Open science: what, why, how.
Stefano Moia
École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland; physiopy (https://github.com/physiopy)
17.01.2023



| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
Stefano Moia
École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland; physiopy (https://github.com/physiopy)
17.01.2023


| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
Open science: what, why, how.
Licenses, containers,
and a bit more around that.
Stefano Moia
École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland; physiopy (https://github.com/physiopy)
17.01.2023


| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
Open science: what, why, how.
Licenses, containers,
and a bit more around that.
and a bit more around that.
Why I really dislike MATLAB.
(And I like python better).
1. Why Open Science and Open Development?
1. Why Open Science and Open Development?
2. Going public: Licenses, releases, DOIs
3. Run your code nth times: Containers
What is Open Science/Development?
To be Open, Science should be publicly available, reusable,
and transparent¹. It should aim at reaching:

Open Source Software Development is the idea of developing a software publicly, sharing it from the beginning of the development, fostering a democratic community of contributors in support of the project, using version control and software testing to improve quality.
1. The Turing Way Community, 2019 (Zenodo). https://the-turing-way.netlify.app (CC-BY 4.0)
- Open Data
- Open Source Software
- Open Hardware
- Open Access (outreach)
- Reproducible results
- Usable artefacts, independently of setting.
- Complete and clear process documentation
Why Open Science/Development?
OS/D makes academic research available to its real funding bodies: citizens, policy-makers, industry.
Well made OS/D guarantees more certain results and reduces the impact of human error.
It also reduces publication bias¹ ².
Ethics
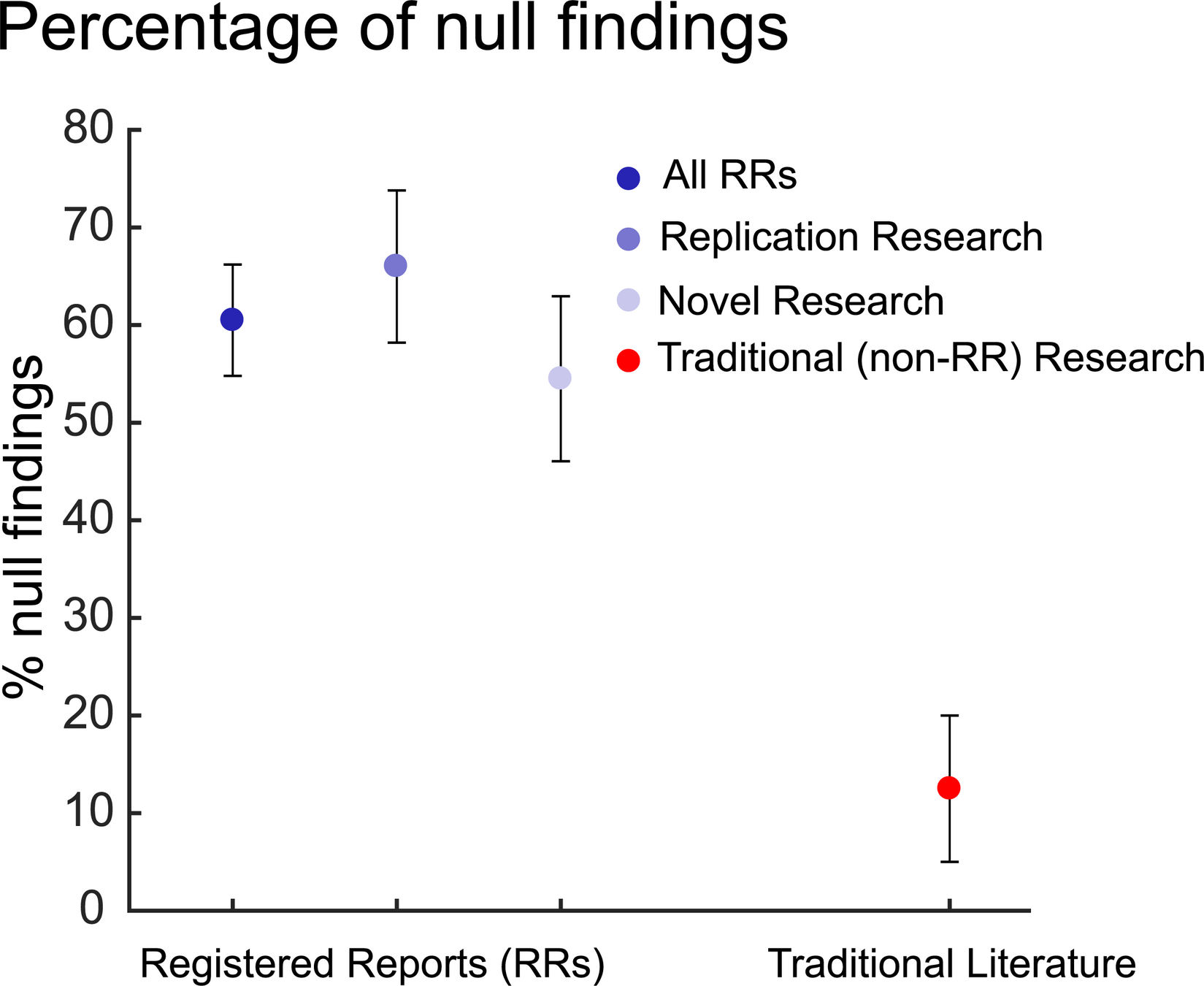
1. Allen & Mehler, 2019 (PLoS Biol) 2. Scheel, Schijen, & Lakens, 2021 (AMPPS)

