Graphs, the Shire, & multimodal imaging
Neuro-X Institute, École polytechnique fédérale de Lausanne, Geneva, Switzerland
Department of Radiology and Medical Informatics (DRIM), Faculty of Medicine, University of Geneva, Geneva, Switzerland
physiopy (https://github.com/physiopy)


| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |


Donostia, 21.11.23
Graphs, the Shire, & multimodal imaging
Donostia, 21.11.23



The Shire
Geneva










Graph Signal Processing
Ortega (2022) Introduction to Graph signal Processing & other material courtesy of Dimitri Van De Ville





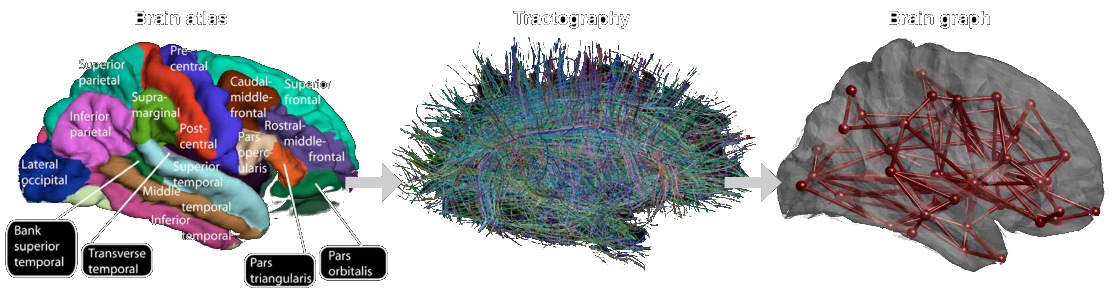
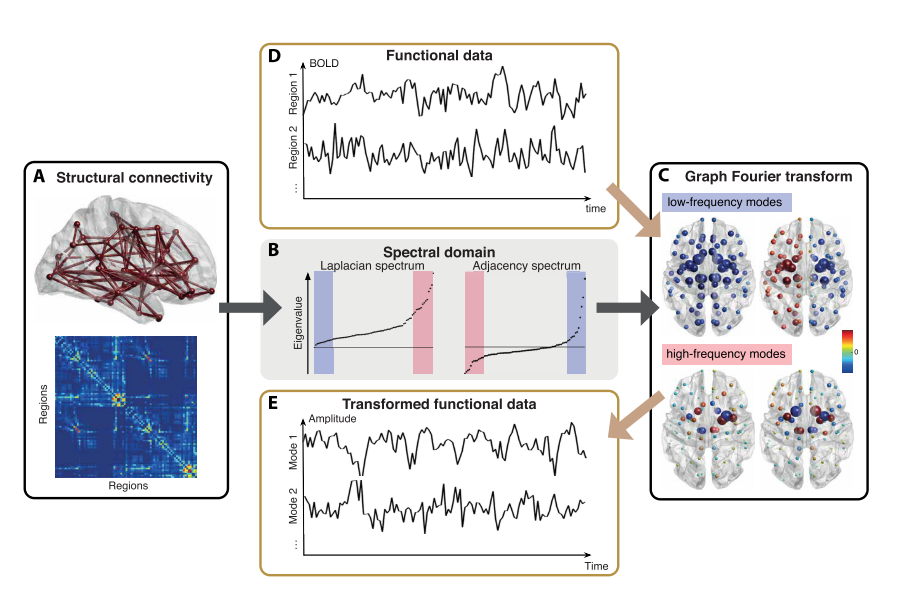
Graph Signal Processing: brain applications
