Field-level inference of primordial non-Gaussianity from galaxy redshift surveys
Hugo SIMON,
PhD student supervised by
Arnaud DE MATTIA and François LANUSSE
CoBALt, 2025/06/30


Field-level inference of primordial non-Gaussianity from galaxy redshift surveys
Hugo SIMON,
PhD student supervised by
Arnaud DE MATTIA and François LANUSSE
CoBALt, 2025/06/30



Optimal extraction of primordial non-Gaussian signal from galaxy redshift survey
Hugo SIMON,
PhD student supervised by
Arnaud DE MATTIA and François LANUSSE
Sesto, 2025/07/17


Field-level inference of primordial non-Gaussianity from galaxy redshift surveys
Hugo SIMON,
PhD student supervised by
Arnaud DE MATTIA and François LANUSSE
CSI, 2025/09/02


Field-level analysis of primordial non-Gaussianity with DESI tracers

Hugo SIMON,
PhD student supervised by
Arnaud DE MATTIA and François LANUSSE
PNG Meeting, 2025/06/18

The universe recipe (so far)
$$\frac{H}{H_0} = \sqrt{\Omega_r + \Omega_b + \Omega_c+ \Omega_\kappa + \Omega_\Lambda}$$
instantaneous expansion rate
energy content
Cosmological principle + Einstein equation
+ Inflation
\(\delta_L \sim \mathcal G(0, \mathcal P)\)
\(\sigma_8:= \sigma[\delta_L * \boldsymbol 1_{r \leq 8}]\)
initial field
primordial power spectrum
std. of fluctuations smoothed at \(8 \text{ Mpc}/h\)

\(\Omega := \{ \Omega_m, \Omega_\Lambda, H_0, \sigma_8, f_\mathrm{NL},...\}\)
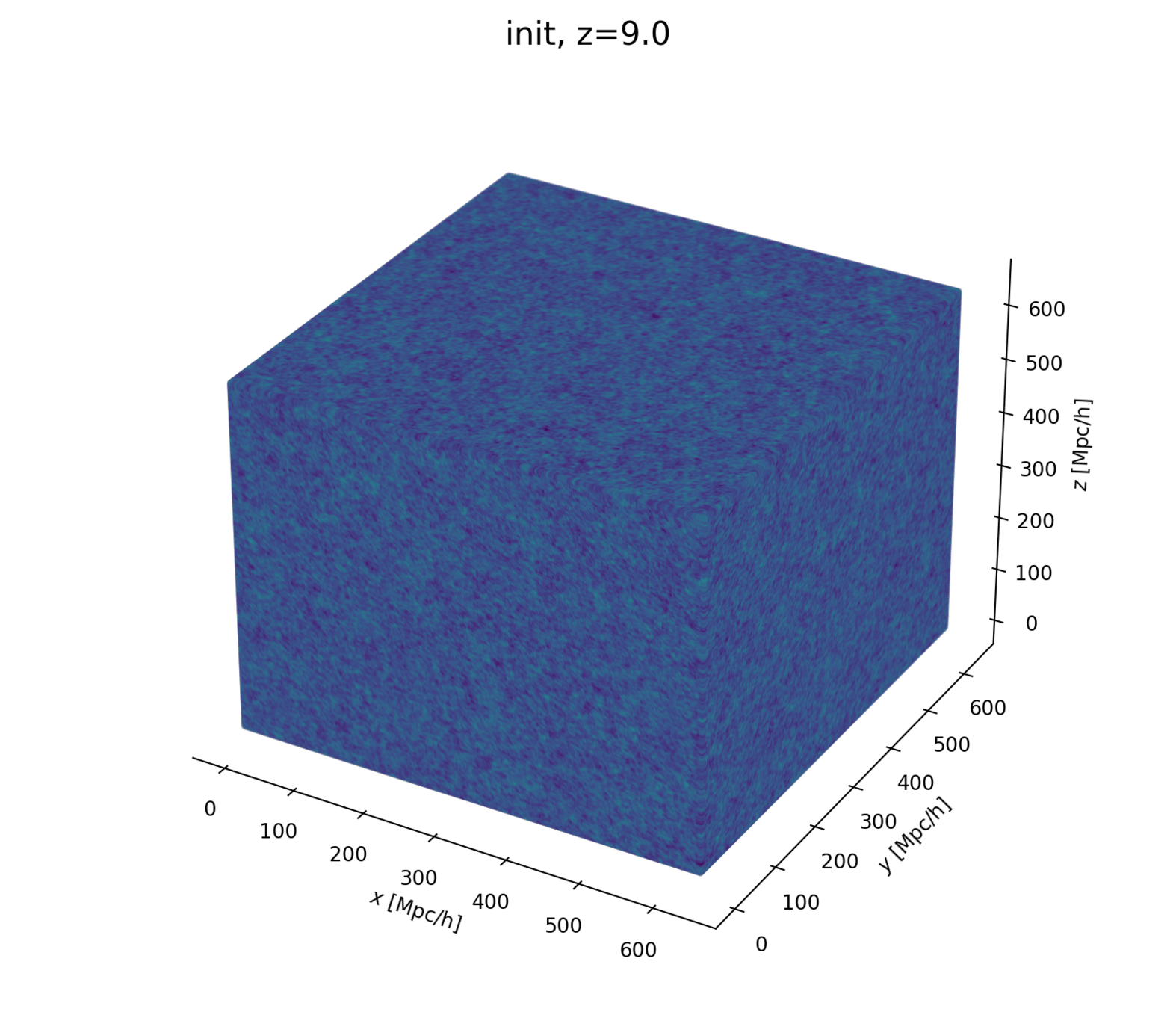
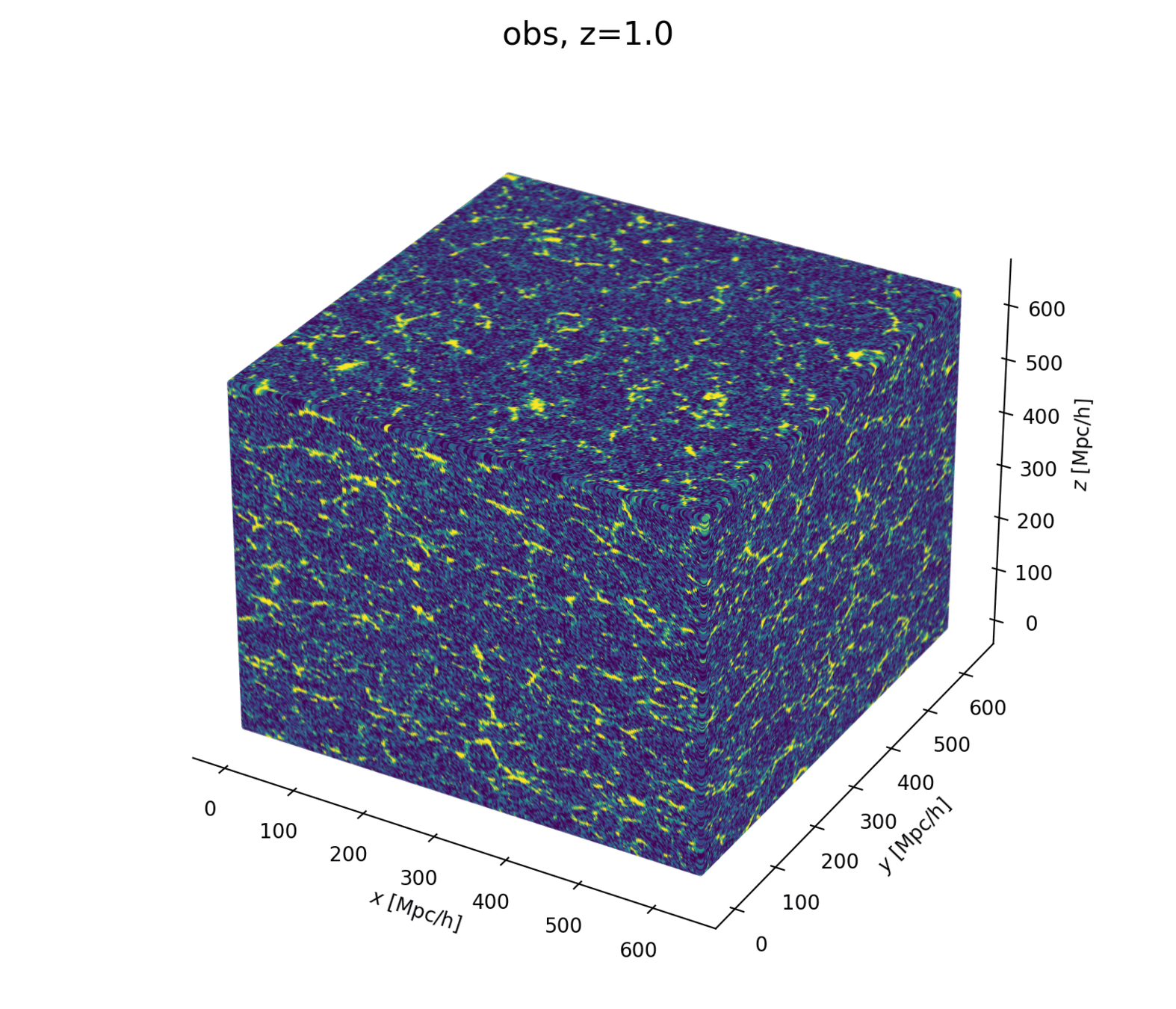
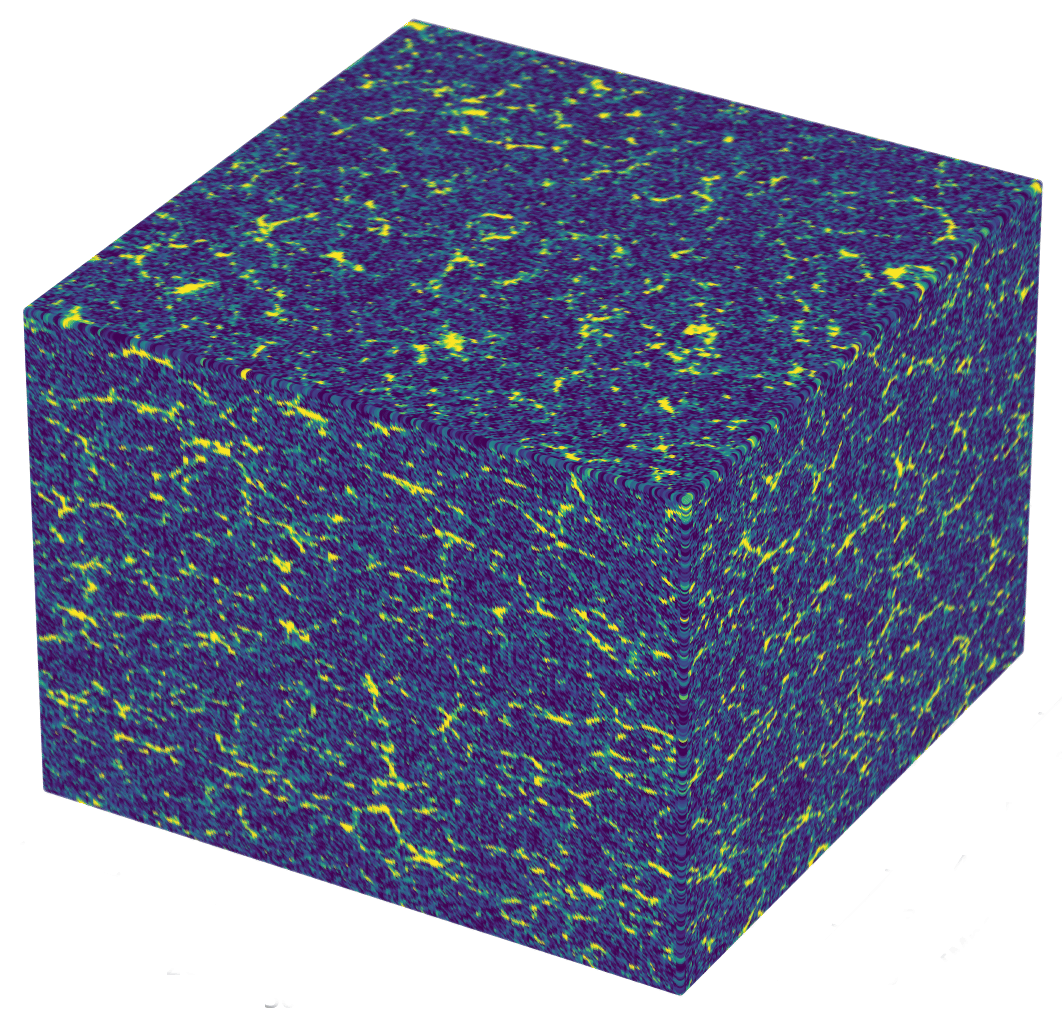
Linear matter spectrum
Structure growth
Cosmological modeling and inference
\(\Omega\)
\(\delta_L\)
\(\delta_g\)
inference

Cosmological inference
\(\Omega := \{ \Omega_m, \Omega_\Lambda, H_0, \sigma_8, f_\mathrm{NL},...\}\)
inference
\(P\)

\(\Omega\)
\(\delta_L\)
\(\delta_g\)

Cosmological inference
\(\Omega := \{ \Omega_m, \Omega_\Lambda, H_0, \sigma_8, f_\mathrm{NL},...\}\)
\(\Omega\)
\(\delta_L\)
\(\delta_g\)


