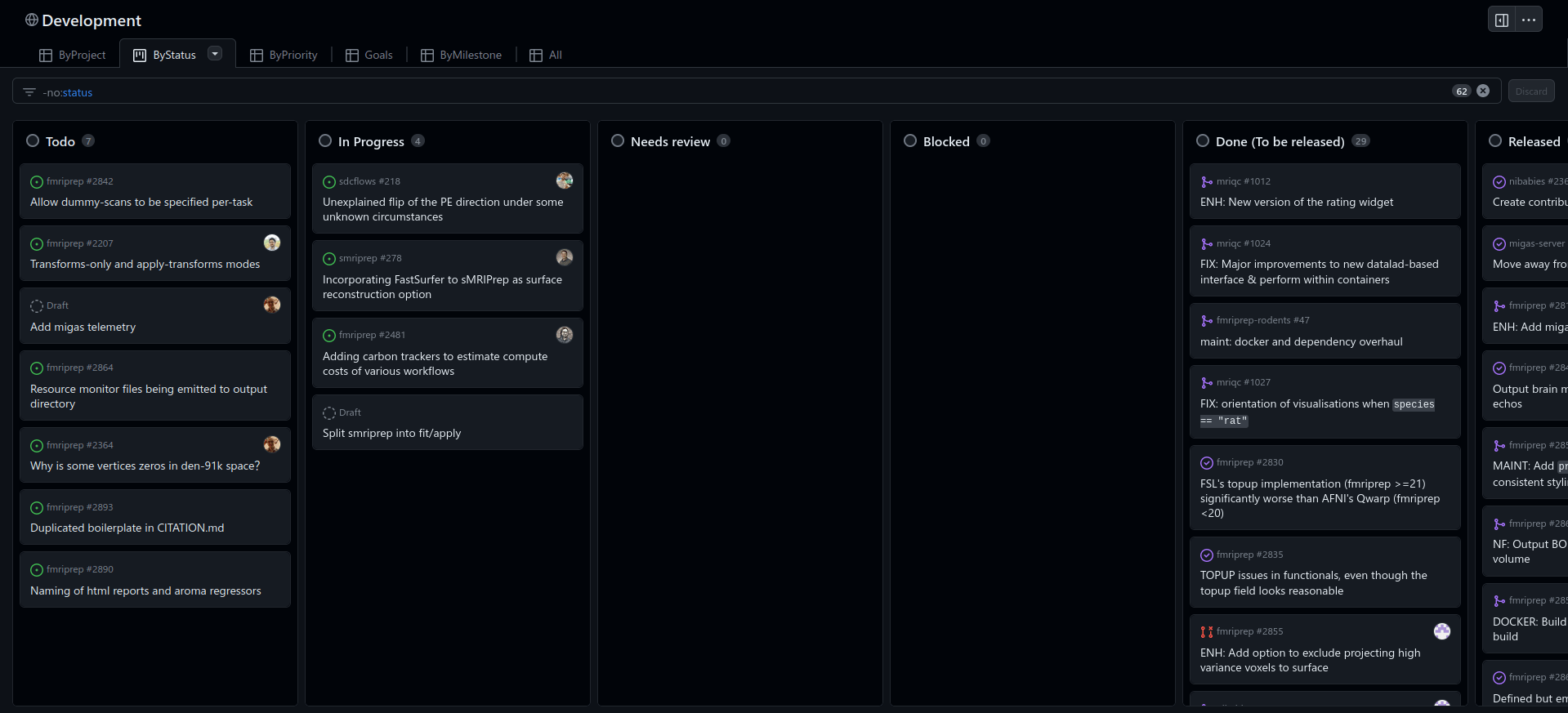
Take home #5
Guide the development of your project using issues, milestones, and labels:
-
it will make your life easier and better organised.
-
it will make your project easier to understand for third parties.


7. Standardisation
(or: reproducible pipelines)


Standard Operating Procedures
https://github.com/TheAxonLab/hcph-sops
Data standards & metadata

1. Gorgolewski, et al., 2016 (Scientific Data) 2. Zwiers, Moia, Oostenweld, 2022, (Front. Neuroinf.)

Data standards & metadata
Containerisation




Docker vs Apptainer


Bootstrap: docker
From: python:3.8.13-slim-buster
%environment
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
%post
# Set install variables, create tmp folder
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
# Prepare repos and install dependencies
pip3 install nigsp[all]
# Final removal of lists and cleanup
rm -rf /var/lib/apt/lists/*
FROM python:3.8.13-slim-buster AS nigspdock
WORKDIR /app
# Prepare environment
COPY .. .
RUN pip3 install .[all]
ENV LANG="en_US.UTF-8" \
LC_ALL="en_US.UTF-8"
CMD nigsp
ARG BUILD_DATE
ARG VCS_REF
ARG VERSION
LABEL org.label-schema.build-date=$BUILD_DATE \
org.label-schema.name="NiGSP" \
org.label-schema.description="NiGSP: python library for Graph Signal Processing on Neuroimaging data" \
org.label-schema.url="https://github.com/miplabch/nigsp" \
org.label-schema.vcs-ref=$VCS_REF \
org.label-schema.vcs-url="https://github.com/miplabch/nigsp" \
org.label-schema.version=$VERSION \
org.label-schema.schema-version="1.0"
Docker
Apptainer
BIDSapps: containers for BIDS pipelines

Take home #6
Adopt community data standards
and add metadata
to improve reusability!


Last take home message:
What you do in your scientific work has an impact on society.
It's not about you.
Open science can help you
make it better.


Thanks to...
...you for the (sustained) attention!
That's all folks!
...the organisers, for having me here
...the Physiopy contributors



| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |

(and for taking care of BHD!)
1. Don't reinvent the wheel, contribute to existing projects!
2. Pay attention to Licencing!
3. Working with VCS allows you to track changes in time, work in parallel, and implement automation.
4. Rely on automation to streamline and ease your job.
5. Different governances set ups have different pros and cons, find your favourite way in the middle!
Take home messages
Any question [/opinions/objections/...]?
Oh, and don't forget!

Oh, and don't forget!