- Novel approach to signal processing, based on graph embedding of timeseries¹.
- Useful for multimodal brain imaging².
- GSP applications can improve subject identification³, improve data smoothing⁴, and reveal structural/functional coupling patterns agreeing with cognitive domain hierarchy and genetic expression⁵.
1. Ortega (2022) Introduction to Graph signal Processing,
3. Griffa et al. (2022) Neuroimage,
5. Preti, Van De Ville (2019) Nat. Commun.
2. Huang et al. (2018) Proc. IEEE,
4. Abramian (2021) Neuroimage,
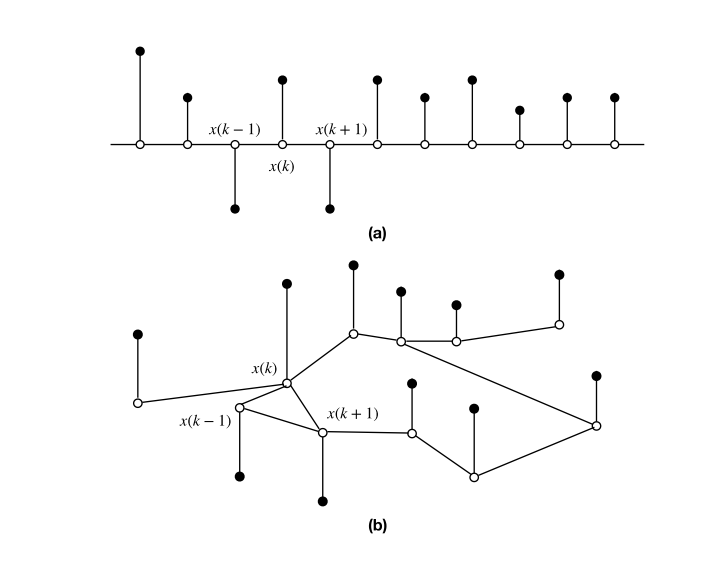
Shuman et al. (2013) IEEE Signal Process Mag
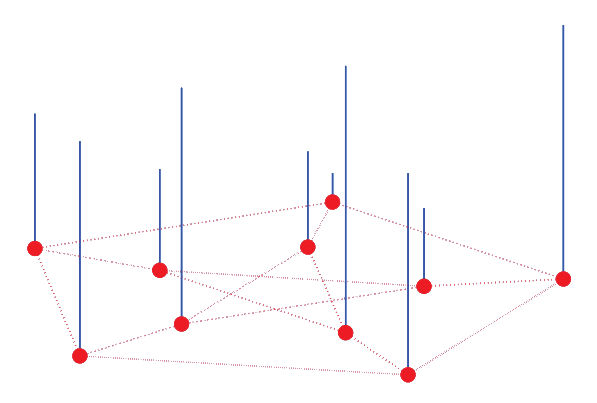
Huang et al. (2018) Proc. IEEE
Huang et al. (2018) Proc. IEEE