inference
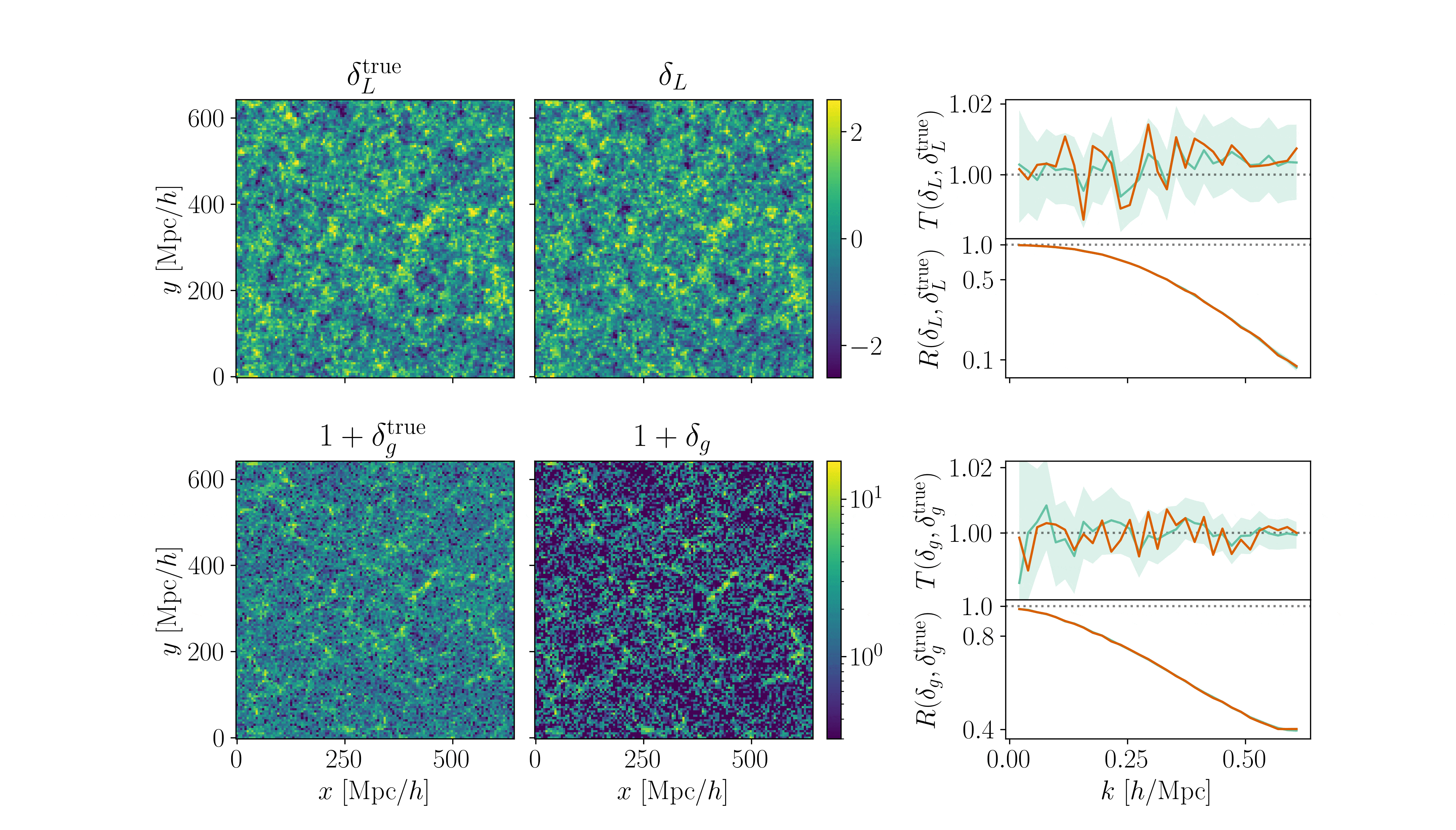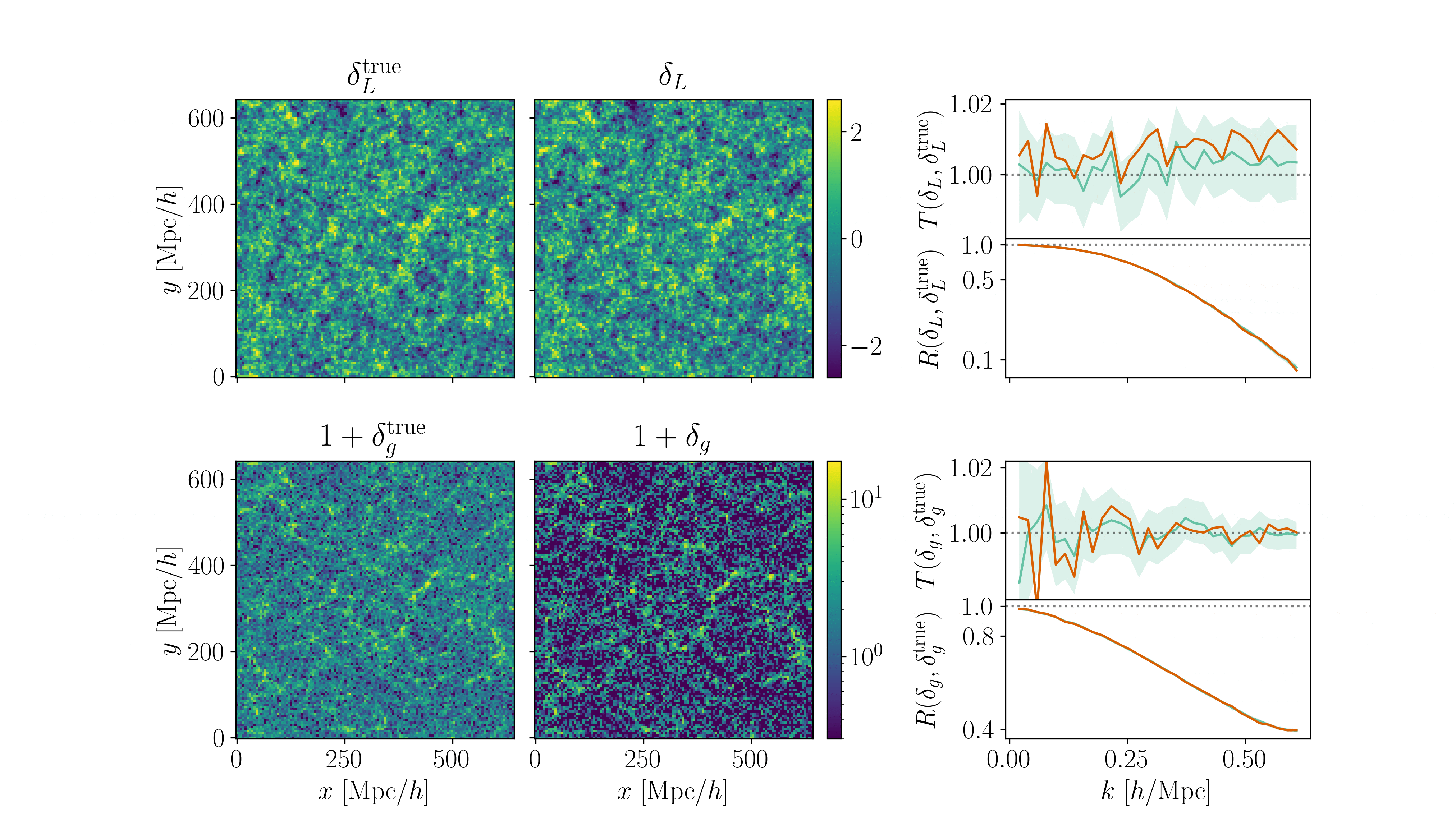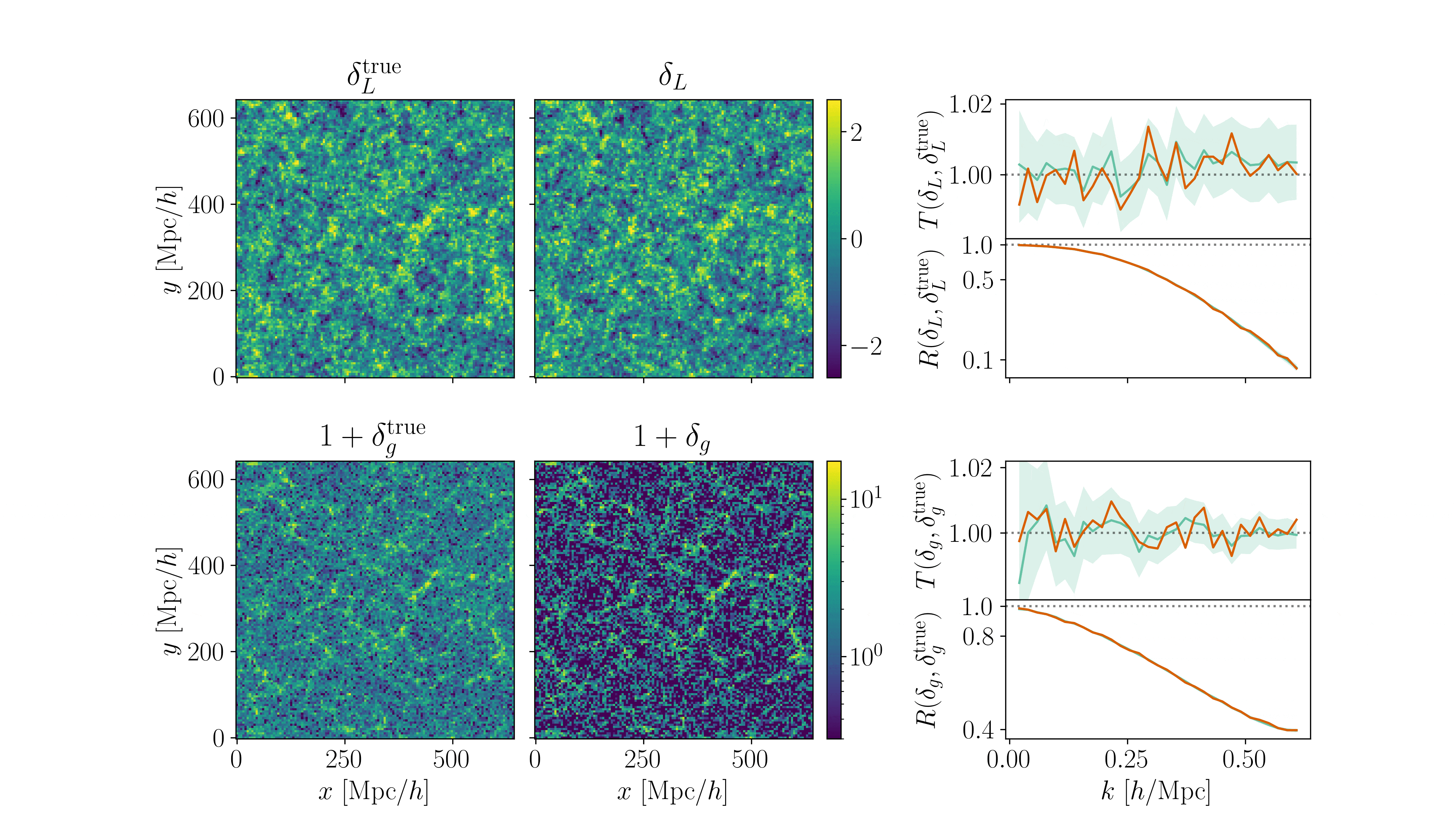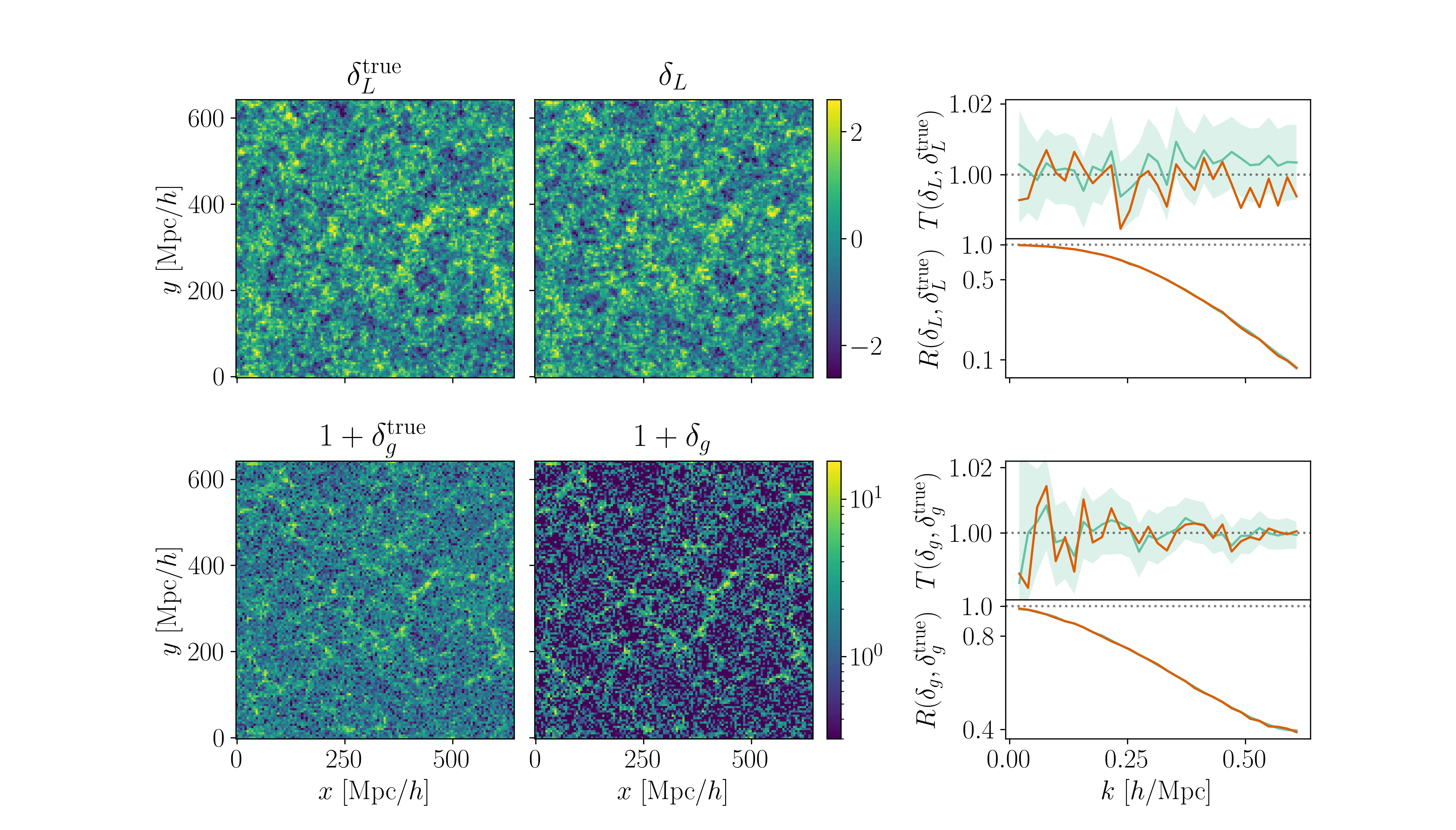

Cosmological inference
inference
\(\Omega := \{ \Omega_m, \Omega_\Lambda, H_0, \sigma_8, f_\mathrm{NL},...\}\)



\(\Omega\)
\(\delta_L\)
\(\delta_g\)
\(P\)

Cosmological inference
\(\Omega := \{ \Omega_m, \Omega_\Lambda, H_0, \sigma_8, f_\mathrm{NL},...\}\)
\(\Omega\)
\(\delta_L\)
\(\delta_g\)


inference


\(128^3\) PM on 8GPU:
4h MCLMC vs. \(\geq\) 80h HMC
Fast & differentiable model with
Bayesian inference
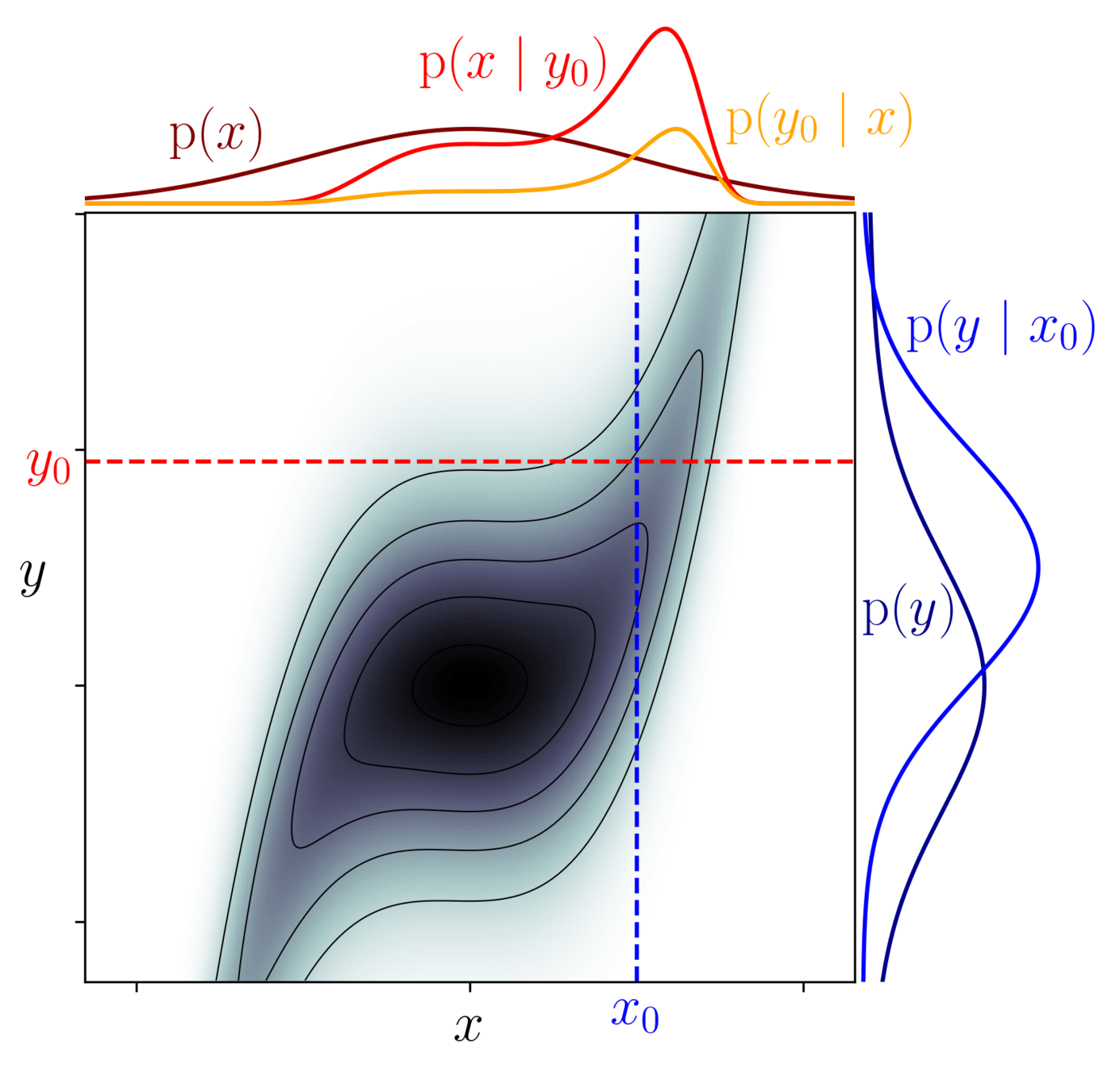
model \(=\begin{cases}x \sim \mathcal N(0,1)\\y\mid x \sim \mathcal N(x^3, 1)\end{cases}\)
Among all possible worlds \(x,y\), restrict to the ones compatible with observation \(y_0\):
$$\underbrace{\mathrm p(x \mid y_0)}_{\text{posterior}} = \frac{\overbrace{\mathrm p(y_0 \mid x)}^{\text{likelihood}}}{\underbrace{\mathrm p(y_0)}_{\text{evidence}}}\underbrace{\mathrm p(x)}_{\text{prior}}$$
\(x\)
\(y\)
condition
\(x\)
\(y\)
\(\mathrm{p}(x,y)\)
\(\mathrm{p}(x \mid y_0)\)
Numerically:
- Simulate a possible world \(x,y\)
- Assess \(x\) credibility by comparing \(y\) to \(y_0\)
- Repeat

Please define model
Model: an observation generator, link between latent and observed variables
\(F\)
\(C\)
\(D\)
\(E\)
\(A\)
\(B\)
\(G\)
Frequentist:
fixed but unknown parameters
Bayesian:
random because unknown
\(A\)
\(B\)
\(C\)
\(F\)
\(D\)
\(E\)
\(G\)
Represented as a DAG which expresses a joint probability factorization, i.e. conditional dependencies between variables
latent param
latent variable
observed variable
Definitions
evidence/prior predictive: $$\mathrm{p}(y) := \int \mathrm{p}(y \mid x) \mathrm{p}(x)\mathrm d x$$
posterior predictive: $$\mathrm{p}(y_1 \mid y_0) := \int \mathrm{p}(y_1 \mid x) \mathrm{p}(x \mid y_0)\mathrm d x$$
\(y\)
\(x\)
Bayes thm
\(\sim\)
\(\underbrace{\mathrm{p}(x \mid y)}_{\text{posterior}}\underbrace{\mathrm{p}(y)}_{\text{evidence}} = \underbrace{\mathrm{p}(y \mid x)}_{\text{likelihood}} \,\underbrace{\mathrm{p}(x)}_{\text{prior}}\)
\(y\)
\(x\)
Marginalization:
\(y\)
\(x\)
\(z\)
\(z\)
\(x\)
\(y\)
\(x\)
\(y\)
\(x\)
Conditioning:
Definitions
\(x\)
\(z\)
Marginalization:
\(y\)
\(x\)
Conditioning:
evidence/prior predictive: $$\mathrm{p}(y) := \int \mathrm{p}(y \mid x) \mathrm{p}(x)\mathrm d x$$
posterior predictive: $$\mathrm{p}(y_1 \mid y_0) := \int \mathrm{p}(y_1 \mid x) \mathrm{p}(x \mid y_0)\mathrm d x$$
\(y\)
\(x\)
Bayes thm
\(\sim\)
\(\underbrace{\mathrm{p}(x \mid y)}_{\text{posterior}}\underbrace{\mathrm{p}(y)}_{\text{evidence}} = \underbrace{\mathrm{p}(y \mid x)}_{\text{likelihood}} \,\underbrace{\mathrm{p}(x)}_{\text{prior}}\)
\(y\)
\(x\)