Why Open Science/Development?
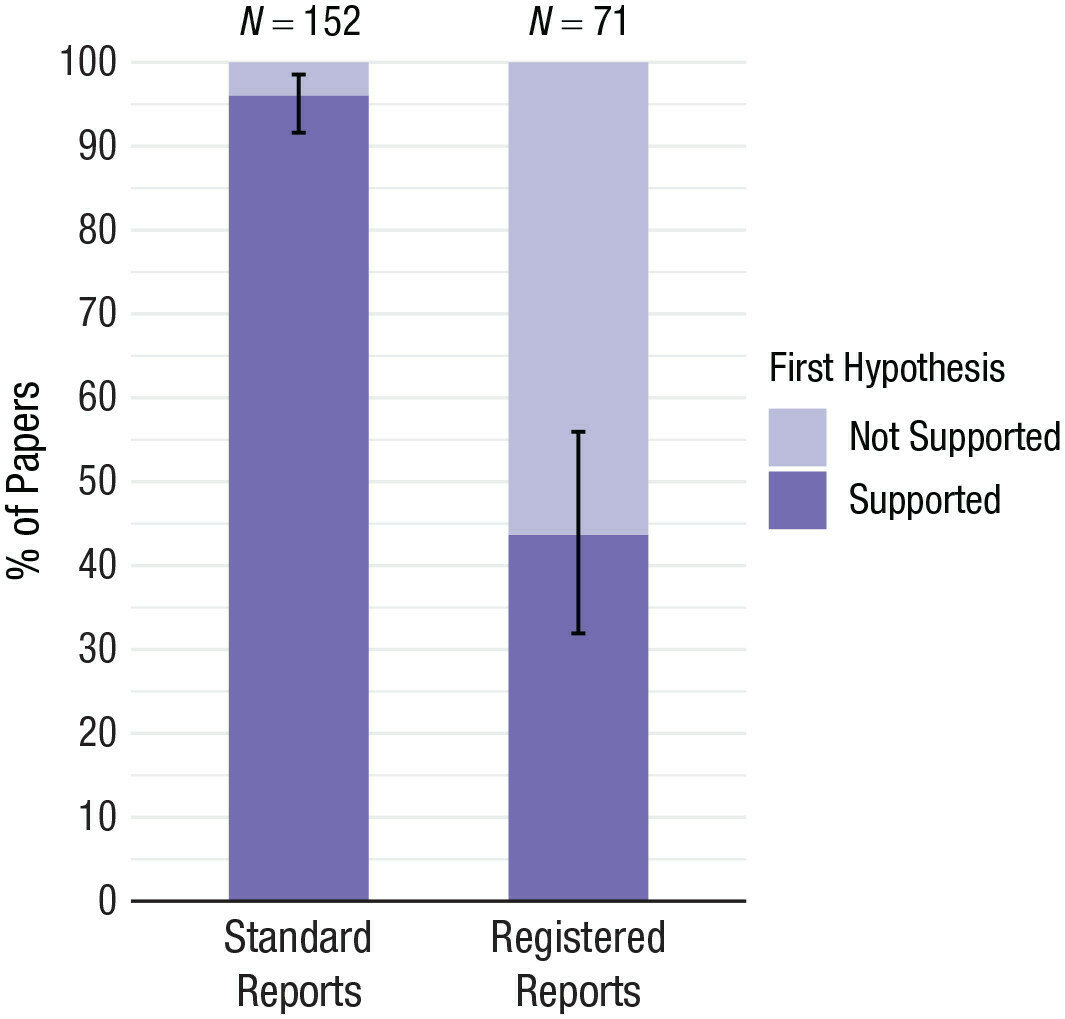
OS/D increases the amount of citable output and the amount of citations¹ ².
Selfishness (career advantage)
1. McKiernan, et al., 2016 (eLife), 2. SpringerNature 2020
Fair warning: it will hardly increase your H-index (current definition).

Why Open Science/Development?
The demand from policy makers is increasing¹.
You have to.
1. McKiernan, et al., 2016 (eLife)

How do we do Open Science?
- Read literature, formulate hypotheses, decide methods, submit a Registered Report (~ Introduction, hypotheses, methods), get through first peer review round.
- Find open development projects, contribute if needed, check licences.
- Decide a (permissive) licence for the artefacts, start open developing code (share it!), use VCS, test and document code, create releases.
- Create containers.
- Collect data, curate them using community schemas (e.g. BIDS), upload them on public databases with embargo ("private time").
- Run analyses in containers, interpret results, write the second part of your Registered Report (~ Results, a posteriori analyses, discussion, conclusions)
- Publish open access.
- Remove embargoes.
- Rinse and repeat.

Take home #1
Make your Science Open.
It will cost time, but it will, eventually, help you.
1. Why Open Science and Open Development?
2. Going public: Licenses, releases, DOIs
3. Run reliable code: Containers
Disclaimer:
I am not a legal expert.
If you ever have any doubts, contact the Technology Transfer Office
of EPFL or UniGE.
License your work
A work that is not licensed is not public (paradox!)
There are n+1 (open source) licences to pick up from.
www.choosealicense.org
The licence should be the first commit you make in a project.
Personal picks for science:
Apache 2.0 and CC-BY-ND-4.0
(consider L-GPLv3.0, and CC-BY-4.0 too)

Understand licensing and ownership
- Check the licence of code, data, and libraries you are "borrowing".
- Pay attention to single vs double licensing (e.g. academic vs commercial).
- Check licence compatibility.
- Remember that institutions might have rights to what their employees do:
- However, they can also help you with licensing and license enforcement.
EPFL is the owner of its employees’ inventions and software. Inventors or authors in case of software have the right to one-third of net revenue resulting from the commercialization of their inventions with some exceptions according to directives.