Graph Signal Processing
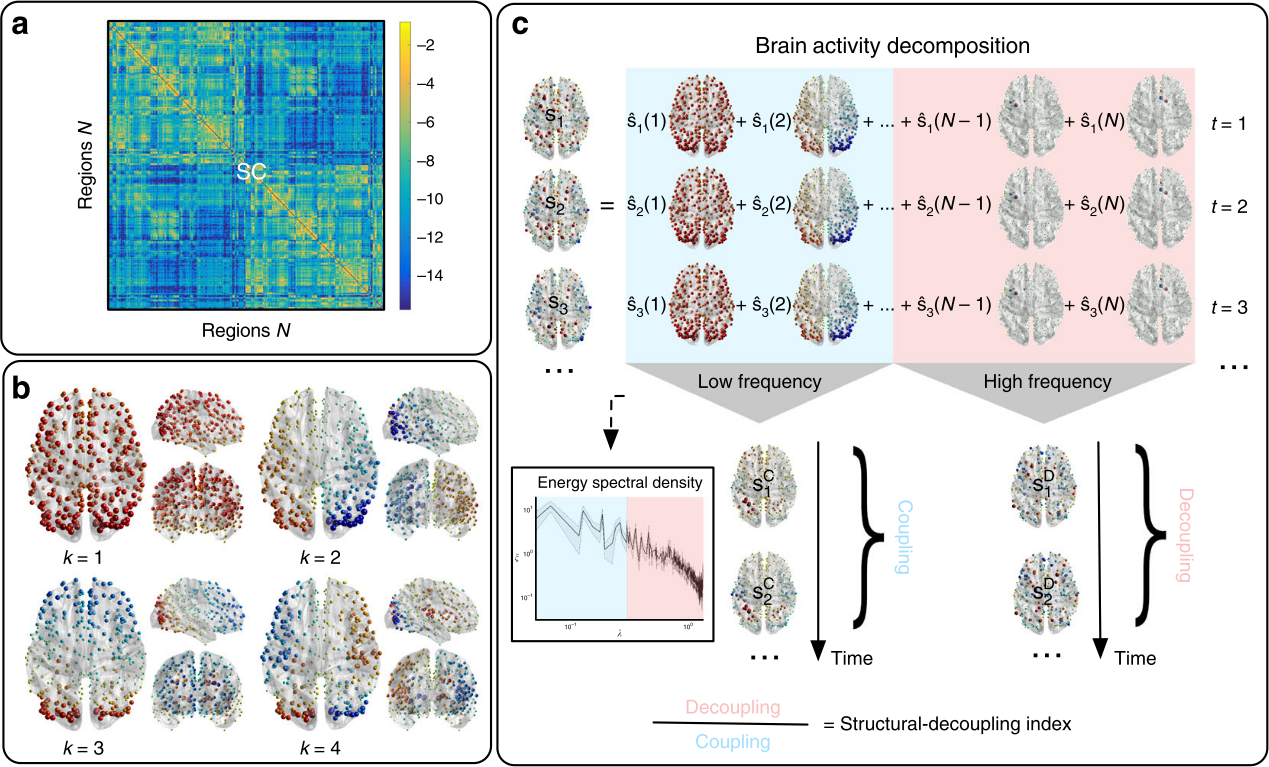
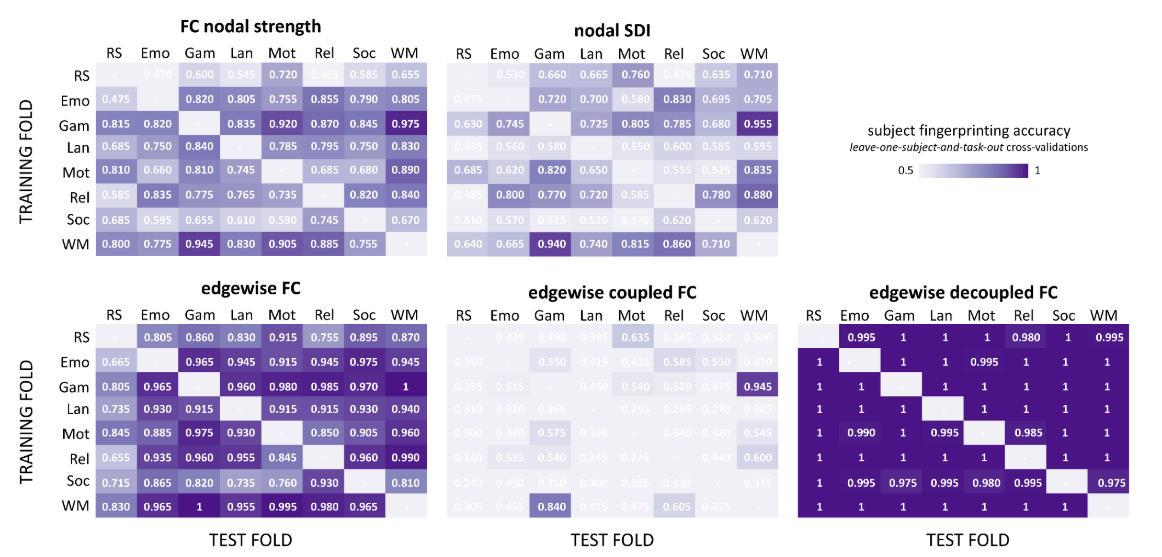