\(s\)
\(\Omega\)
Cosmology is (sometimes) hard
Probabilistic Programming: Wish List
\(\begin{cases}x,y,z \text{ samples}\\\mathrm{p}(x,y,z)\\ U= -\log \mathrm{p}\\\nabla U,\nabla^2 U,\dots\end{cases}\)
1. Modeling
e.g. in JAX with \(\texttt{NumPyro}\), \(\texttt{JaxCosmo}\), \(\texttt{JaxPM}\), \(\texttt{DISCO-EB}\)...
2. DAG compilation
PPLs e.g. \(\texttt{NumPyro}\), \(\texttt{Stan}\), \(\texttt{PyMC}\)...
3. Inference
e.g. MCMC or VI with \(\texttt{NumPyro}\), \(\texttt{Stan}\), \(\texttt{PyMC}\), \(\texttt{emcee}\)...
4. Extract and viz
e.g. KDE with \(\texttt{GetDist}\), \(\texttt{GMM-MI}\), \(\texttt{mean()}\), \(\texttt{std()}\)...
Physics job
Stats job
Stats job
auto!
\(x\)
\(z\)
\(y\)
\(x\)
\(y\)
1
2
3
4

Field-level inference
Summary stat inference
\(\Omega\)
\(s\)
\(\delta_g\)
\(\Omega\)
\(\delta_L\)
\(s\)
marginalize
condition
marginalize
\(\Omega\)
\(s\)
\(\delta_g\)
\(\Omega\)
\(\delta_L\)
condition
Two approaches to cosmological inference
Cosmo model
\(\mathrm{p}(\Omega,s)\)
\(\mathrm{p}(\Omega \mid s)\)
\(\Omega\)
\(\delta_g\)
\(\mathrm{p}(\Omega,\delta_L,\delta_g, s)= \mathrm{p}(s \mid \delta_g) \, \mathrm{p}(\delta_g \mid \Omega,\delta_L)\, \mathrm{p}(\delta_L \mid \Omega)\, \mathrm{p}(\Omega)\)
\(\mathrm{p}(\Omega,\delta_L \mid \delta_g)\)
\(\mathrm{p}(\Omega \mid \delta_g)\)
\(\delta_g\)
\(\Omega\)
\(\delta_L\)
\(s\)
Two approaches to cosmological inference
Cosmo model
Problem:
- \(s\) is too simple \(\implies\) lossy compression
- \(s\) is too complex \(\implies\) intractable marginalization
The Problem:
- high-dimensional integral $$\mathrm{p}(\Omega \mid \delta_g) = \int \mathrm{p}(\Omega, \delta_L \mid \delta_g) \;\mathrm d \delta_L$$
- To probe scales of \(15\ \mathrm{Mpc}/h\) in DESI volume, \(\operatorname{dim}(\delta_L) \simeq 1024^3\)
The Promise:
- "lossless" explicit inference
Field-level inference
Summary stat inference
We gotta pump this information up
- Field-level
- CNN...
- WST...
- Halo, Peak, Void, Split, Hole...
- 3PCF, Bispectrum
- 2PCF, Power spectrum
- 1D-PDF

$$0-$$
$$\mathrm H(\delta_g)-$$












- At large scales, matter density field almost Gaussian so power spectrum is almost lossless compression, and is relatively tractable
- To prospect smaller non-Gaussian scales, let's use:
We gotta pump this information up
- Field-level
- CNN, GNN...
- WST, 1D-PDFs, Holes...
- Peak, Void, Split, Cluster...
- 3PCF, Bispectrum
- 2PCF, Power spectrum

$$0-$$
$$\mathrm H(\delta_g)-$$













- At large scales, matter density field is Gaussian so power spectrum is lossless compression, and is relatively tractable
- To prospect smaller non-Gaussian scales, let's add:
- all the data
- learn the stat
- multiscale count
- object correlations
- more correlations
- standard analysis
Bayes, information flavor

$$\boldsymbol{H}(X\mid Y_1)$$
$$\boldsymbol{H}(X)$$
$$\boldsymbol{H}( Y_1)$$
$$\boldsymbol{I}(X; Y_1)$$
$$\boldsymbol{H}( Y_2)$$
$$\boldsymbol{I}(X\mid Y_1; Y_2)$$
$$\boldsymbol{H}(X\mid Y_1,Y_2)$$
\(\boldsymbol{H}(X)\) = missing information on \(X\) = amount of bits to communicate \(X\)
$$\boldsymbol{H}(X\mid Y_1)$$
$$\begin{align*}\operatorname{\boldsymbol{H}}(X\mid Y) &= \boldsymbol{H}(Y \mid X) + \boldsymbol{H}(X) - \boldsymbol{H}(Y)\\&= \boldsymbol{H}(X) - \boldsymbol{I}(X;Y) \leq \boldsymbol{H}(X)\end{align*}$$
The summary stats question in cosmology
$$\boxed{\min_s \operatorname{\mathrm{H}}(\Omega\mid s(\delta_g))} = \mathrm{H}(\Omega) - \max_s \mathrm{I}(\Omega ; s(\delta_g))$$
$$\mathrm{H}(\Omega)$$
$$\mathrm{H}(\delta_g)$$
$$\mathrm{H}(\mathcal s_1)$$
$$\mathrm{H}(\mathcal s_2)$$
$$\mathrm{H}(\mathcal P)$$
non-Gaussianities
relevant stat
(low info but high mutual info)
irrelevant stat
(high info but low mutual info)
also a relevant stat
(high info and mutual info)
Which stats are relevant for cosmo inference?
-
Prior on
- Cosmology \(\Omega\)
- Initial field \(\delta_L\)
- EFT Lagrangian parameters \(b\)
(Dark matter-galaxy connection)
-
LSS formation: 2LPT or PM
(BullFrog or FastPM) - Apply galaxy bias
- Redshift-Space Distortions
- Observational noise
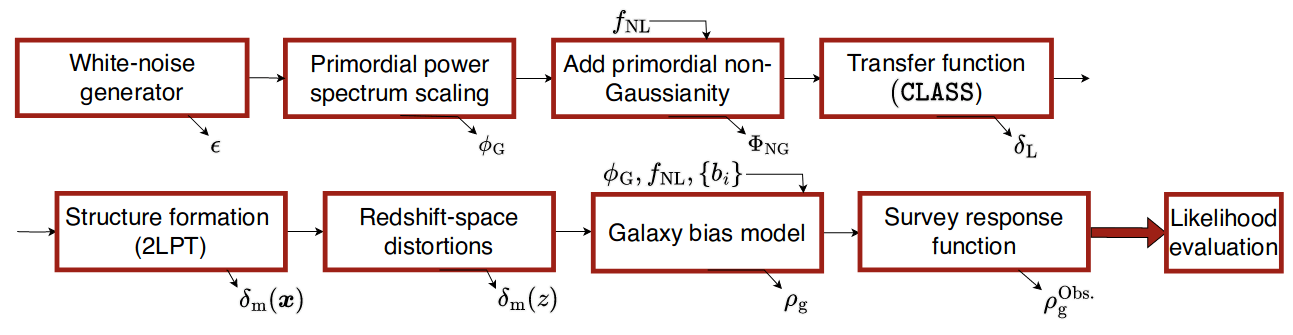
Field-Level Modeling
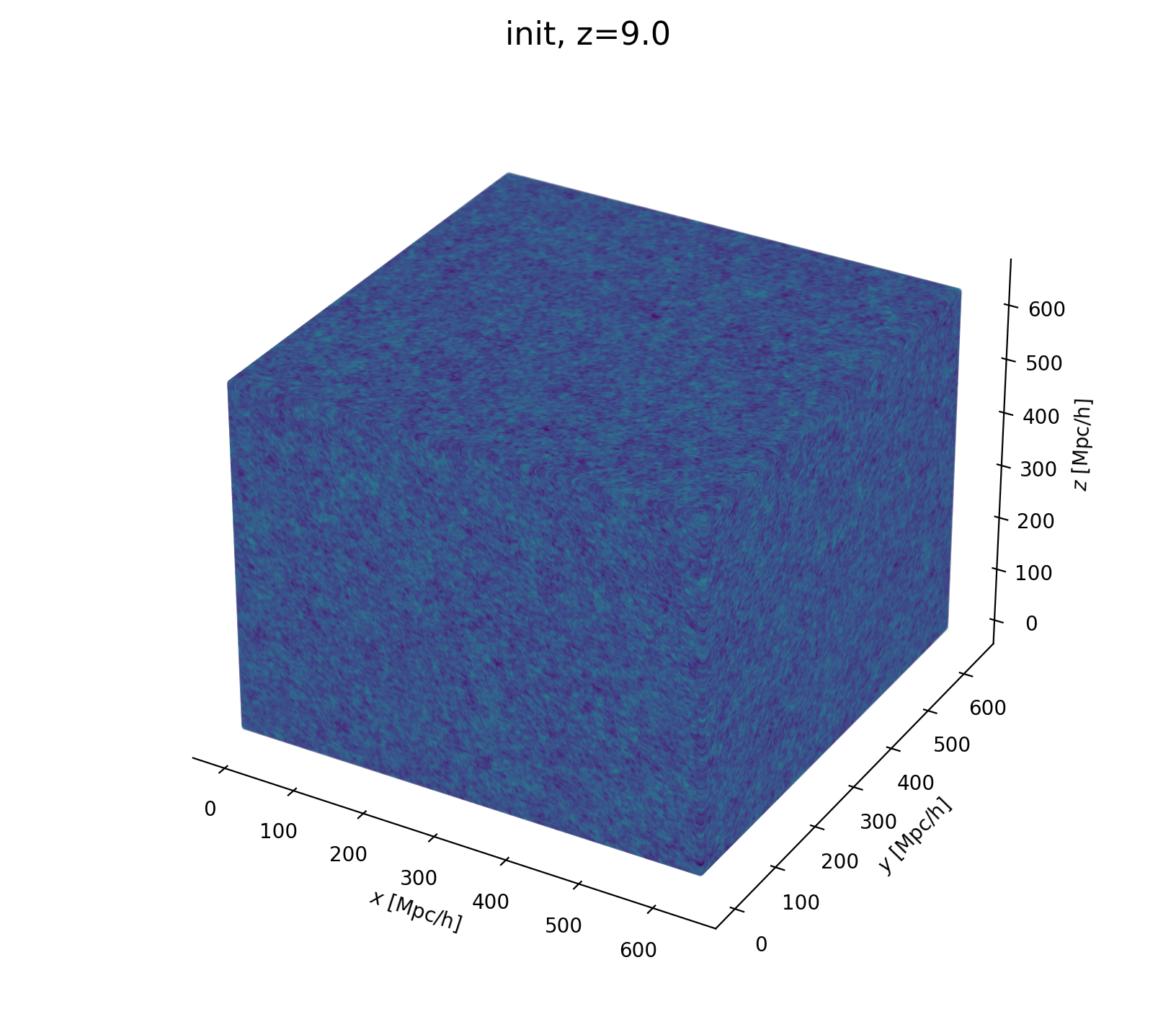
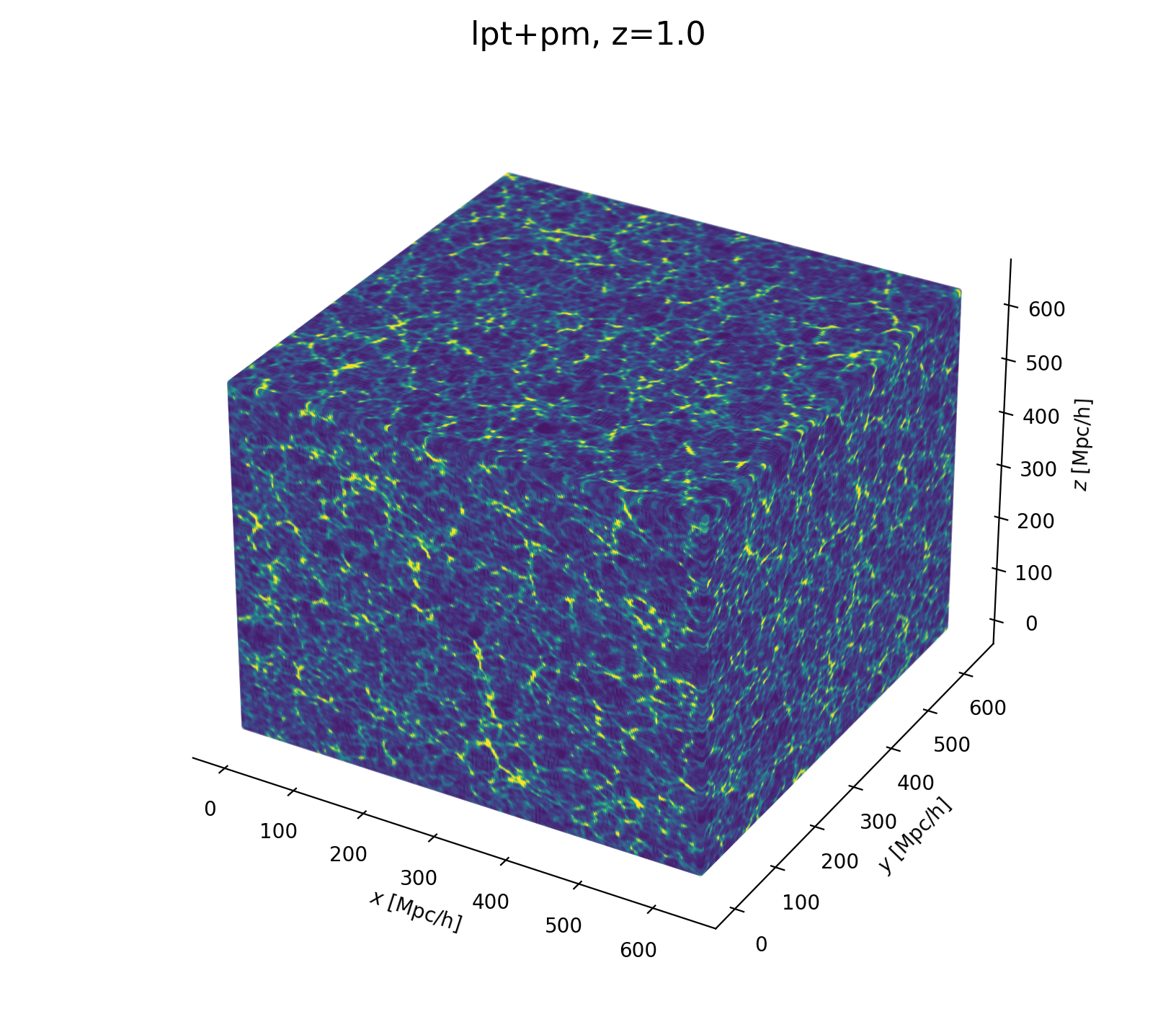
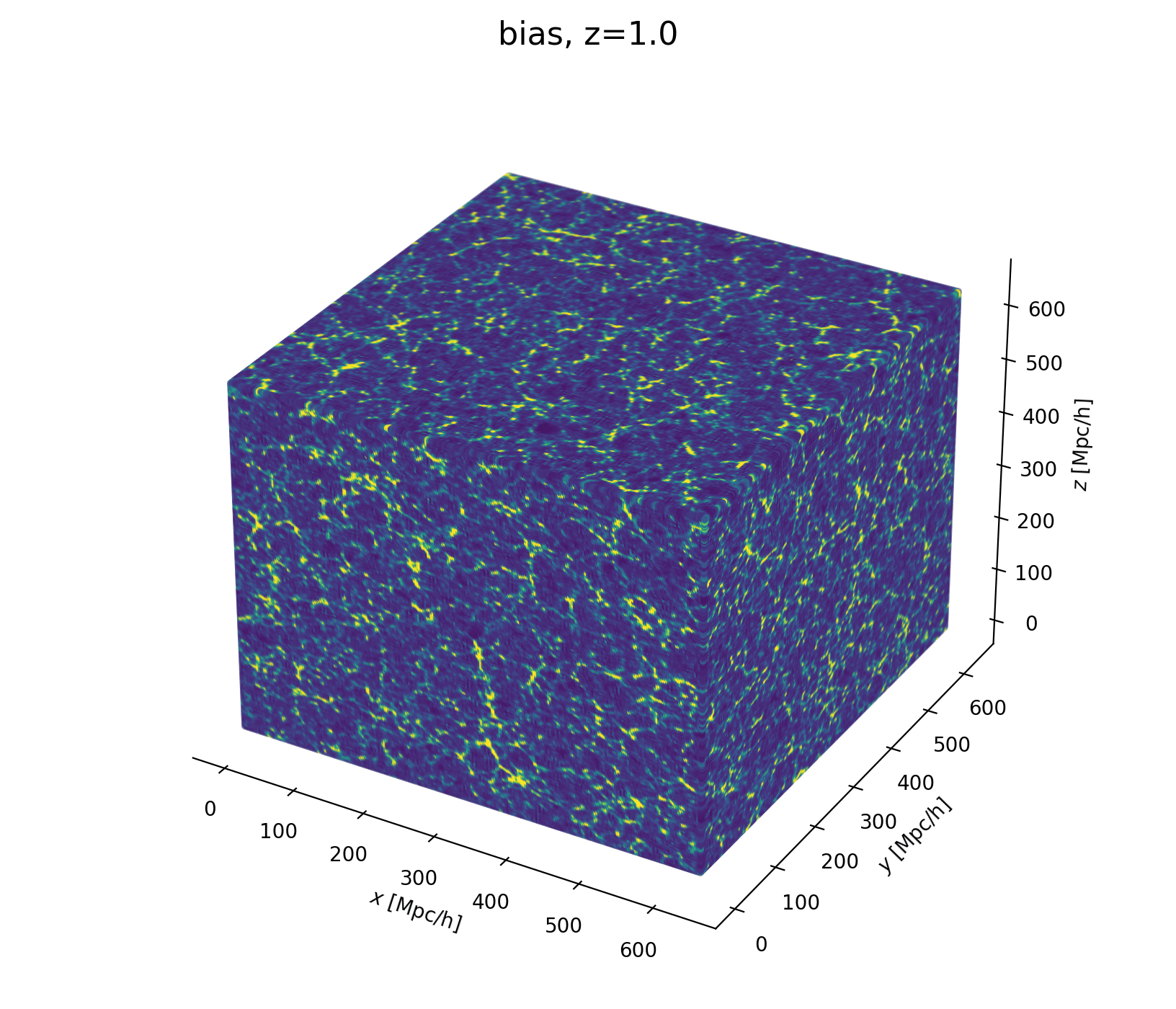
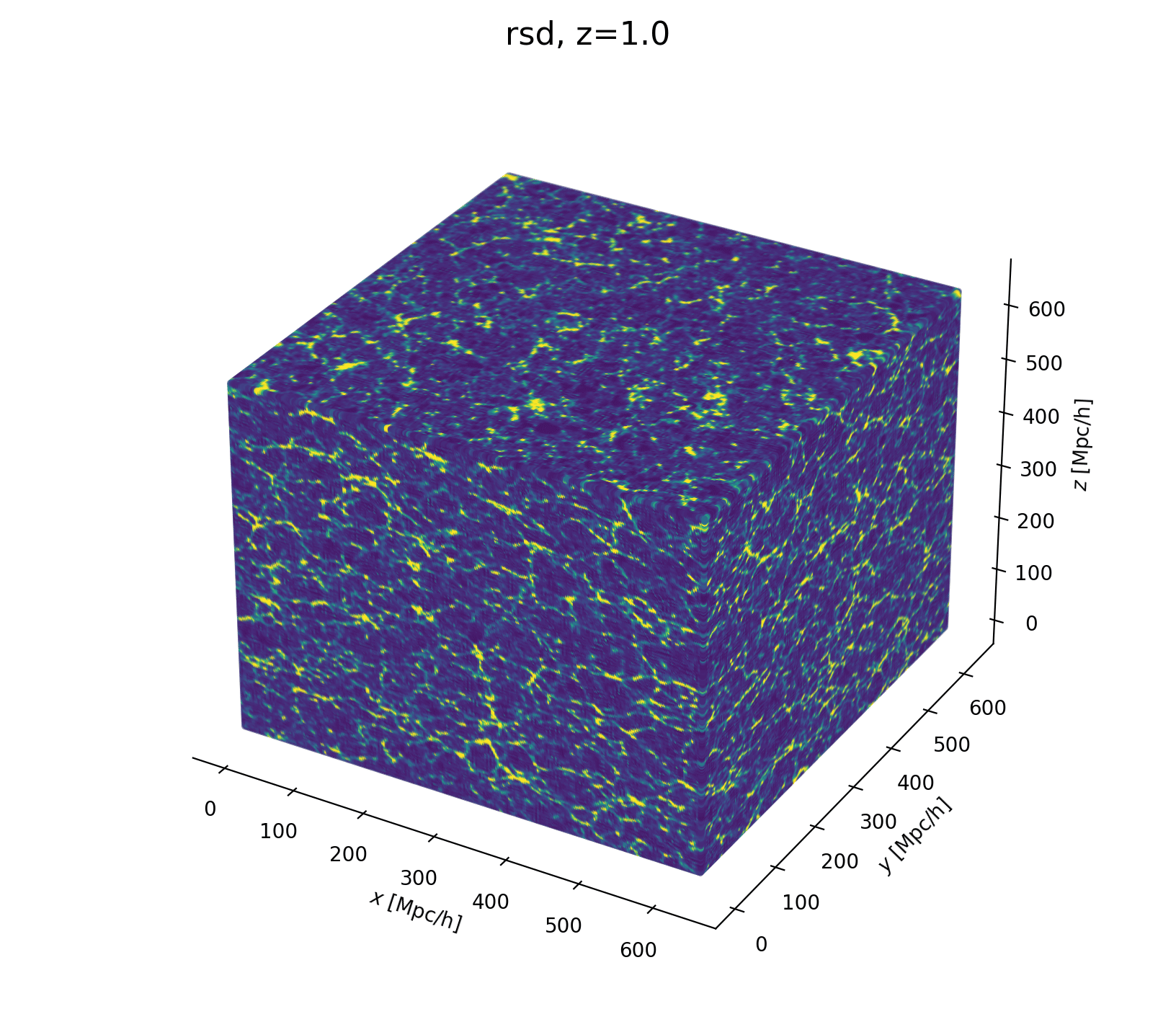
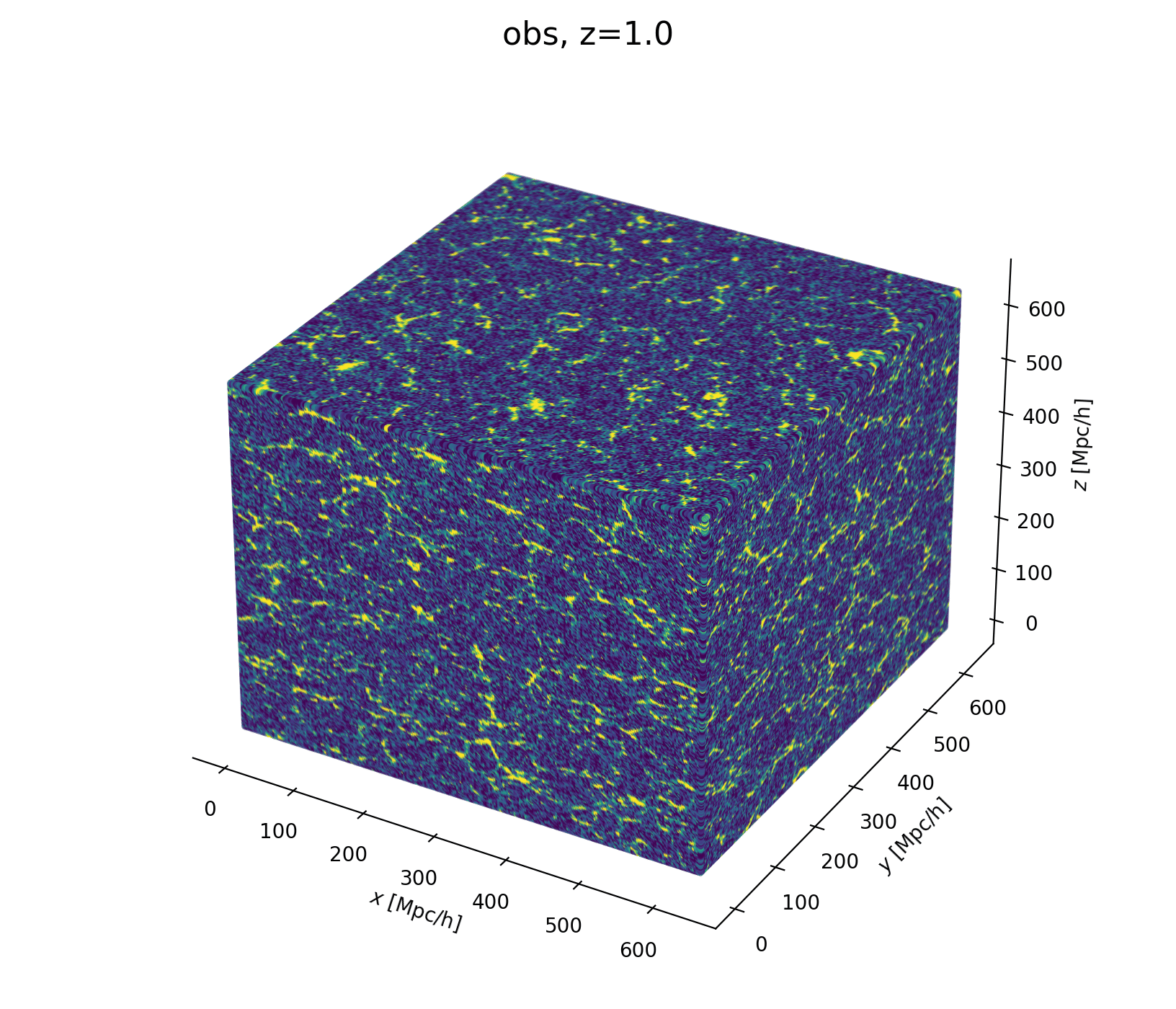
Fast and differentiable model thanks to (\(\texttt{NumPyro}\) and \(\texttt{JaxPM}\))