Licence compatibility
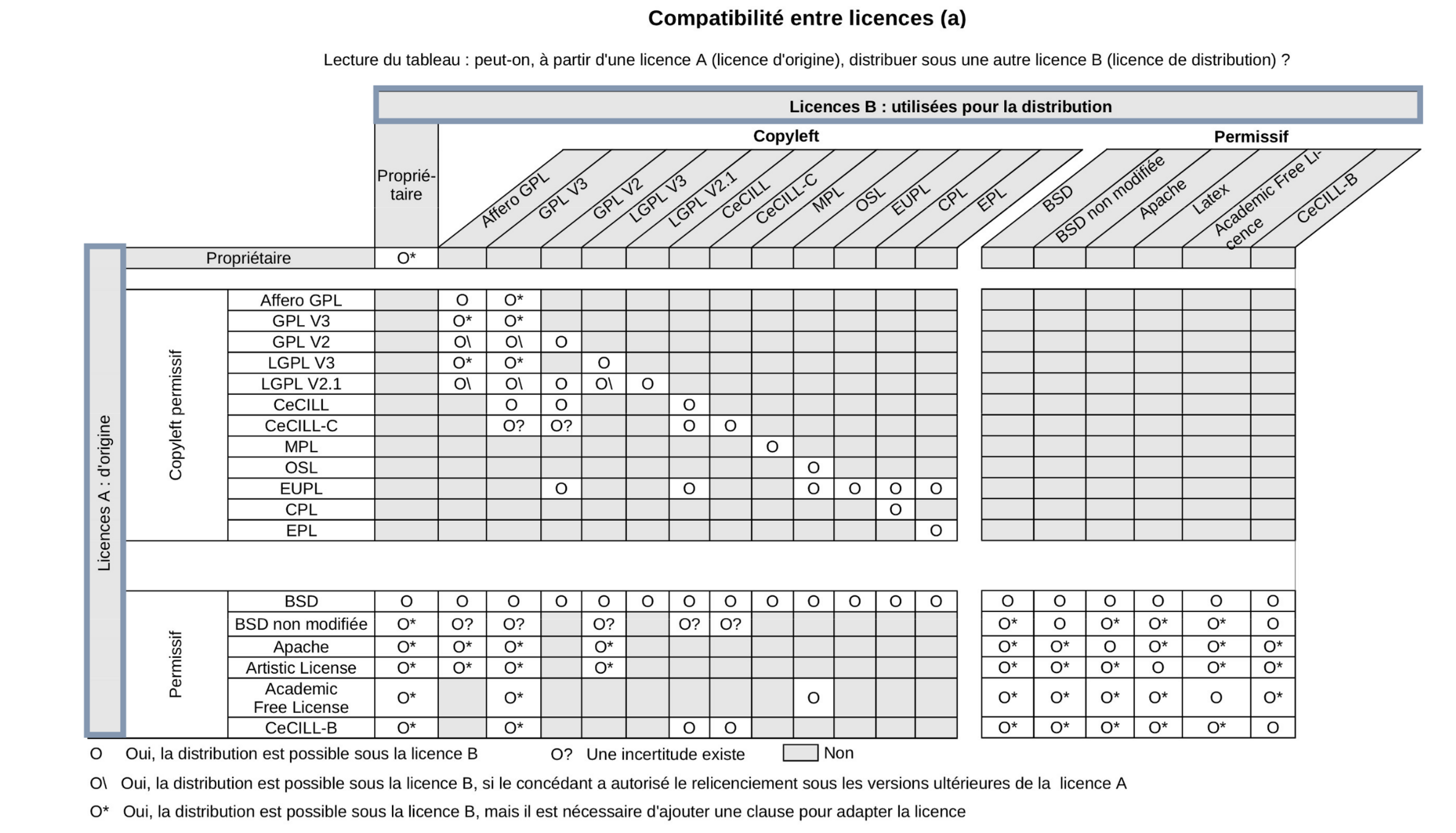
© Sebastien Adams, I WANT TO DISTRIBUTE MY SOFTWARE DEVELOPMENTS. HOW TO DEFINE AN OPEN LICENSING STRATEGY?
© Benjamin Jean (2011), Option libre. Du bon usage des licences libres.

License your work in the right way
- Put a copy of the licence or a link to it as close as possible to "borrowed" material, if not in it.
- If any license requires its adoption for derivatives (e.g. GPL), you must licence your work with the same licence.
- You can ask the original authors to change their licence (e.g. GPL to L-GPL) or give you special permissions.
- Remember to add licences disclaimers in all of your files.

[...]
if __name__ == "__main__":
_main(sys.argv[1:])
"""
Copyright 2022, Stefano Moia & EPFL.
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
"""
License your work in the right way


[...]
License your work in the right way
MATLAB users:
- If you include external functions/scripts/libraries, your work is considered a derivative. Report licence, authors, and origin of the code inside them and respect their licence.
- Alternatively, don't include anything but state requirements / create install scripts.
- If you are releasing a build, the build is considered a derivative.
Python users:
- If you copy-paste code, your work is a derivative.
- Imports are trickier:
- Technically, GPL or © licences triggers on import.
- Practically, it's a really grey area. Make those imports optional, and specify their licences as clearly as possible.