Griffa et al. 2021 (bioRxiv), Preti et al. 2019 (Nat. Commun.)




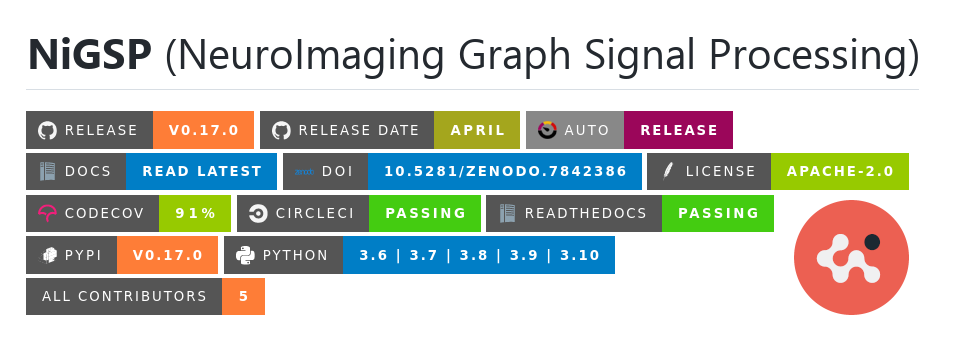
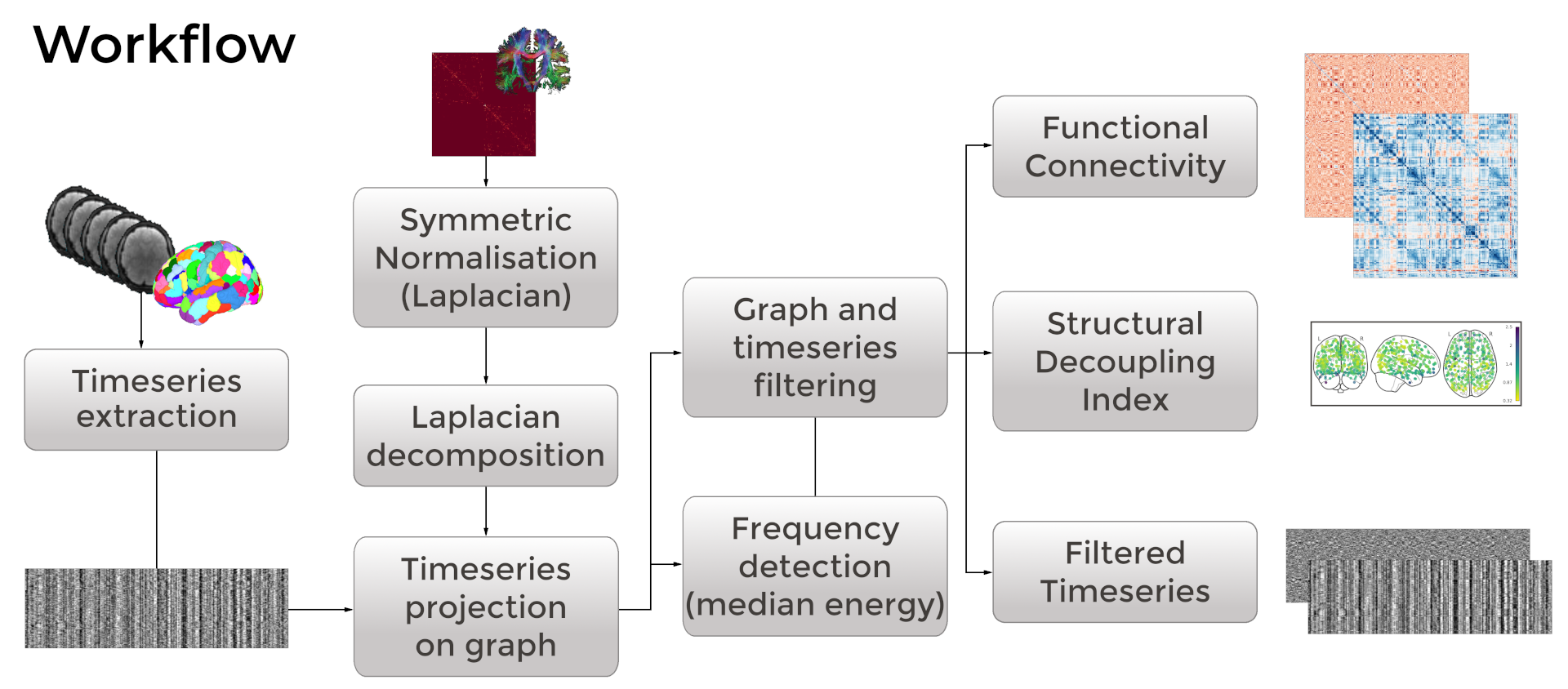
NiGSP