Field-Level Modeling
Fast and differentiable model thanks to (\(\texttt{NumPyro}\) and \(\texttt{JaxPM}\))

gradients,
they make me:
How to N-body-differentiate?







\((\boldsymbol q, \boldsymbol p)\)
\(\delta(\boldsymbol x)\)
\(\delta(\boldsymbol k)\)
paint*
read*
fft*
ifft*
fft*
*: differentiable, e.g. with via \(\texttt{JaxPM}\), in \(\mathcal O(n \log n)\)
apply forces
to move particles
solve Vlasov-Poisson
to compute forces
\(\begin{cases}\dot {\boldsymbol q} \propto \boldsymbol p\\ \dot{\boldsymbol p} = \boldsymbol f \end{cases}\)
\(\begin{cases}\nabla^2 \phi \propto \delta\\ \boldsymbol f = -\nabla \phi \end{cases} \implies \boldsymbol f \propto \frac{i\boldsymbol k}{k^2} \delta\)
Baryonic Acoustic Oscillations
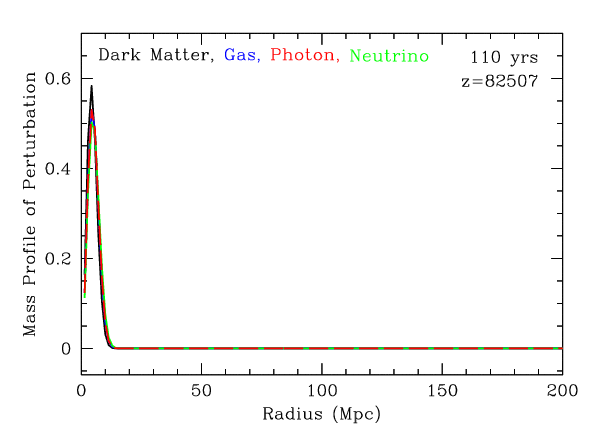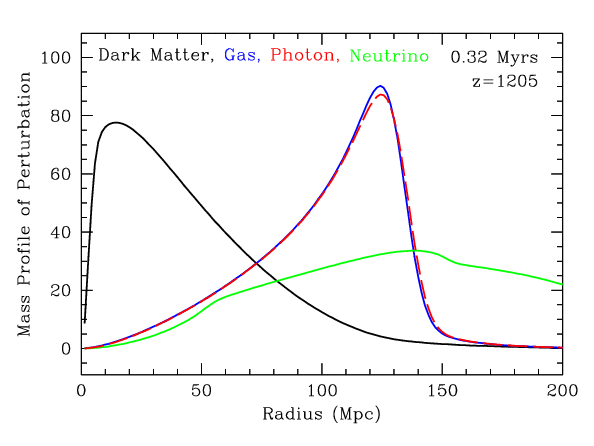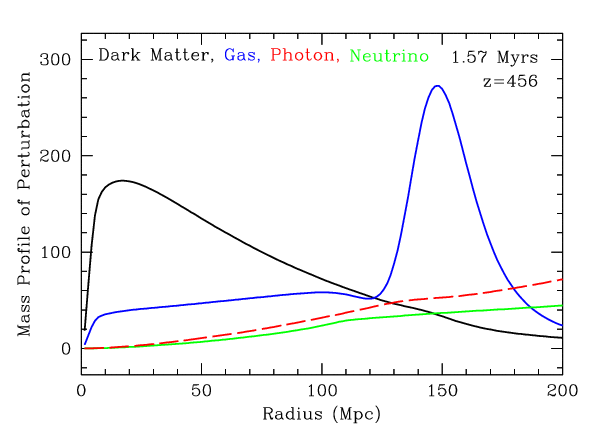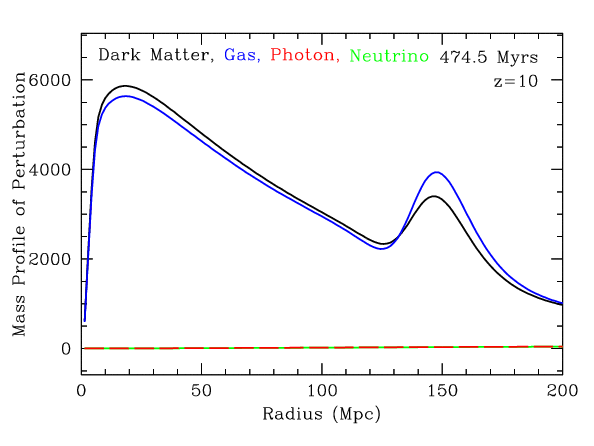
Radial mass profile in \(\mathrm{cMph}\) of initially point-like overdensity at origin
(Infinite Impulse Response)
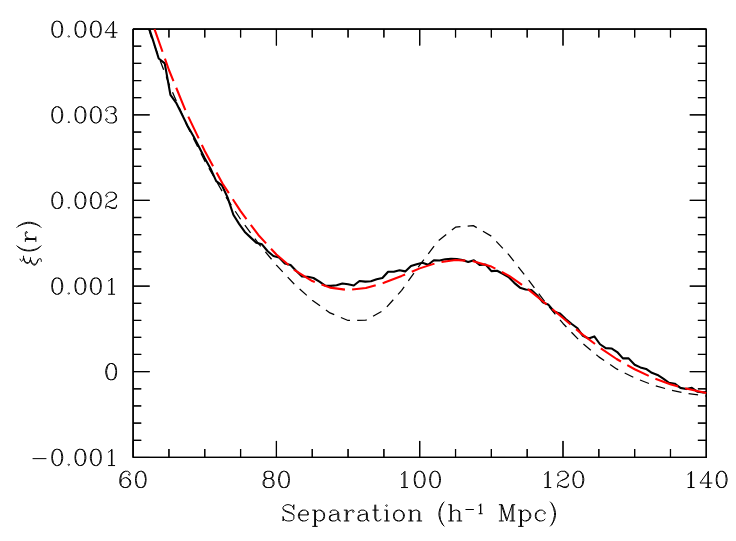
\(z=0.3\)
BAO observation
$$\begin{cases}\frac{\Delta \theta^\mathrm{fid}}{\Delta \theta} = \frac{D_A}{D_A^\mathrm{fid}}\frac{r_d^\mathrm{fid}}{r_d}=: \alpha_\perp\\\frac{\Delta z^\mathrm{fid}}{\Delta z} = \frac{D_H}{D_H^\mathrm{fid}}\frac{r_d^\mathrm{fid}}{r_d}=: \alpha_\parallel\end{cases}$$
\(r_d\)
\(r_d\)
\(\Delta \theta\)
\(\Delta z\)


\(D_A(z)\)
\(\Delta \theta = \frac{r_d}{D_A}\)

BAO observation

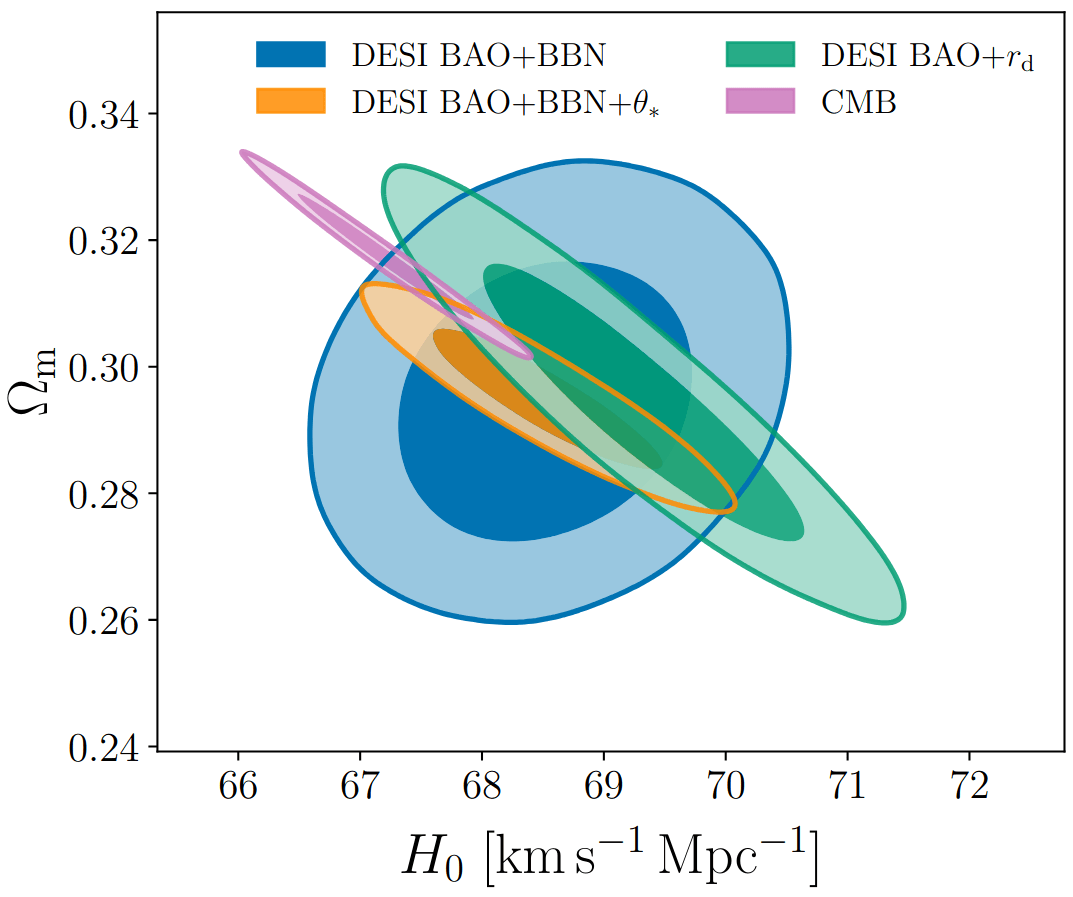
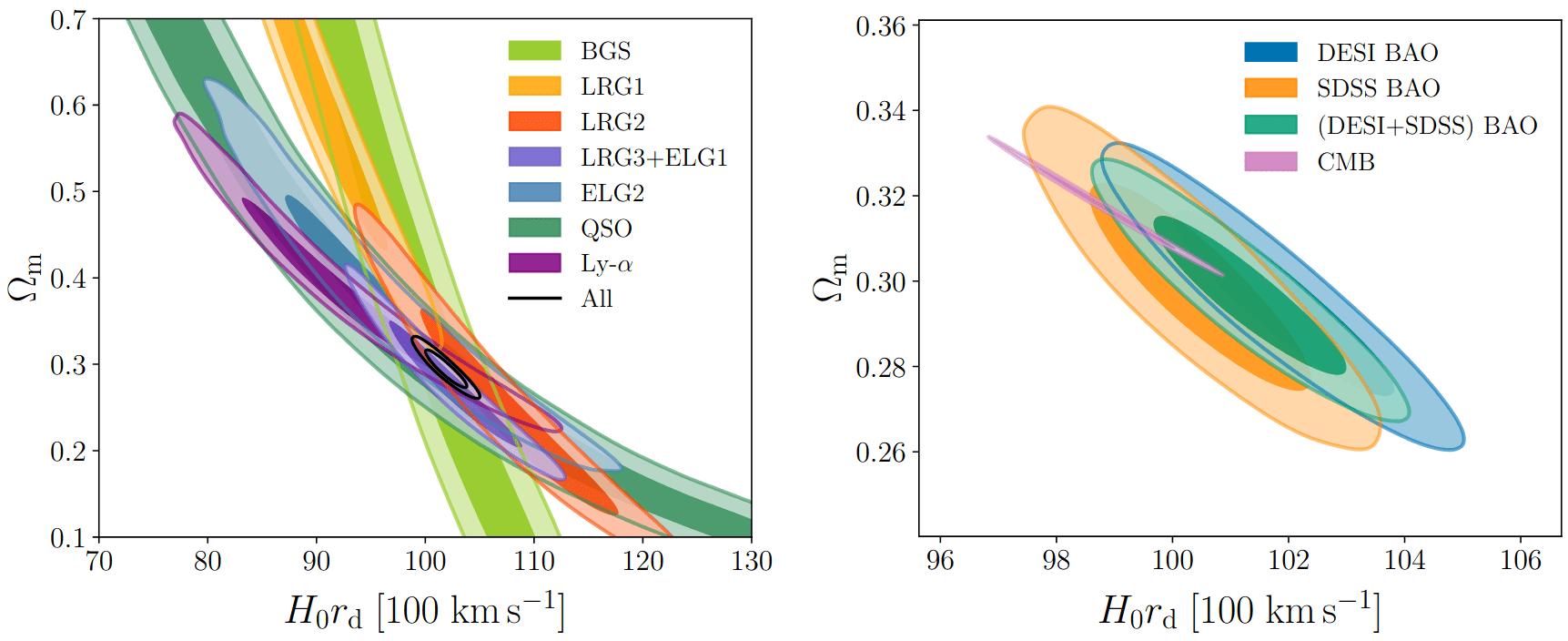
Measuring \(\alpha_\perp(z)\) and \(\alpha_\parallel(z)\) for multiple \(z\) constraints \(\Omega_m\) and \(H_0 r_d\)