Take home #2
Licensing is as complicated as it is important.
Double check licenses of borrowed material, report them in your own work
for licence tracking.
Remember that institutions have rights on your work, but can should help you with licensing.
Make releases
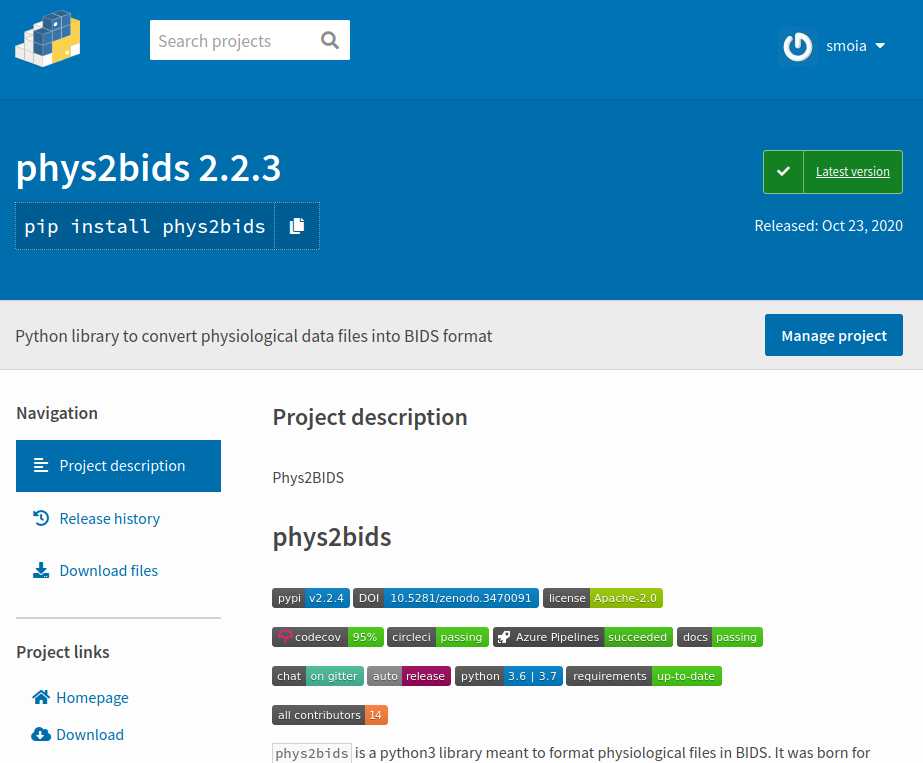
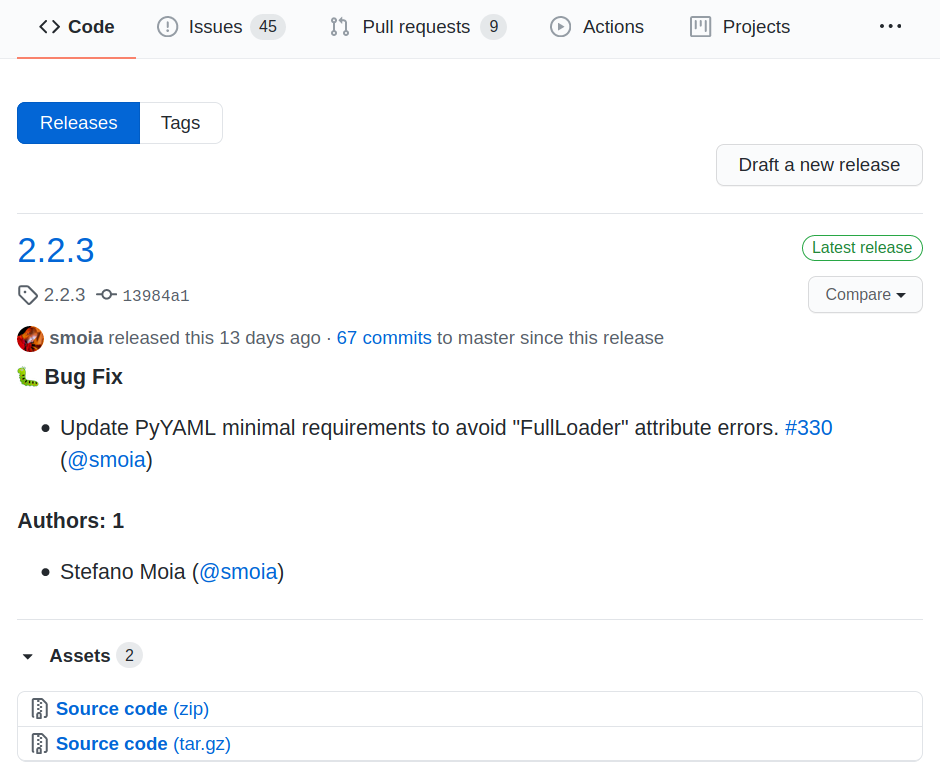
Releases make your work easier to retrieve (and cite).
Imagine them as hard links to a certain moment in time.
(e.g. paper #1 vs paper #2).
You can create, package, and distribute releases, all automagically through CI/CDs.
MATLAB users:
You can have to create, package, and distribute releases manually.

MATLAB packaging

*feat Giulia's laptop
Make your project identifiable (and citable)
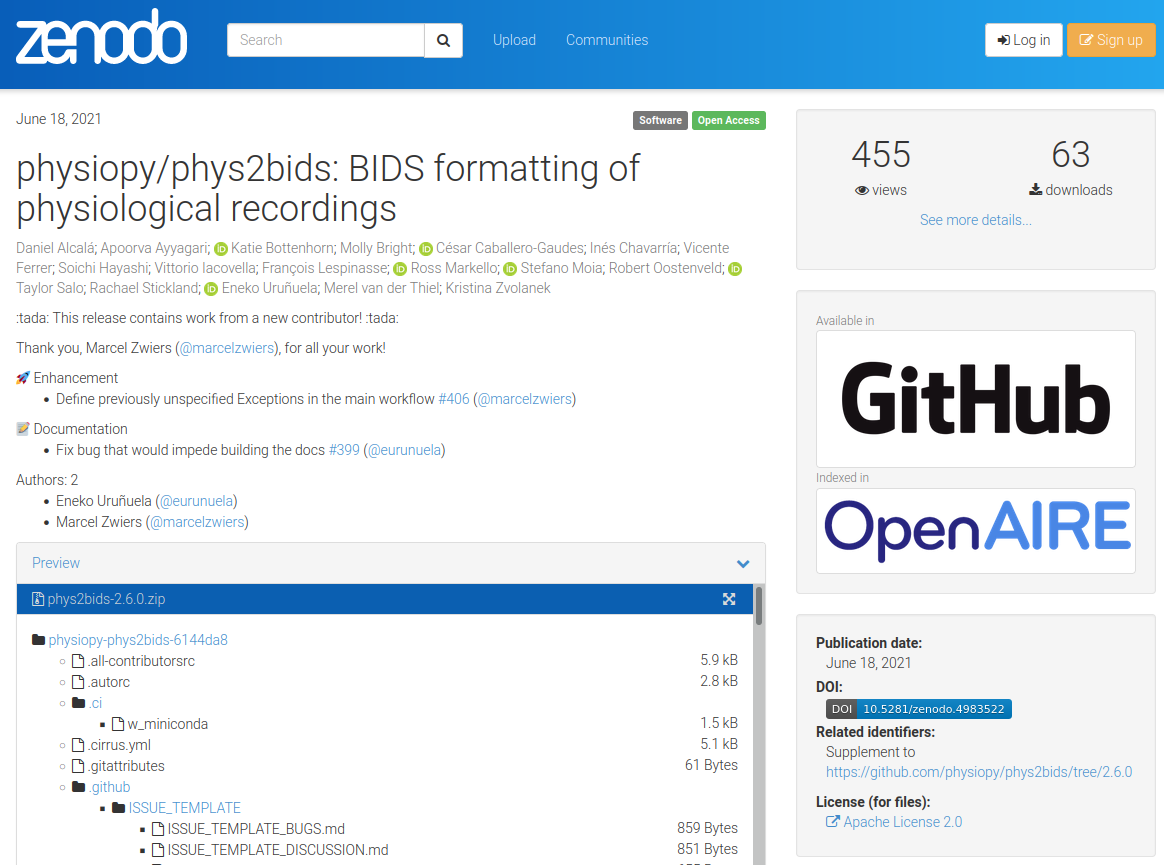
{
"license": "Apache-2.0",
"title": "physiopy/phys2bids: BIDS formatting of physiological recordings",
"upload_type": "software",
"creators": [
[...]
{
"orcid": "0000-0002-7796-8795",
"affiliation": "Florida International University",
"name": "Katie Bottenhorn"
},
[...]
],
"access_right": "open"
}
Publish!
If your project is software related, think about publishing it.
While there are various journals that can be targeted for a software publication, JOSS is free and completely integrated in GitHub.