NiGSP is an open development python based module to apply
Graph Signal Processing, with emphasis on neuroimaging applications (atlas based)
It offers a set of function for filter creations, filtering, visualisation,
and metrics computations
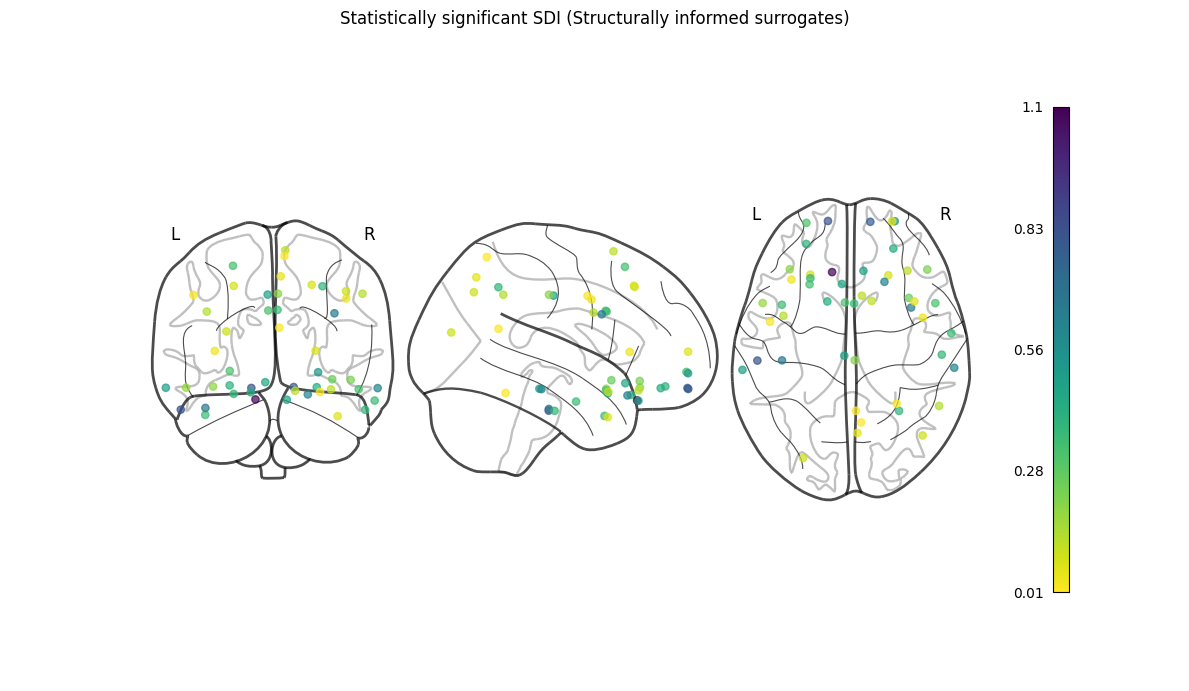
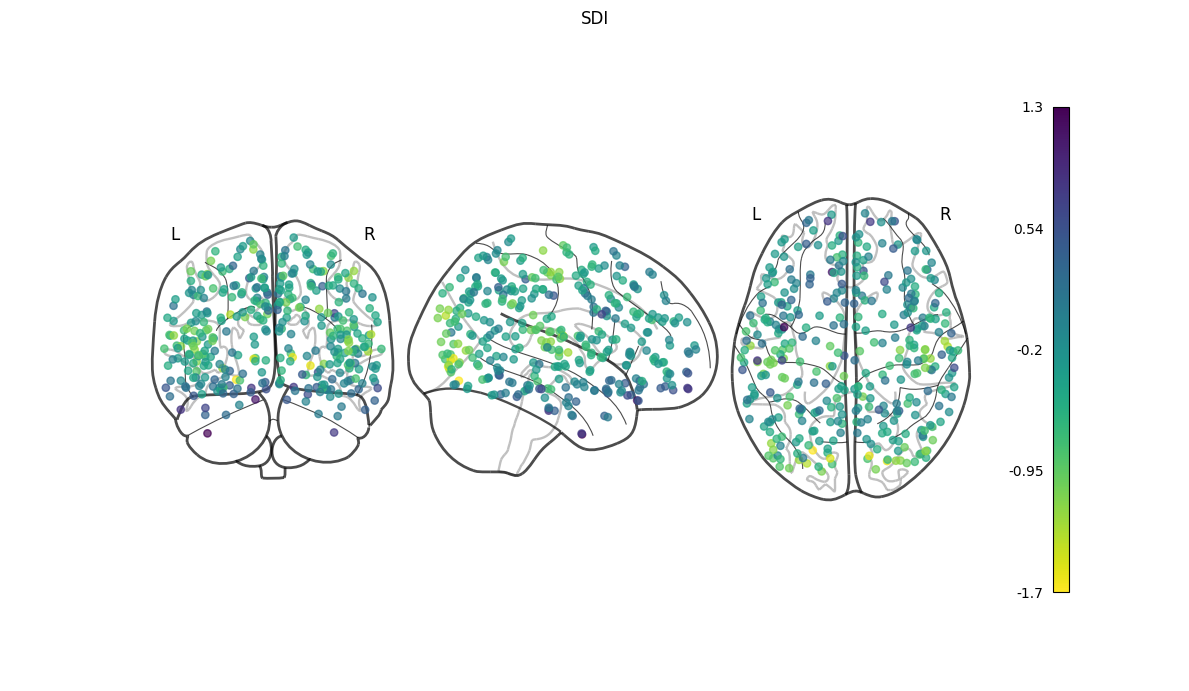
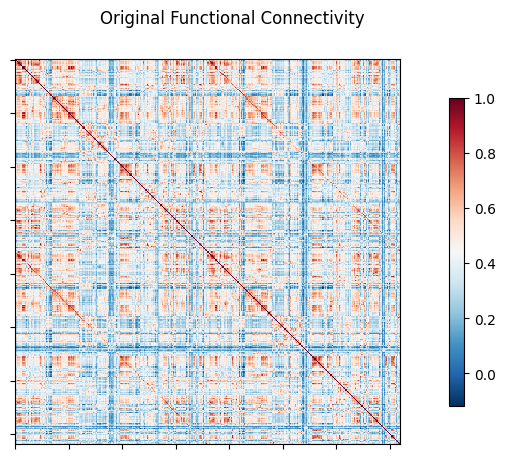
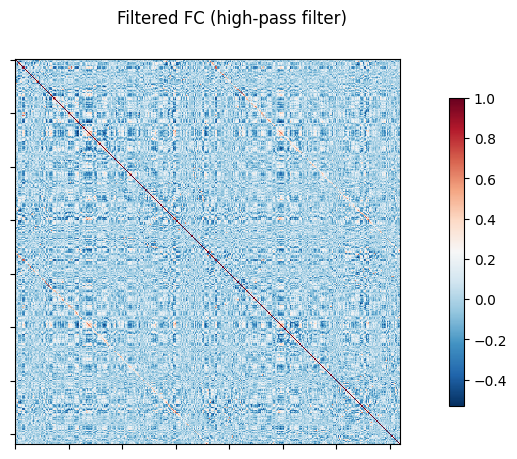