\(\alpha_\mathrm{iso}\)
\(\alpha_\mathrm{AP}\)

\(\alpha_\parallel\)
\(\alpha_\perp\)
$$\begin{cases}\frac{\Delta \theta^\mathrm{fid}}{\Delta \theta} = \frac{D_A}{D_A^\mathrm{fid}}\frac{r_d^\mathrm{fid}}{r_d}=: \alpha_\perp\\\frac{\Delta z^\mathrm{fid}}{\Delta z} = \frac{D_H}{D_H^\mathrm{fid}}\frac{r_d^\mathrm{fid}}{r_d}=: \alpha_\parallel\end{cases}$$
Make Acoustic oscillations Sharp Again
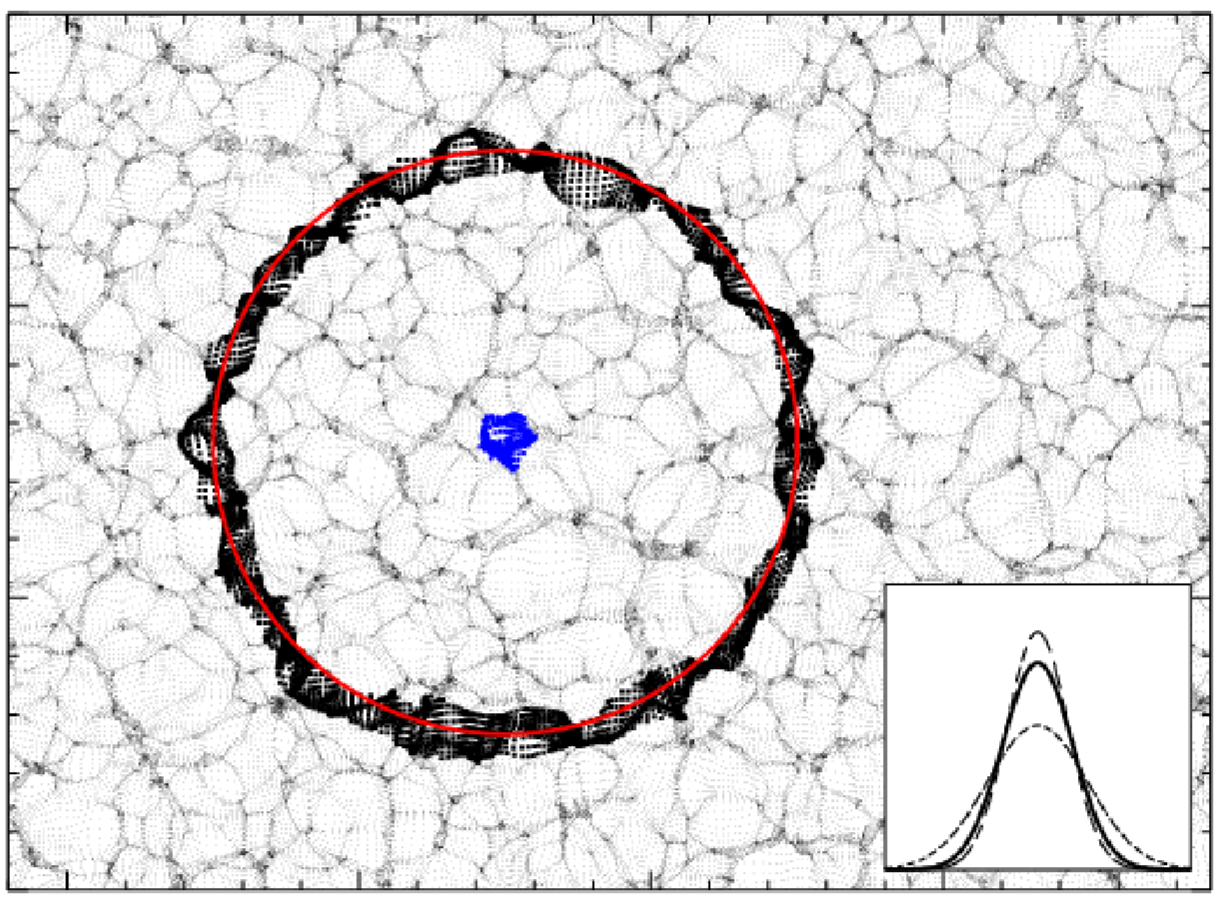
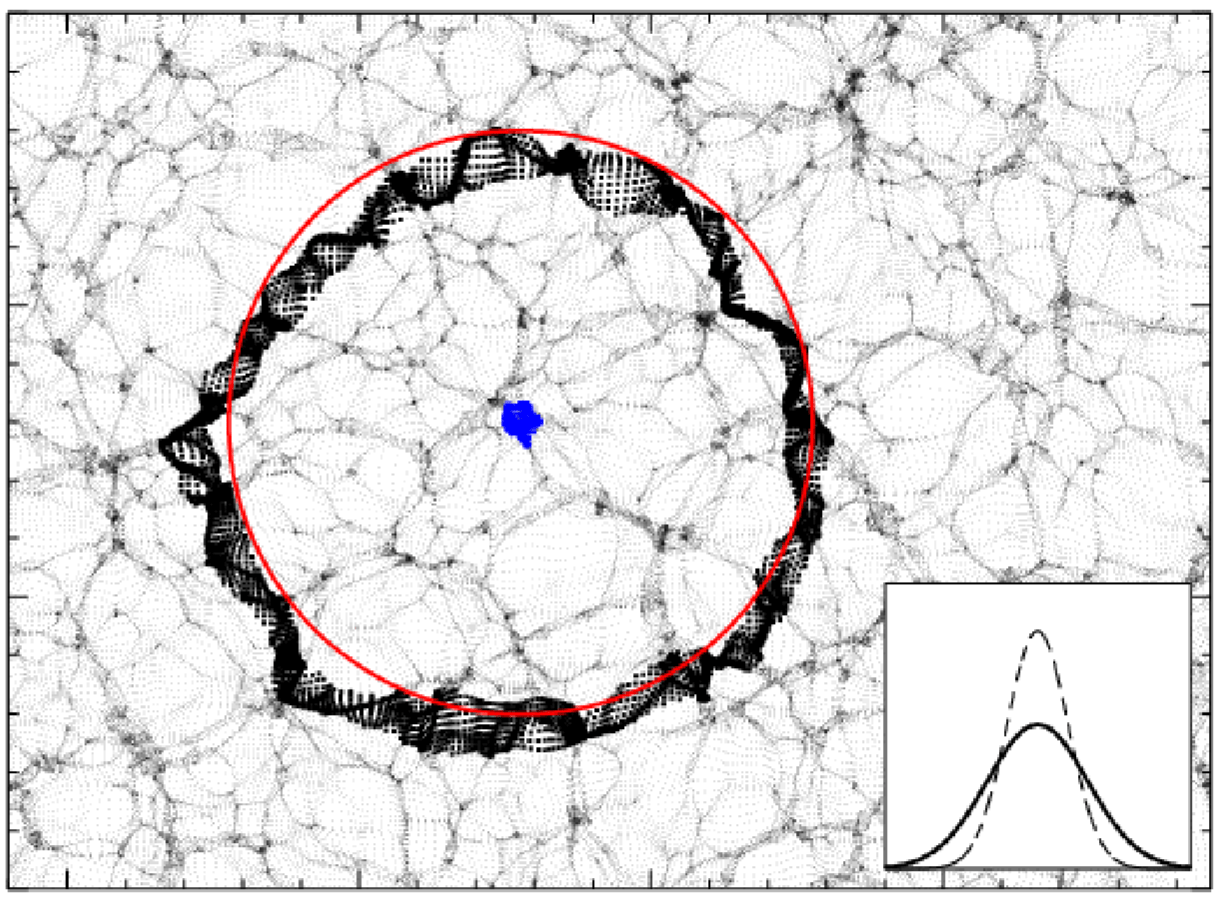
Pictural BAO pre-recon
Pictural BAO post-recon
Field-level Inference
- \(\mathcal H(\boldsymbol q, \boldsymbol p) = U(\boldsymbol q) + \frac 1 2 \boldsymbol p^\top M^{-1} \boldsymbol p\)
- samples canonical ensemble $$\mathrm p_\text{C}(\boldsymbol q, \boldsymbol p) \propto e^{-\mathcal H(\boldsymbol q, \boldsymbol p)} \propto \mathrm p(\boldsymbol q)\,\mathcal N(\boldsymbol 0, M)$$