Take home #3
Go public:
-
licence your work,
-
release it,
-
assign a DOI.
1. Why Open Science and Open Development?
2. Going public: Licenses, releases, DOIs
3. Run reliable code: Containers
Replicable, Robust, Reproducible, Generalisable

The Turing Way Community, & Scriberia, 2022 (Zenodo). Illustrations from The Turing Way (CC-BY 4.0)
Guaranteeing reproducibility is important for "reusable, transparent" research.

Really Replicable?
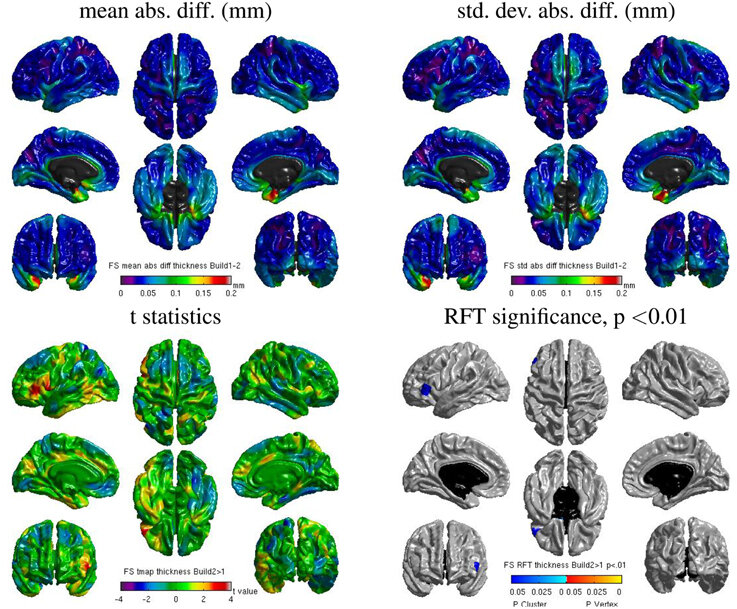
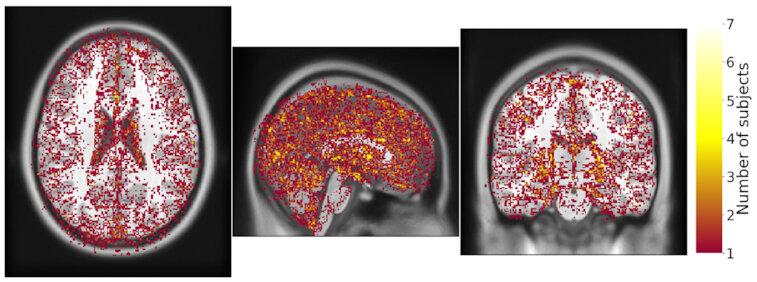
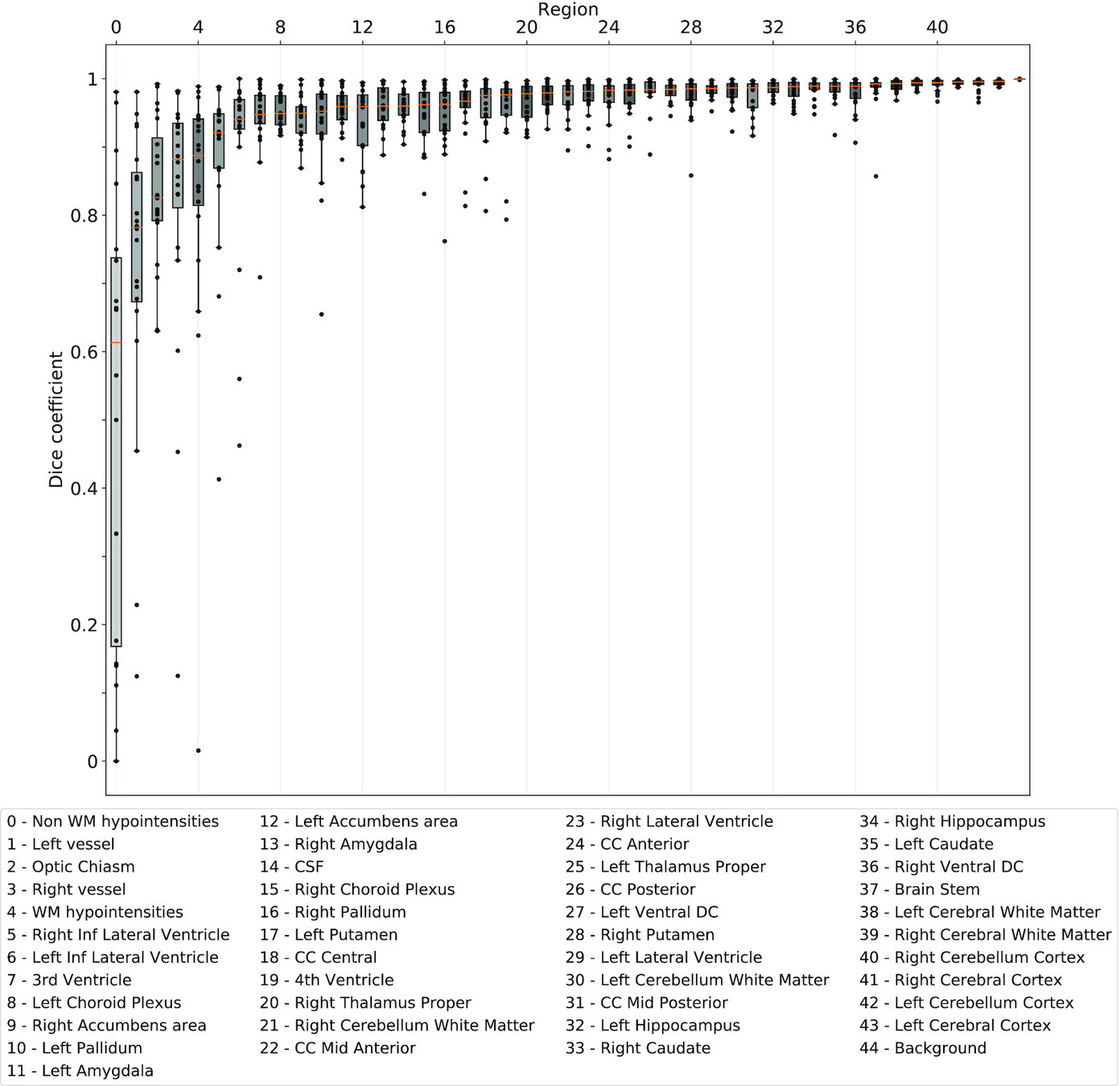
Same hardware, two Freesurfer builds (different glibc version)
Difference in estimated cortical tickness.¹
Same hardware, same FSL version, two glibc versions
Difference in estimated tissue segmentation.²
Same hardware, two Freesurfer builds (two glibc versions)
Difference in estimated parcellation.²
1. Glatard, et al., 2015 (Front. Neuroinform.) 2. Ali, et al., 2021 (Gigascience)