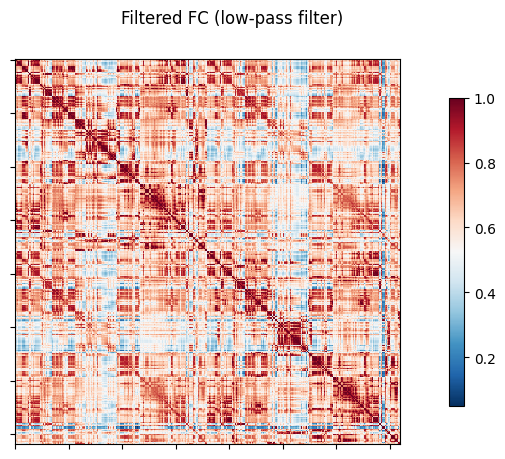
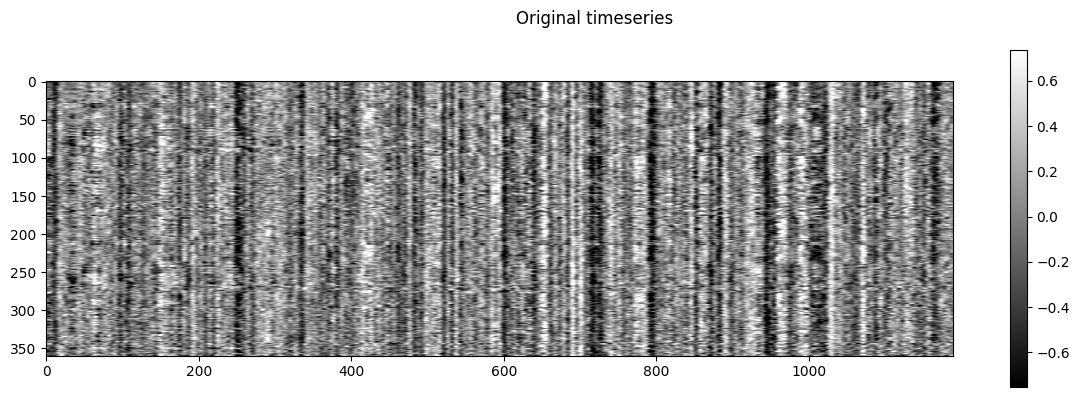
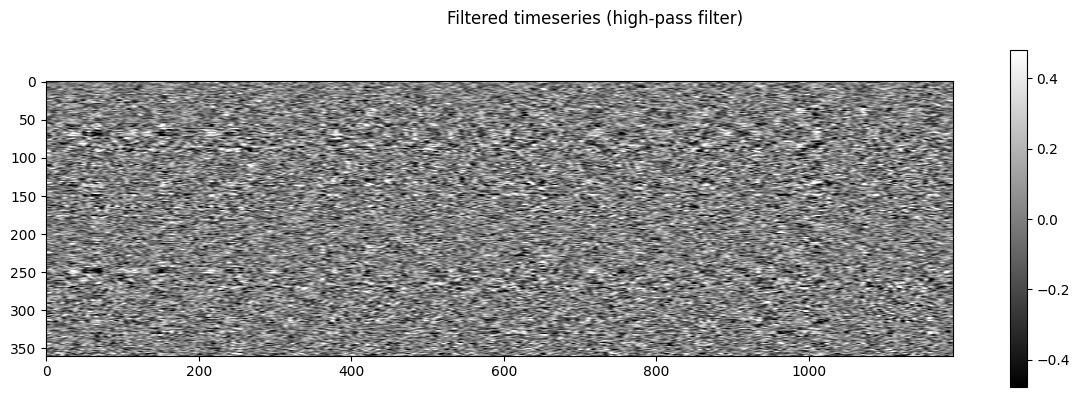
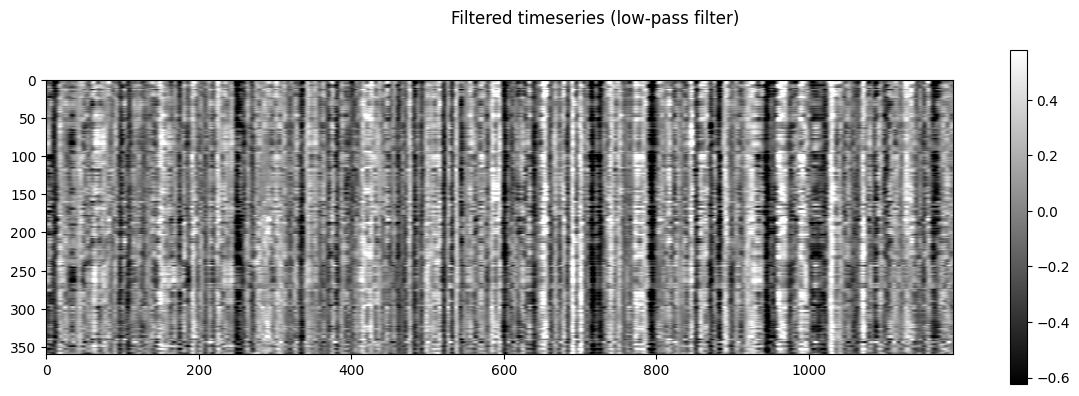