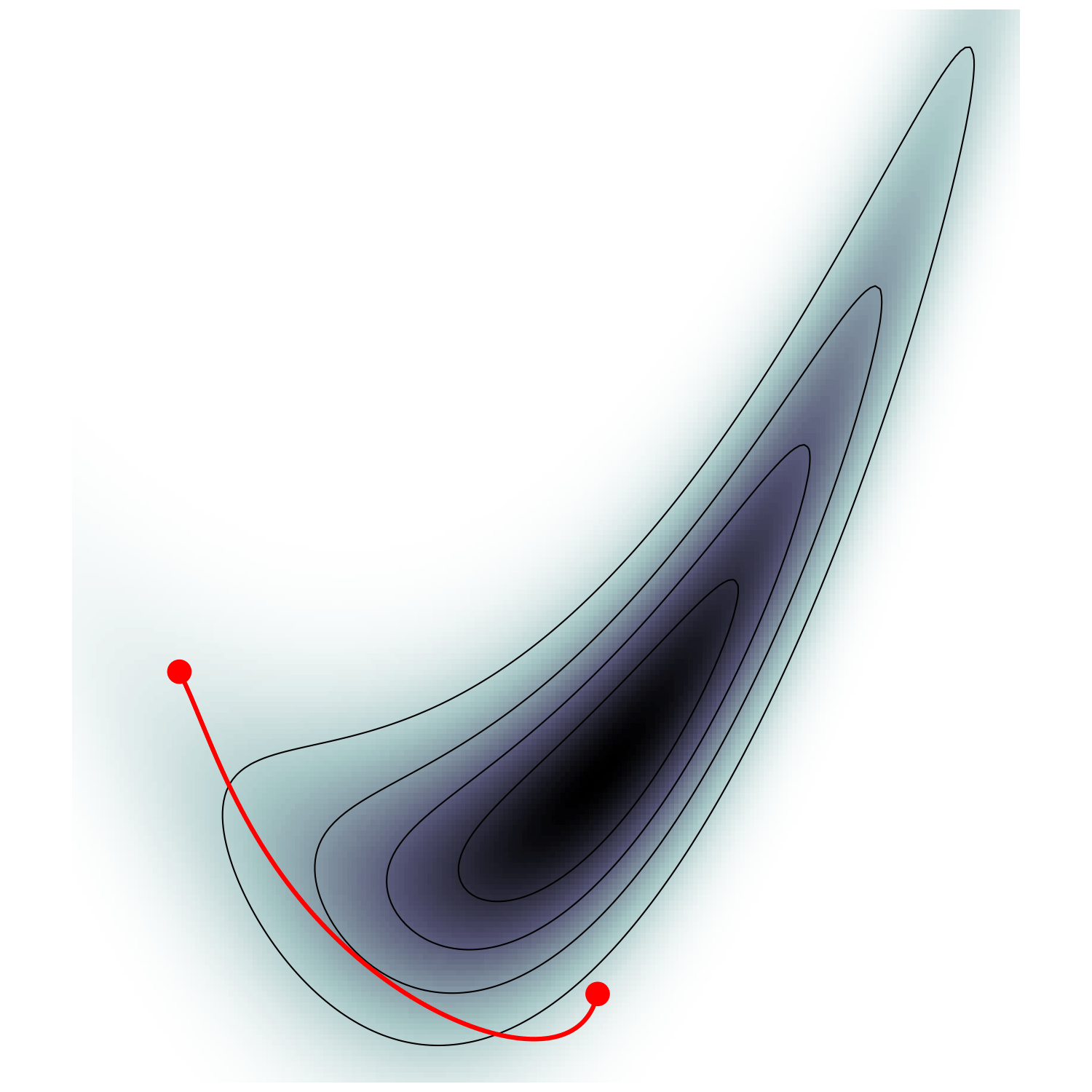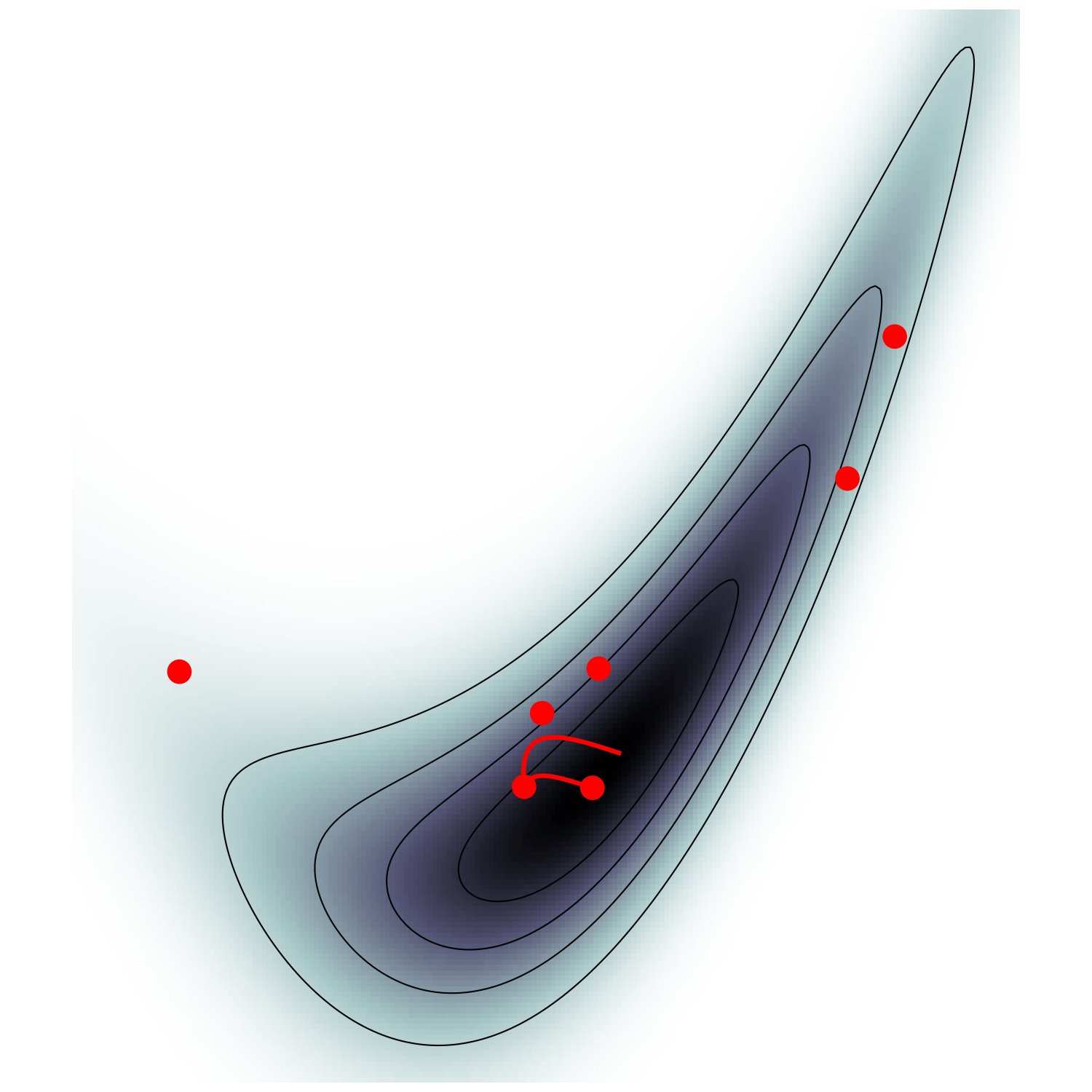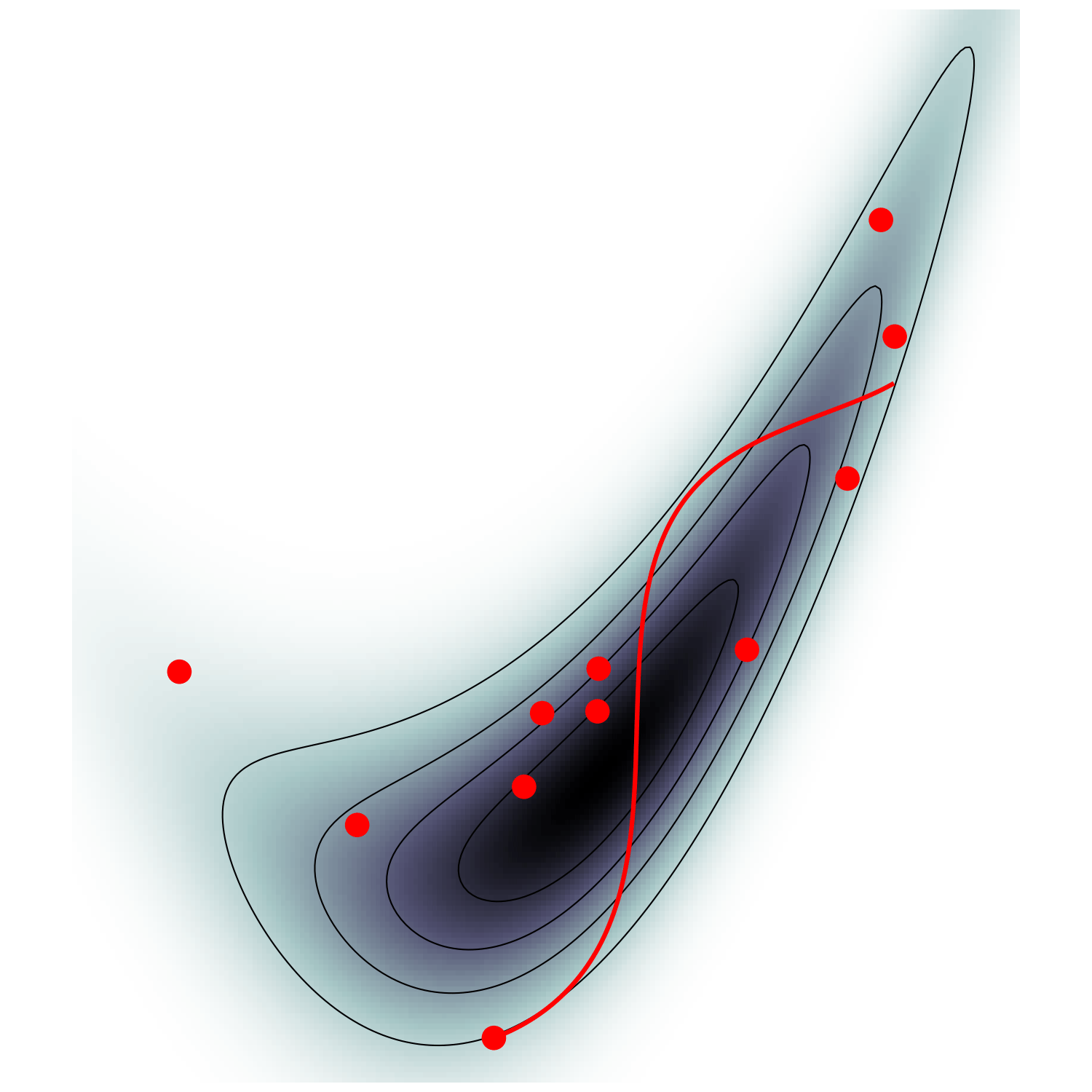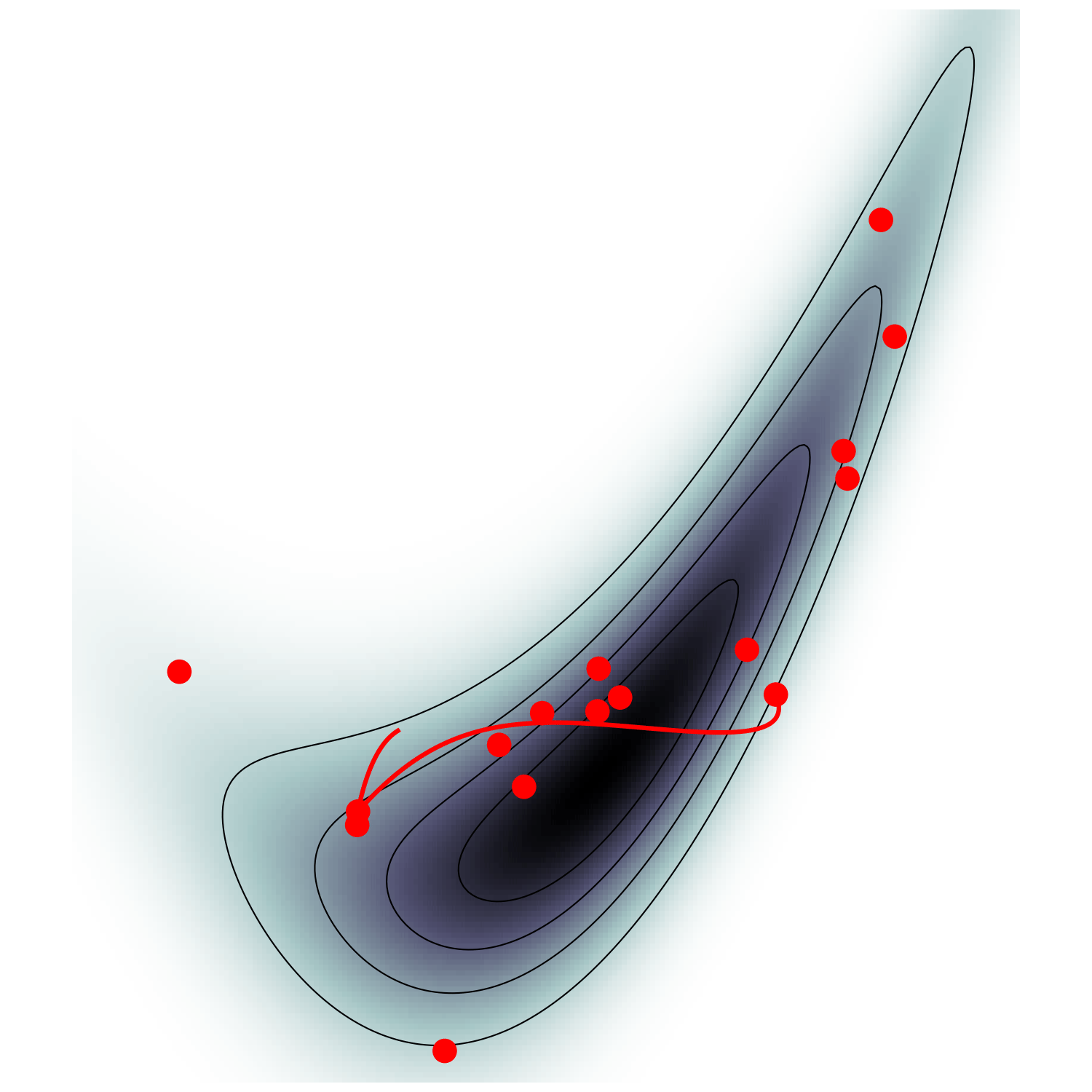
HMC (e.g. Neal2011)
Inferring jointly cosmology, bias parameters, and initial matter field
MCMC sampling
High-dimensional sampling is hard
𝓐 𝓭𝓻𝓾𝓷𝓴 𝓶𝓪𝓷 𝔀𝓲𝓵𝓵 𝓯𝓲𝓷𝓭 𝓱𝓲𝓼 𝔀𝓪𝔂 𝓱𝓸𝓶𝓮, 𝓫𝓾𝓽 𝓪 𝓭𝓻𝓾𝓷𝓴 𝓫𝓲𝓻𝓭 𝓶𝓪𝔂 𝓰𝓮𝓽 𝓵𝓸𝓼𝓽 𝓯𝓸𝓻𝓮𝓿𝓮𝓻 (\(\mathrm p \approx 0.66\))
🌸 𝓢𝓱𝓲𝔃𝓾𝓸 𝓚𝓪𝓴𝓾𝓽𝓪𝓷𝓲
\(-\nabla\)
\(d \approx 1\)
🏠
🚶♀️
To maintain constant move-away probability, step-size \(\simeq d^{-1/2}\)
\(d \gg 1\)

🪺
🐦
Canonical MCMC samplers
Recipe😋 to sample from \(\mathrm p \propto e^{-U}\)
- take particle with position \(\boldsymbol q\), momentum \(\boldsymbol p\), mass matrix \(M\), and Hamiltonian $$\mathcal H(\boldsymbol q, \boldsymbol p) = U(\boldsymbol q) + \frac 1 2 \boldsymbol p^\top M^{-1} \boldsymbol p$$
- follow Hamiltonian dynamics during time \(L\)
$$\begin{cases} \dot {{\boldsymbol q}} = \partial_{\boldsymbol p}\mathcal H = M^{-1}{{\boldsymbol p}}\\ \dot {{\boldsymbol p}} = -\partial_{\boldsymbol q}\mathcal H = - \nabla U(\boldsymbol q) \end{cases}$$and refresh momentum \(\boldsymbol p \sim \mathcal N(\boldsymbol 0,M)\)
- usually, perform Metropolis adjustment
- this samples canonical ensemble $$\mathrm p_\text{C}(\boldsymbol q, \boldsymbol p) \propto e^{-\mathcal H(\boldsymbol q, \boldsymbol p)} \propto \mathrm p(\boldsymbol q)\,\mathcal N(\boldsymbol 0, M)$$
gradient guides particle toward high density sets
scales poorly with dimension
must average over all energy levels