Containerisation

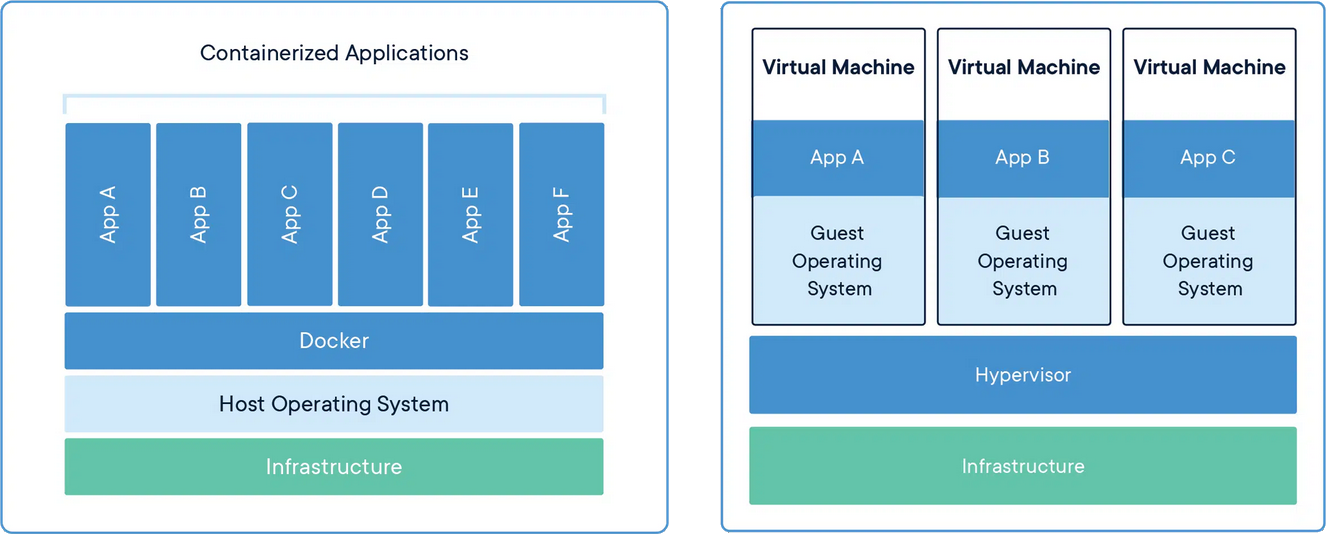


Docker vs Apptainer

Docker:
- Targeting Laptops: better OS support
- Offers public hub to share built containers
- Docker containers can be built in Singularity

Apptainer:
- Built for HPCs (Unix only), maintained by the Linux Foundation
- Easier syntax
- Supports Docker containers


Docker vs Apptainer


Bootstrap: docker
From: python:3.8.13-slim-buster
%environment
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
%post
# Set install variables, create tmp folder
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
# Prepare repos and install dependencies
pip3 install nigsp[all]
# Final removal of lists and cleanup
rm -rf /var/lib/apt/lists/*
FROM python:3.8.13-slim-buster AS nigspdock
WORKDIR /app
# Prepare environment
COPY .. .
RUN pip3 install .[all]
ENV LANG="en_US.UTF-8" \
LC_ALL="en_US.UTF-8"
CMD nigsp
ARG BUILD_DATE
ARG VCS_REF
ARG VERSION
LABEL org.label-schema.build-date=$BUILD_DATE \
org.label-schema.name="NiGSP" \
org.label-schema.description="NiGSP: python library for Graph Signal Processing on Neuroimaging data" \
org.label-schema.url="https://github.com/miplabch/nigsp" \
org.label-schema.vcs-ref=$VCS_REF \
org.label-schema.vcs-url="https://github.com/miplabch/nigsp" \
org.label-schema.version=$VERSION \
org.label-schema.schema-version="1.0"
Docker
Apptainer
Apptainer example: complete data analysis.