NiGSP: workflow
nigsp -f timeseries.nii.gz -s sc_mtx.tsv -a atlas.nii.gz --informed-surrogates -n 1000Based on Preti, Van De Ville (2019) Nat. Commun. and Griffa et al. (2022) Neuroimage




Features

- Digital Object Identifier
- Automatic output of citations
- all-contributors authorship
- Apache 2.0 licence
- Thoroughly tested
- Graphic identity



NiGSP: workflow extension

- Common data loading
- Different central "workflow" for different analyses/publications
- Common data visualisation and data export
- Wrapper for BIDS-compatible reading
- Easy to run at subject or group level
- Improved performance




Graph Signal Processing and lesions
1. Egger et al. 2021 (Stroke); 2. Egger et al. in prep.; * Morishita, Inoue, 2016 (Neurol. Med.-Chir.)
Subcortical brain lesions (e.g. stroke) may affect structural connectivity (SC) by disrupting white matter traits.

*
However, the extent of its impact might be underrated by current tractography methods, especially in the acute phase of stroke¹.
Instead, the portion of SC interested by the lesion only can be computed and removed from the global SC¹. Doing so increases agreement between SC and prognosis¹ or SC and behavioural scores²






GSP-based diffusivity model
Total
Unaffected
Affected



Brain GSP community


physiopy
Raw data
BIDSification
physiological data preprocessing
phys. denoising
phys. imaging
Data acquisition
Process description & knowledge base
QC/QA



physiopy
Raw data
phys2bids
peakdet
phys2denoise
phys. imaging
Data acquisition
physiopy's documentation
physioQC



Sneak peak
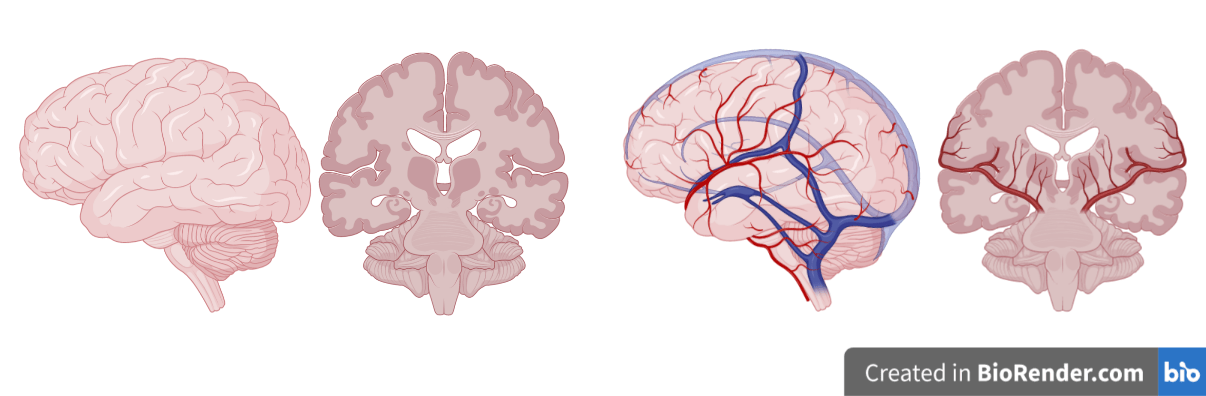




- Vascular connectivity
- Impact of macro-angioarchitecture on BOLD signal dinamics
- BOLD (and ASL) vascular inpainting
- Structure-informed signal decomposition
Thanks to...
...the MIP:Lab @ EPFL
...you for the (sustained) attention!
That's all folks!
...the Physiopy contributors





Graphs, Milka, & diffusion models
By Stefano Moia
Graphs, Milka, & diffusion models
- 70