Hamiltonian Monte Carlo (e.g. Neal2011)
Microcanonical MCMC samplers
Recipe😋 to sample from \(\mathrm p \propto e^{-U}\)
-
take particle with position \(\boldsymbol q\), momentum \(\boldsymbol p\), mass matrix \(M\), and Hamiltonian $$\mathcal H(\boldsymbol q, \boldsymbol p) = \frac {\boldsymbol p^\top M^{-1} \boldsymbol p} {2 m(\boldsymbol q)} - \frac{m(\boldsymbol q)}{2} \quad ; \quad m=e^{-U/(d-1)}$$
-
follow Hamiltonian dynamics during time \(L\)
$$\begin{cases} \dot{\boldsymbol q} = M^{-1/2} \boldsymbol u\\ \dot{\boldsymbol u} = -(I - \boldsymbol u \boldsymbol u^\top) M^{-1/2} \nabla U(\boldsymbol q) / (d-1) \end{cases}$$ and refresh \(\boldsymbol u \leftarrow \boldsymbol z/ \lvert \boldsymbol z \rvert \quad ; \quad \boldsymbol z \sim \mathcal N(\boldsymbol 0,I)\)
-
usually, perform Metropolis adjustment
- this samples microcanonical/isokinetic ensemble $$\mathrm p_\text{MC}(\boldsymbol q, \boldsymbol u) \propto \delta(H(\boldsymbol q, \boldsymbol u)) \propto \mathrm p (\boldsymbol q) \delta(|\boldsymbol u|^2 - 1)$$
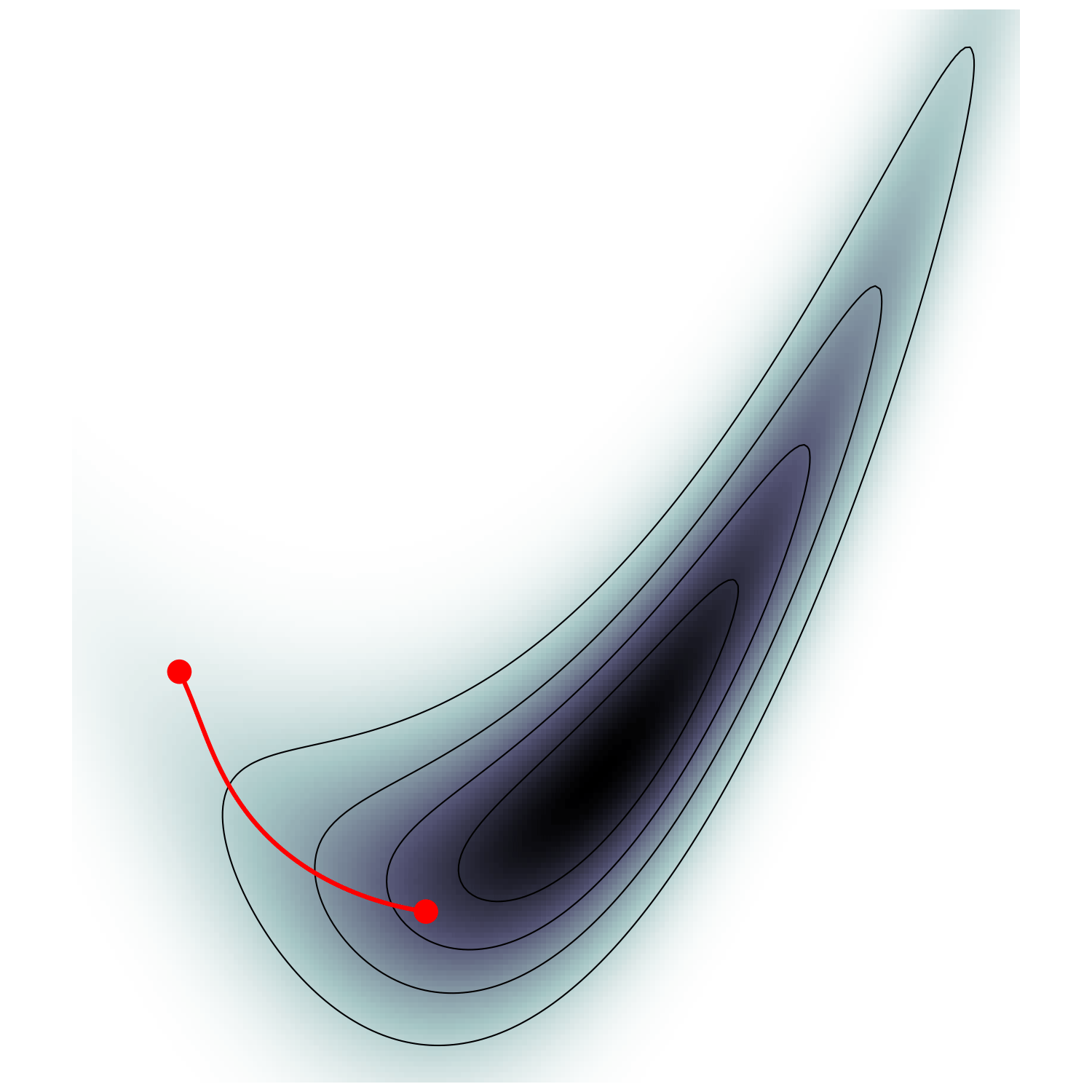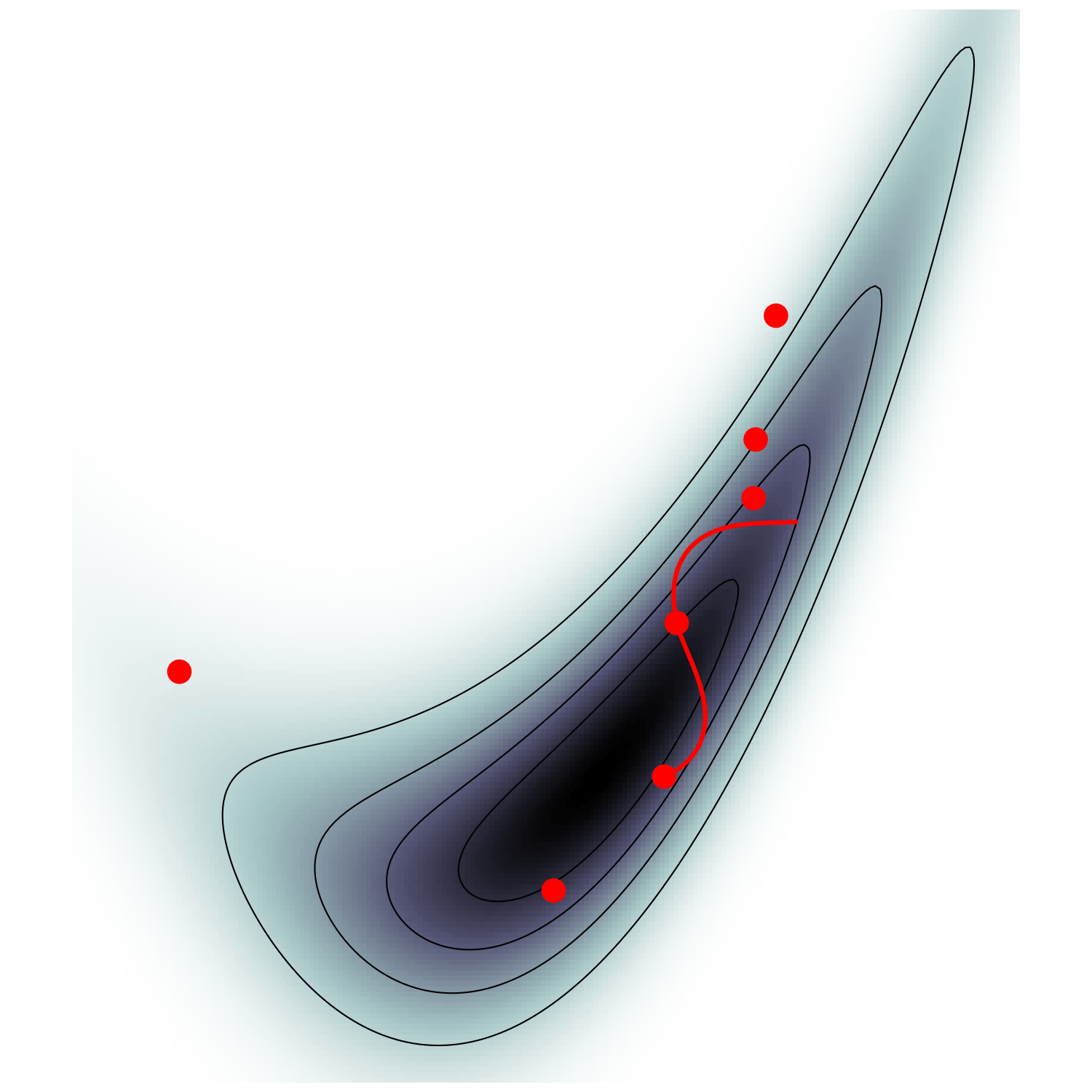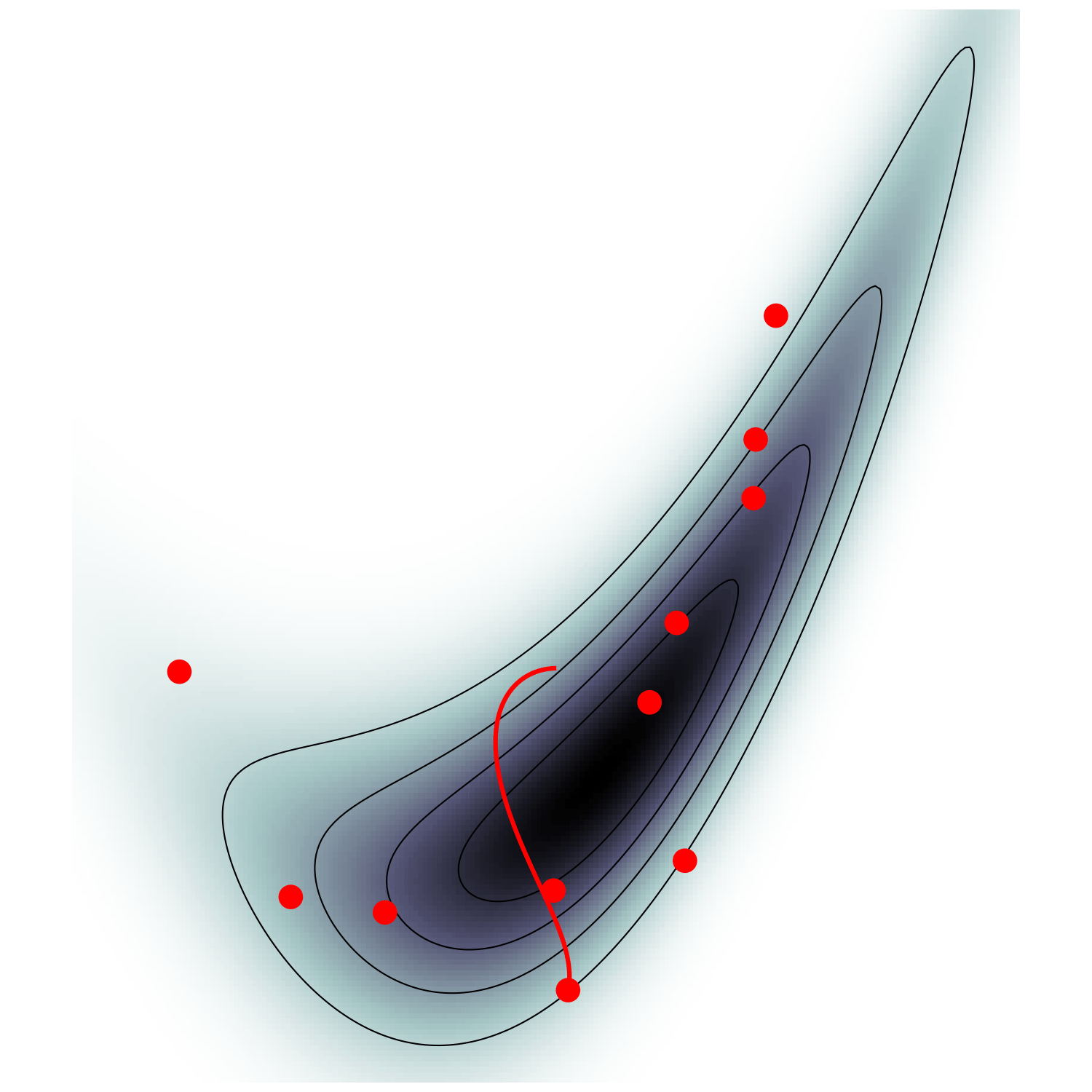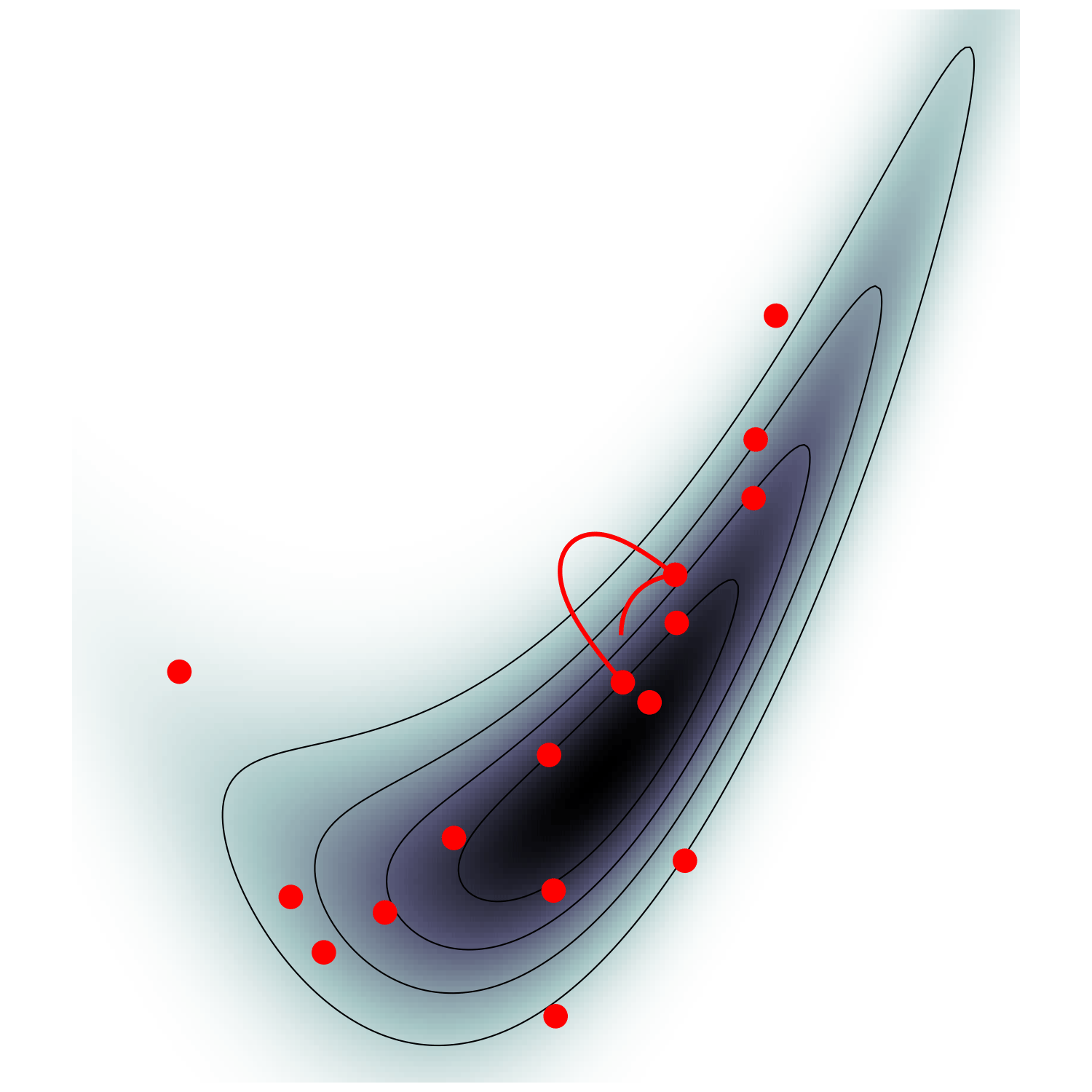
single energy/speed level
let's try avoiding that
gradient guides particle toward high density sets
MicroCanonical HMC (Robnik+2022)
Canonical/Microcanonical MCMC samplers
- \(\mathcal H(\boldsymbol q, \boldsymbol p) = \frac {\boldsymbol p^\top M^{-1} \boldsymbol p} {2 m(\boldsymbol q)} - \frac{m(\boldsymbol q)}{2} \quad ; \quad m:=e^{-U/(d-1)}\)
- samples microcanonical/isokinetic ensemble $$\mathrm p_\text{MC}(\boldsymbol q, \boldsymbol p) \propto \delta(\mathcal H(\boldsymbol q, \boldsymbol p)) \propto \mathrm p (\boldsymbol q) \delta(\dot{\boldsymbol q}^\top M \dot{\boldsymbol q} - 1)$$

Hamiltonian Monte Carlo (e.g. Neal2011)

MicroCanonical HMC (Robnik+2022)
- \(\mathcal H(\boldsymbol q, \boldsymbol p) = U(\boldsymbol q) + \frac 1 2 \boldsymbol p^\top M^{-1} \boldsymbol p\)
- samples canonical ensemble $$\mathrm p_\text{C}(\boldsymbol q, \boldsymbol p) \propto e^{-\mathcal H(\boldsymbol q, \boldsymbol p)} \propto \mathrm p(\boldsymbol q)\,\mathcal N(\boldsymbol 0, M)$$
What it looks like
Inferring jointly cosmology, bias parameters, and initial matter field allows full universe history reconstruction
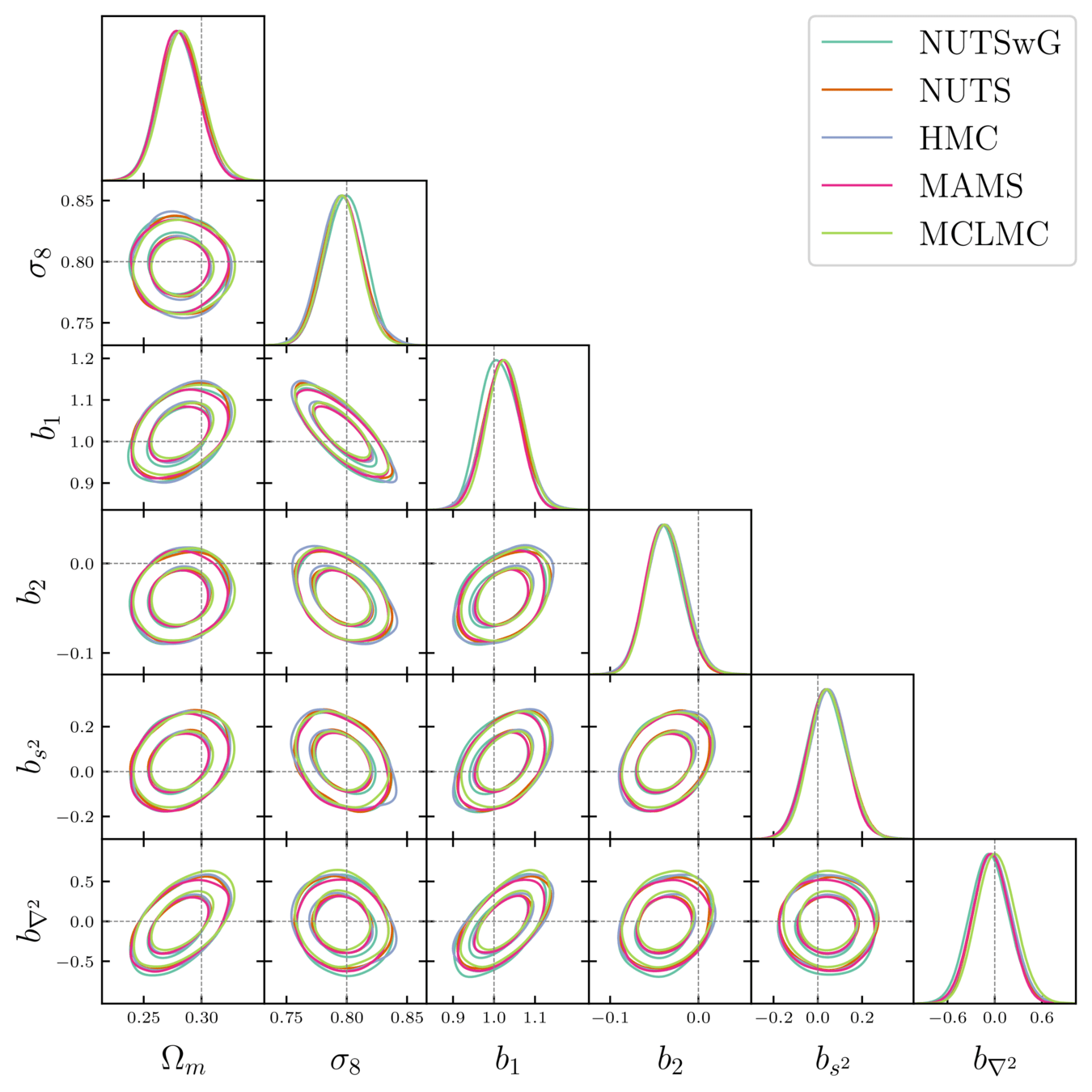