Bootstrap: docker
From: ubuntu:20.04
%environment
# export templateloc=/usr/share/afni/atlases
export AFNIPATH=/opt/afni-AFNI_22.3.07
export AFNI_PLUGINPATH="$AFNIPATH"
export templateloc=/usr/share/afni/atlases
export AFNI_AUTOGZIP=YES
export AFNI_COMPRESSOR=GZIP
export ANTSPATH="/opt/ants-2.4.2/bin"
export ANTSSCRIPTS="/opt/ants-2.4.2/Scripts"
export C3DPATH="/opt/convert3d-1.0.0"
export FSLDIR="/opt/fsl-6.0.6.2"
source ${FSLDIR}/etc/fslconf/fsl.sh
export FSLOUTPUTTYPE="NIFTI_GZ"
export FSLMULTIFILEQUIT="TRUE"
export FSLTCLSH="$FSLDIR/bin/fsltclsh"
export FSLWISH="$FSLDIR/bin/fslwish"
export FSLLOCKDIR=""
export FSLMACHINELIST=""
export FSLREMOTECALL=""
export FSLGECUDAQ="cuda.q"
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
export R_LIBS="/usr/lib/R"
export LD_LIBRARY_PATH="/opt/ants-2.4.2/lib:$LD_LIBRARY_PATH"
export PREPROCPATH="/opt/preprocessing"
export PATH="$AFNIPATH:$ANTSPATH:$ANTSSCRIPTS:$C3DPATH/bin:$FSLDIR/bin:$PREPROCPATH:$PREPROCPATH/00.pipelines:$PATH"
%post
# Set install variables, create tmp folder
export TMPDIR="/tmp/general_preproc_build_$( date -u +"%F_%H-%M-%S" )"
[[ -d ${TMPDIR} ]] && rm -rf ${TMPDIR}
mkdir -p ${TMPDIR}
export DEBIAN_FRONTEND=noninteractive
export TZ=Europe/Brussels
apt update -qq
apt install -y -q --no-install-recommends ca-certificates dirmngr gnupg
# Prepare repos and install dependencies
apt-key adv --keyserver keyserver.ubuntu.com --recv-keys C9A7585B49D51698710F3A115E25F516B04C661B
apt-key adv --keyserver keyserver.ubuntu.com --recv-keys 6E12762B81063D17BDDD3142F142A4D99F16EB04
echo "deb https://ppa.launchpadcontent.net/marutter/rrutter4.0/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb-src https://ppa.launchpadcontent.net/marutter/rrutter4.0/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb https://ppa.launchpadcontent.net/c2d4u.team/c2d4u4.0+/ubuntu focal main" | tee -a /etc/apt/sources.list
echo "deb-src https://ppa.launchpadcontent.net/c2d4u.team/c2d4u4.0+/ubuntu focal main" | tee -a /etc/apt/sources.list
apt update -qq
apt install -y -q --no-install-recommends \
bc \
build-essential \
bzip2 \
cmake \
curl \
dc \
file \
freeglut3-dev \
g++ \
gcc \
git \
less \
libcurl4-openssl-dev \
libeigen3-dev \
libexpat1-dev \
libf2c2-dev \
libfftw3-3 \
libfftw3-dev \
libgdal-dev \
libgfortran4 \
libgfortran-8-dev \
libglew-dev \
libgl1-mesa-dev \
libgl1-mesa-dri \
libgl1-mesa-glx \
libglib2.0-dev \
libglu1-mesa-dev \
libglw1-mesa \
libgomp1 \
libgsl-dev \
libgts-dev \
libjpeg8-dev \
liblapack3 \
libopenblas-dev \
libmotif-dev \
libnetpbm10-dev \
libnode-dev \
libpng16-16 \
libpng-dev \
libquadmath0 \
libtiff5 \
libtiff5-dev \
libudunits2-dev \
libxext-dev \
libxi-dev \
libxm4 \
libxmhtml-dev \
libxml2-dev \
libxmu-dev \
libxmu-headers \
libxpm-dev \
libxt-dev \
m4 \
make \
mesa-common-dev \
nano \
r-base-dev \
rsync \
tcsh \
python3-distutils \
python3-pip \
python3-rpy2 \
python-is-python3 \
qhull-bin \
xvfb \
zlib1g-dev
# Install AFNI
mkdir -p ${TMPDIR}/afni
cd ${TMPDIR}/afni || exit 1
ln -s /usr/lib/x86_64-linux-gnu/libgsl.so.23 /usr/lib/x86_64-linux-gnu/libgsl.so.19
ln -s /usr/lib/x86_64-linux-gnu/libXp.so.6 /usr/lib/x86_64-linux-gnu/libXp.so
git clone https://github.com/afni/afni.git source
cd source || exit 1
git fetch --tags
git -c advice.detachedHead=false checkout AFNI_22.3.07
cd src || exit 1
cp other_builds/Makefile.linux_ubuntu_16_64_glw_local_shared Makefile
make itall
mv linux_ubuntu_16_64_glw_local_shared /opt/afni-AFNI_22.3.07
export PATH="/opt/afni-AFNI_22.3.07:$PATH"
export R_LIBS="/usr/lib/R"
rPkgsInstall -pkgs ALL
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/afni
# Install ANTs
mkdir -p ${TMPDIR}/ants/build
git clone https://github.com/ANTsX/ANTs.git ${TMPDIR}/ants/source
cd ${TMPDIR}/ants/source || exit 1
git fetch --tags
git -c advice.detachedHead=false checkout v2.4.2
cd ${TMPDIR}/ants/build || exit 1
cmake -DCMAKE_INSTALL_PREFIX=/opt/ants-2.4.2 -DBUILD_SHARED_LIBS=ON -DBUILD_TESTING=OFF ${TMPDIR}/ants/source
make -j 10
mkdir -p /opt/ants-2.4.2
cd ANTS-build || exit 1
make install
mv ../../source/Scripts/ /opt/ants-2.4.2
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/ants
# Install C3D
echo "Downloading Convert3D ..."
mkdir -p /opt/convert3d-1.0.0
curl -fsSL https://sourceforge.net/projects/c3d/files/c3d/1.0.0/c3d-1.0.0-Linux-x86_64.tar.gz/download \
| tar -xz -C /opt/convert3d-1.0.0 --strip-components 1
# Install FSL
mkdir -p ${TMPDIR}/fsl
cd ${TMPDIR}/fsl || exit 1
curl -fL https://fsl.fmrib.ox.ac.uk/fsldownloads/fslinstaller.py --output ./fslinstaller.py
chmod +x fslinstaller.py
python3 fslinstaller.py -d /opt/fsl-6.0.6.2 -V 6.0.6.2
# echo "Installing FSL conda environment ..."
# bash /opt/fsl-6.0.6.2/etc/fslconf/fslpython_install.sh -f /opt/fsl-6.0.6.2
cd ${TMPDIR} || exit 1
rm -rf ${TMPDIR}/fsl
# Clone EuskalIBUR preprocessing.
git clone https://github.com/smoia/EuskalIBUR_preproc.git /opt/preprocessing
apt install -y -q csvtool
# Install PYTHON things.
pip3 install pip==22.3.1 setuptools==65.5.1 wheel==0.38.4
# Install wxPython in a particular way.
pip3 install --no-cache -f https://extras.wxpython.org/wxPython4/extras/linux/gtk3/ubuntu-20.04 wxpython==4.2.0
# Install datalad, fsleyes, nilearn, peakdet, phys2cvr.
pip3 install \
annexremote==1.6.0 \
boto==2.49.0 \
certifi==2022.12.7 \
cffi==1.15.1 \
chardet==4.0.0 \
charset-normalizer==2.1.1 \
contourpy==1.0.6 \
cryptography==38.0.4 \
cycler==0.11.0 \
datalad==0.17.10 \
dill==0.3.6 \
distro==1.8.0 \
fasteners==0.18 \
fonttools==4.38.0 \
fsleyes==1.5.0 \
fsleyes-props==1.8.2 \
fsleyes-widgets==0.12.3 \
fslpy==3.10.0 \
h5py==3.7.0 \
humanize==4.4.0 \
idna==3.4 \
importlib-metadata==5.1.0 \
iso8601==1.1.0 \
jaraco.classes==3.2.3 \
jeepney==0.8.0 \
Jinja2==3.1.2 \
joblib==1.2.0 \
keyring==23.11.0 \
keyrings.alt==4.2.0 \
kiwisolver==1.4.4 \
lxml==4.9.2 \
MarkupSafe==2.1.1 \
matplotlib==3.6.2 \
more-itertools==9.0.0 \
msgpack==1.0.4 \
nibabel==4.0.2 \
nilearn==0.9.2 \
numpy==1.23.5 \
packaging==22.0 \
pandas==1.5.2 \
patool==1.12 \
peakdet==0.2.0rc1 \
phys2cvr==0.16.0 \
Pillow==9.3.0 \
platformdirs==2.6.0 \
pycparser==2.21 \
PyOpenGL==3.1.6 \
pyparsing==2.4.7 \
python-dateutil==2.8.2 \
python-gitlab==3.12.0 \
pytz==2022.6 \
requests==2.28.1 \
requests-toolbelt==0.10.1 \
scikit-learn==1.2.0 \
scipy==1.9.3 \
SecretStorage==3.3.3 \
simplejson==3.18.0 \
six==1.16.0 \
threadpoolctl==3.1.0 \
tqdm==4.64.1 \
urllib3==1.26.13 \
Whoosh==2.7.4 \
zipp==3.11.0
# Final removal of lists and cleanup
cd /tmp || exit 1
rm -rf ${TMPDIR}
rm -rf /var/lib/apt/lists/*Containers in action


apptainer build --sandbox container.img docker://afni/afni_dev_base:AFNI_22.2.12
apptainer shell -w -e -f --no-home container.img
apptainer build --sandbox container.sif recipe.def
apptainer exec -e -f --no-home -B /some/place:/tmp
-B /some/place/elsewhere:/scripts \
-B /another/place/:/data \
container.sif /scripts/run_batch_analysis.sh sub-001 ses-01apptainer exec docker://ghcr.io/apptainer/lolcow cowsay "Hello $USER!"Try it now on the server!
Easily build a neuroimaging container

Easily build a neuroimaging container

BIDSapps: containers for BIDS pipelines

Take home #4
Don't think that because your study "works", it's reproducible.
Create (or adopt) containers and share them with your code to maximise the reproducibility of your analyses.
(And to stop asking Dimitri to install software)
That's all folks!
Thank you!




Any question [/opinions/objections/...]?
Oh, and don't forget!

Oh, and don't forget!

Open science (MIP:Lab meeting 17.01.2023)
By Stefano Moia
Open science (MIP:Lab meeting 17.01.2023)
CC-BY 4.0 Stefano Moia, 2023. Images are property of the original authors and should be shared following their respective licences. This presentation is otherwise licensed under CC BY 4.0. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/
- 108