- Different samplers and strategies used for field-level (e.g. Lavaux+2018, Bayer+2023). Additional comparisons required.
- We provide a consistent benchmark for field-level from galaxy surveys. Build upon \(\texttt{NumPyro}\) and \(\texttt{BlackJAX}\).
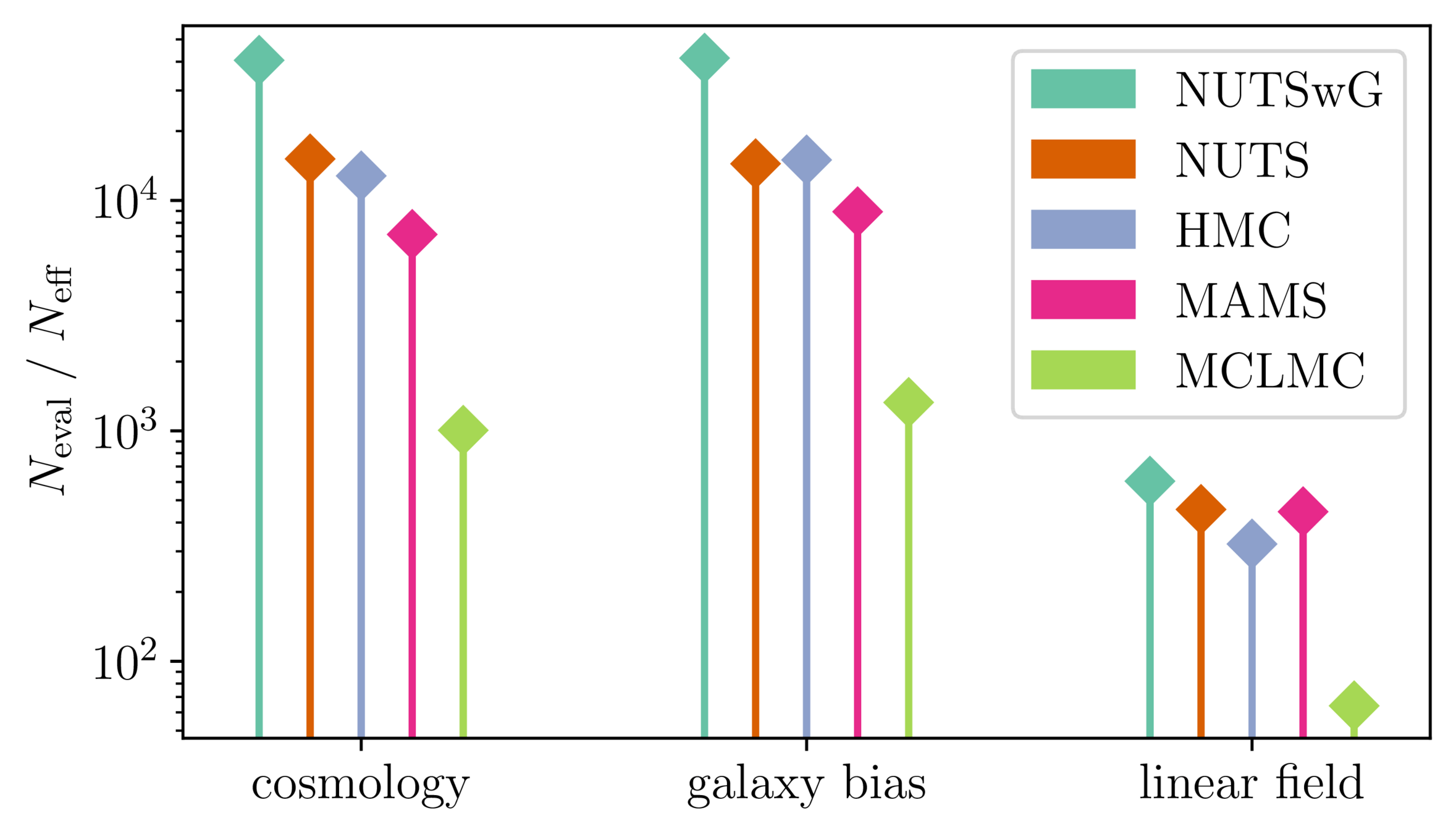
Samplers comparison
= NUTS within Gibbs
= auto-tuned HMC
= adjusted MCHMC
= unadjusted Langevin MCHMC
10 times less evaluations required
Unadjusted microcanonical sampler outperforms any adjusted sampler
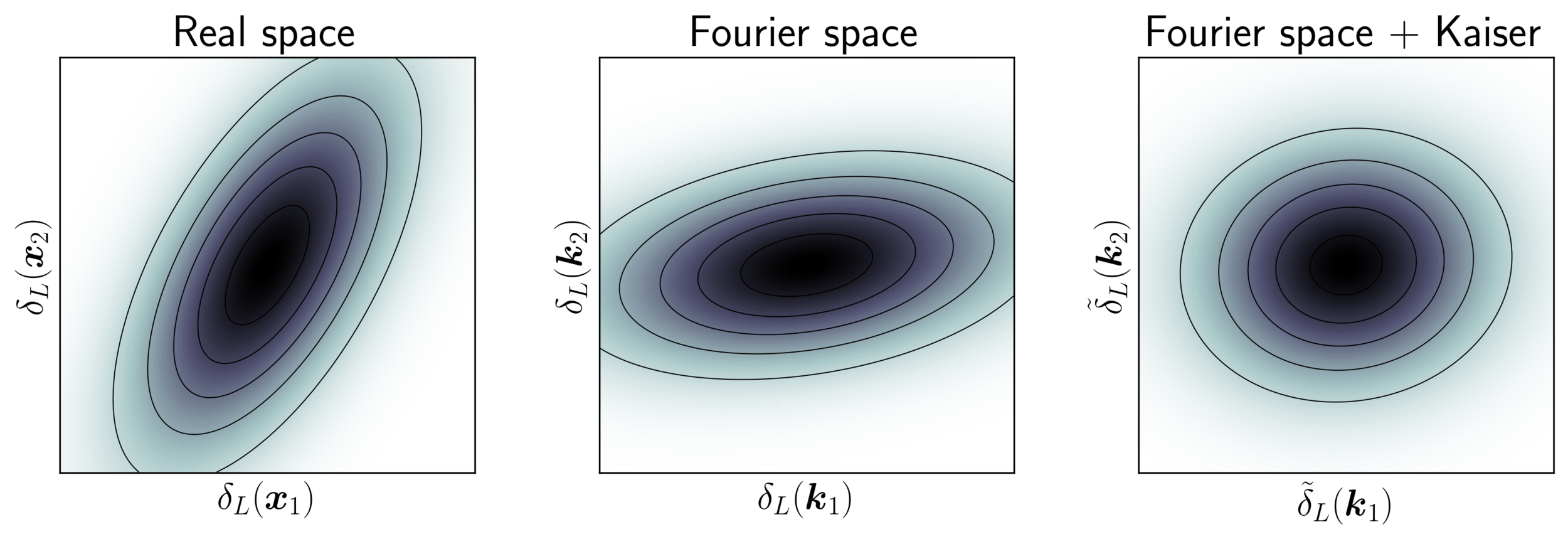
Model preconditioning
- Sampling is easier when target density is isotropic Gaussian
- The model is reparametrized assuming a tractable Kaiser model:
linear growth + linear Eulerian bias + flat sky RSD + Gaussian noise
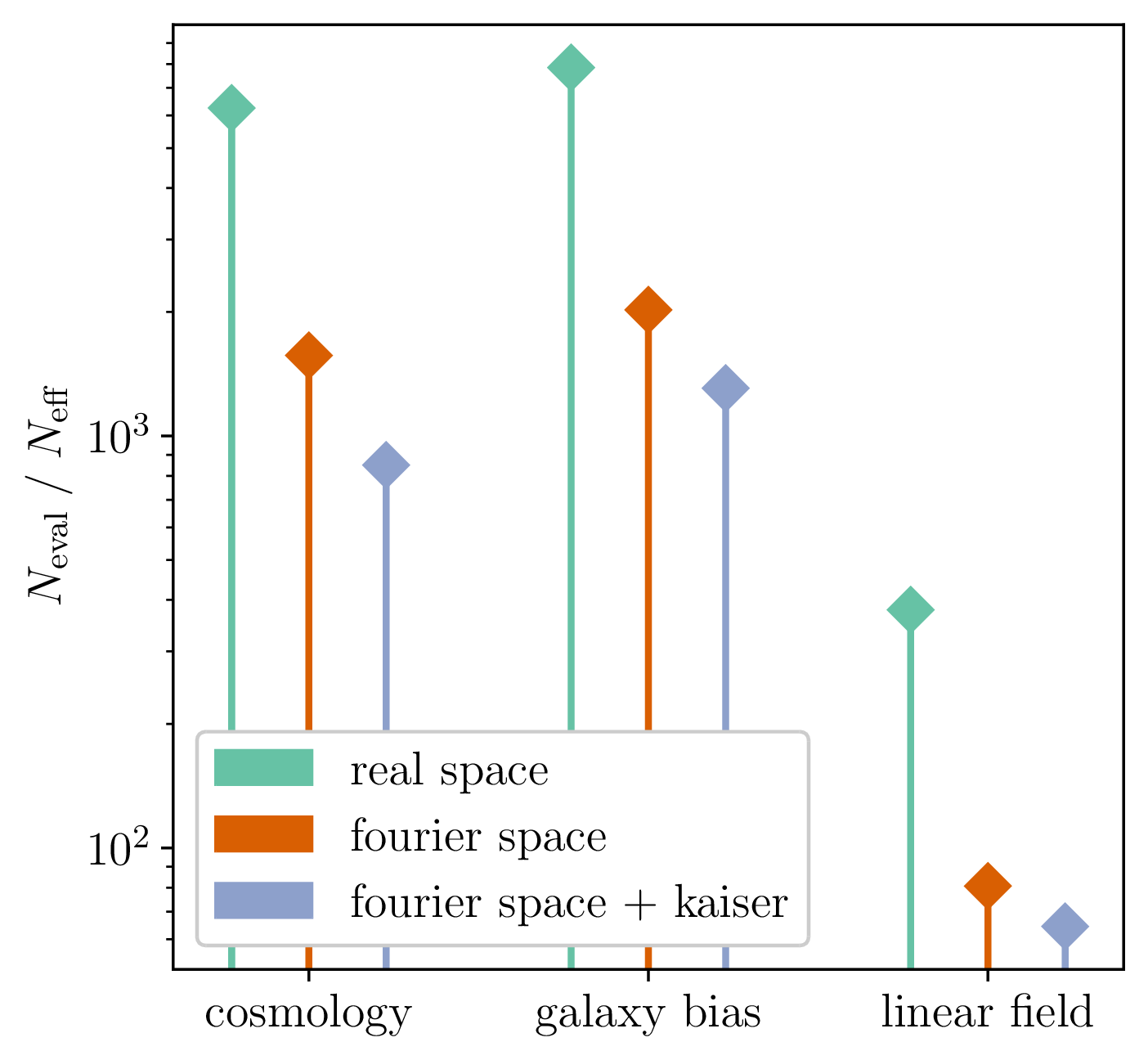
10 times less evaluations required


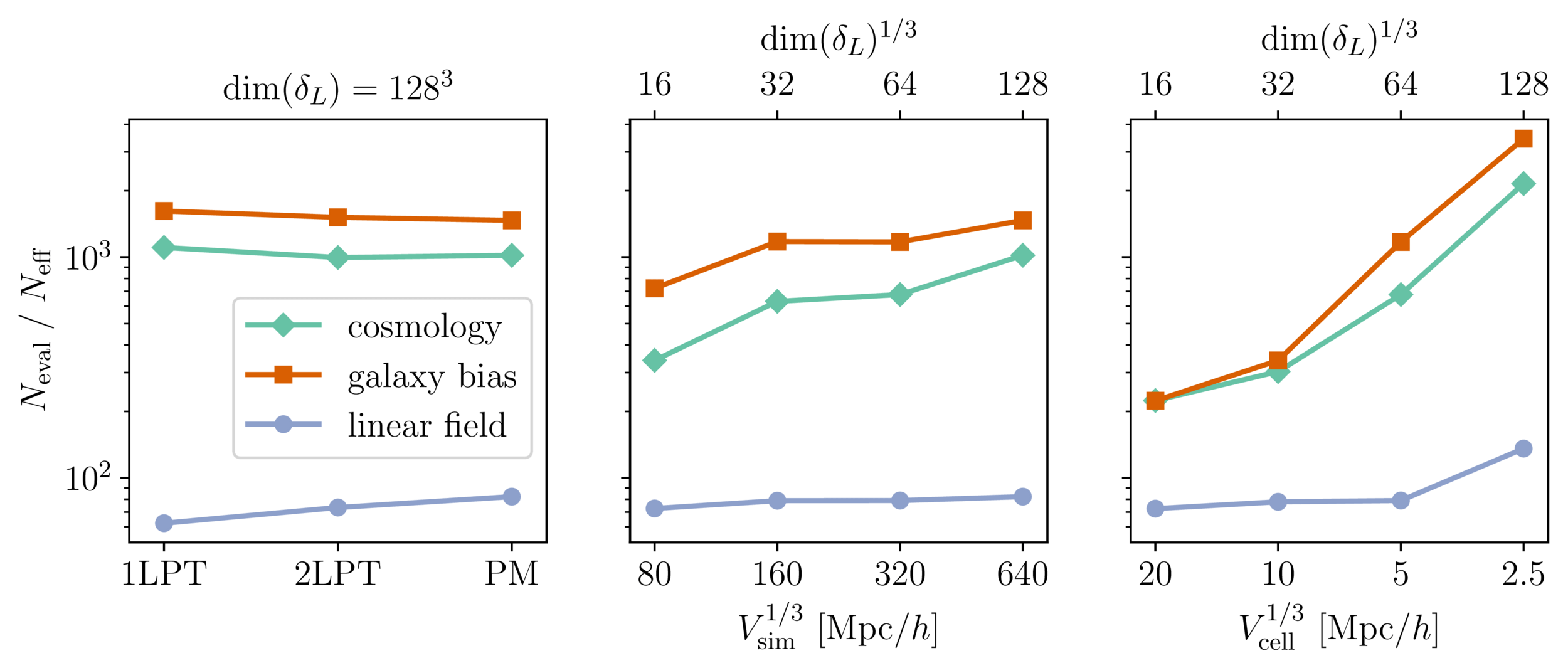
Benchmark results
- Promising for future inferences, going multi-GPU using JaxDecomp
- Code readily available at github.com/hsimonfroy/benchmark-field-level
\(128^3\) PM on 8GPU:
4h MCLMC vs. \(\geq\)80h NUTS
Mildly dependent with respect to formation model and volume
Probing smaller scales could be harder
Handling bias in unadjusted MCLMC
- Microcanonical dynamics \(\implies\) energy should not vary
- Numerical integration yields quantifiable errors that can be linked to bias
- Stepsize can be tuned to ensure controlled bias, see Robnik+2024
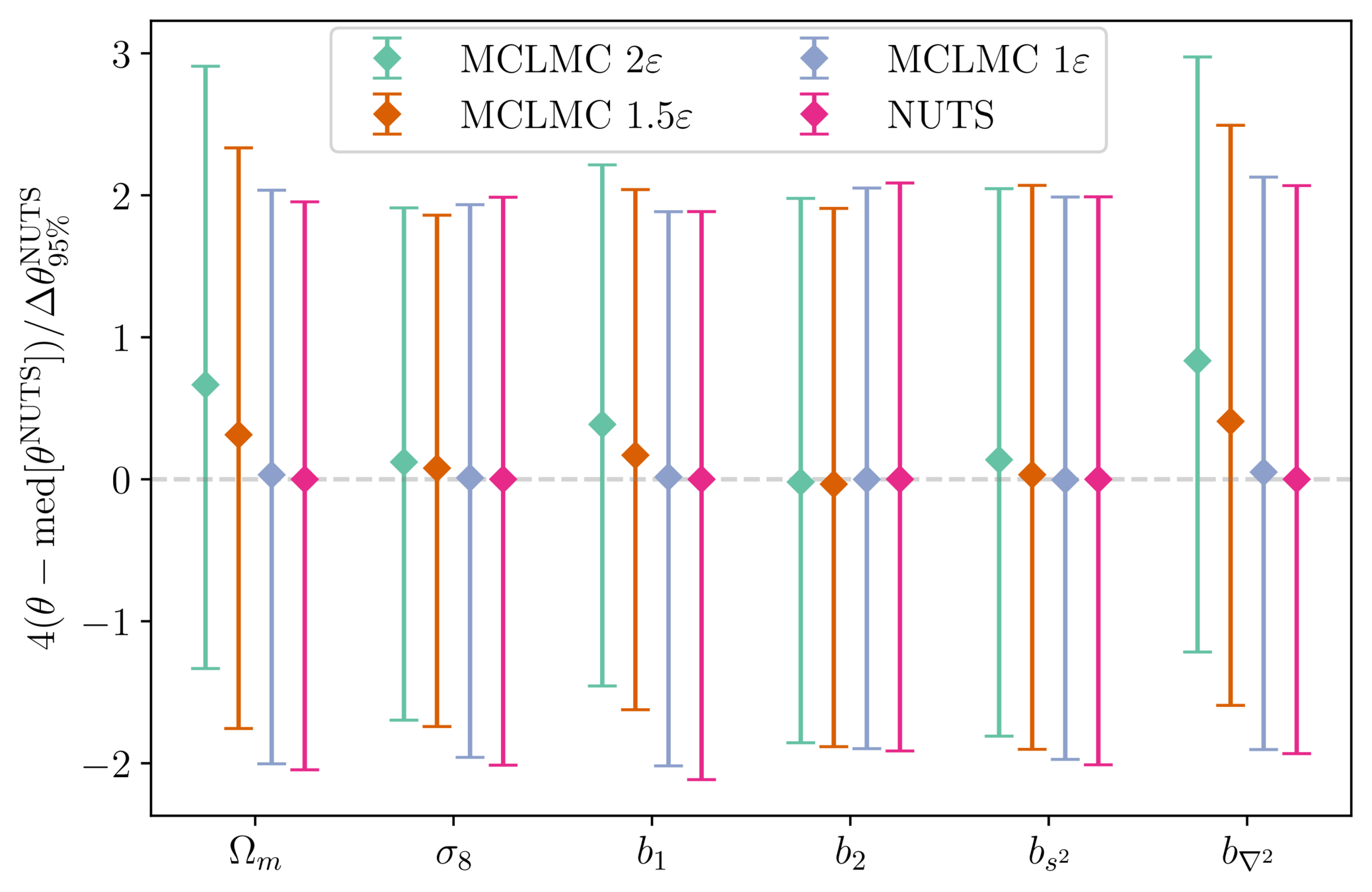
reducing stepsize rapidly brings bias under Monte Carlo error
-
Effective Sample Size (ESS)
- number of i.i.d. samples that yield same statistical power.
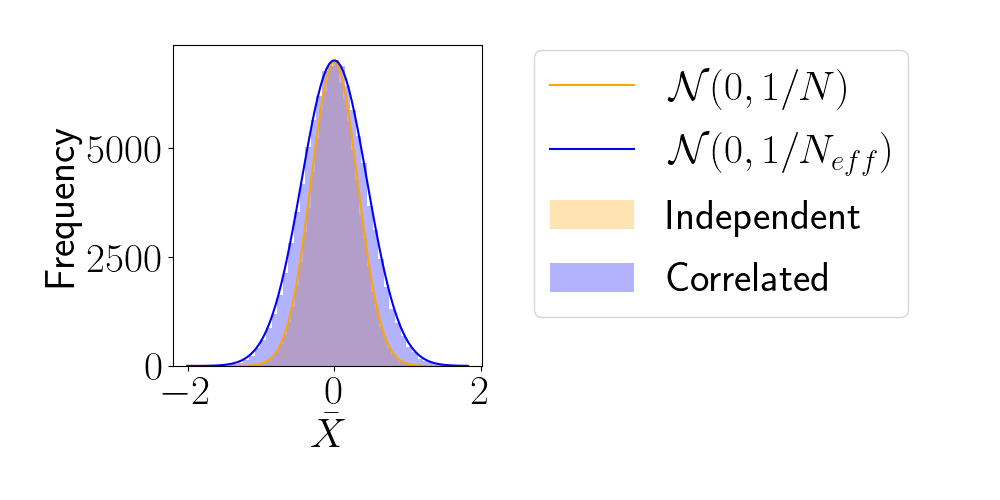
- For sample sequence of size \(N\) and autocorrelation \(\rho\) $$N_\textrm{eff} = \frac{N}{1+2 \sum_{t=1}^{+\infty}\rho_t}$$so aim for as less correlated sample as possible.
- Main limiting computational factor is model evaluation (e.g. N-body), so characterize MCMC efficiency by \(N_\text{eval} / N_\text{eff}\)
How to compare samplers?
Primordial Non-Gaussianity from galaxies
Local-type PNG is constrained by the induced scale-dependent bias
\(\phi_{\mathrm{NL}}=\phi+{\color{purple}f_{\mathrm{NL}}}\phi^{2}\)
\(\delta(\boldsymbol k)\simeq\left(b_{1}+ b_\phi {\color{purple}f_\mathrm{NL}}k^{-2} \right) \delta_L(\boldsymbol k)\)
Field-level modeling of PNG
$$\begin{align*}w_g&=1+{\color{purple}b_{1}}\,\delta_{\rm L}+{\color{purple}b_{2}}\delta_{\rm L}^{2}+{\color{purple}b_{s^2}}s^{2}+ {\color{purple}b_{\nabla^2}} \nabla^2 \delta _{\rm L}\\&\quad\quad\! + {\color{purple}b_\phi f_{\rm NL}} \phi + {\color{purple} b_{\phi\delta} f_{\rm NL}} \phi \delta_{\rm L}\\\Delta \boldsymbol q_\parallel &= H^{-1} \dot{\boldsymbol q}_\parallel + {\color{purple}b_{\nabla_\parallel}} \nabla_\parallel \delta_\mathrm{L}\end{align*}$$
\(\phi_{\mathrm{NL}}=\phi+{\color{purple}f_{\mathrm{NL}}}\phi^{2}\)
\(\boldsymbol q_\mathrm{LPT} \simeq \boldsymbol q_\mathrm{in} + \Psi_\mathrm{LPT}(\boldsymbol q_\mathrm{in}, z(\boldsymbol q_\mathrm{in}))\)
one-shot 2LPT light-cone
\(n_g^\mathrm{obs}(\boldsymbol q) = (1+\delta_g(\boldsymbol q))\, {\color{purple}\bar n_g(\,r)}\, {\color{blue}W(\boldsymbol q)}\, {\color{purple}\beta_i} {\color{green}T^i(\theta)}\)
RIC relax + selection + imag. templates
\(\delta_g \sim \mathcal N(\delta_g^\mathrm{det}, \sigma^2)\) with
\(\sigma(k) = {\color{purple}\sigma_0}(1+{\color{purple}\sigma_2} k^2 + {\color{purple}\sigma_{\mu2}}(k\mu)^2)\)
EFT-based modeling, many scale cuts alleviating discretization effects (see Stadler+2024)
A word on integral constraints
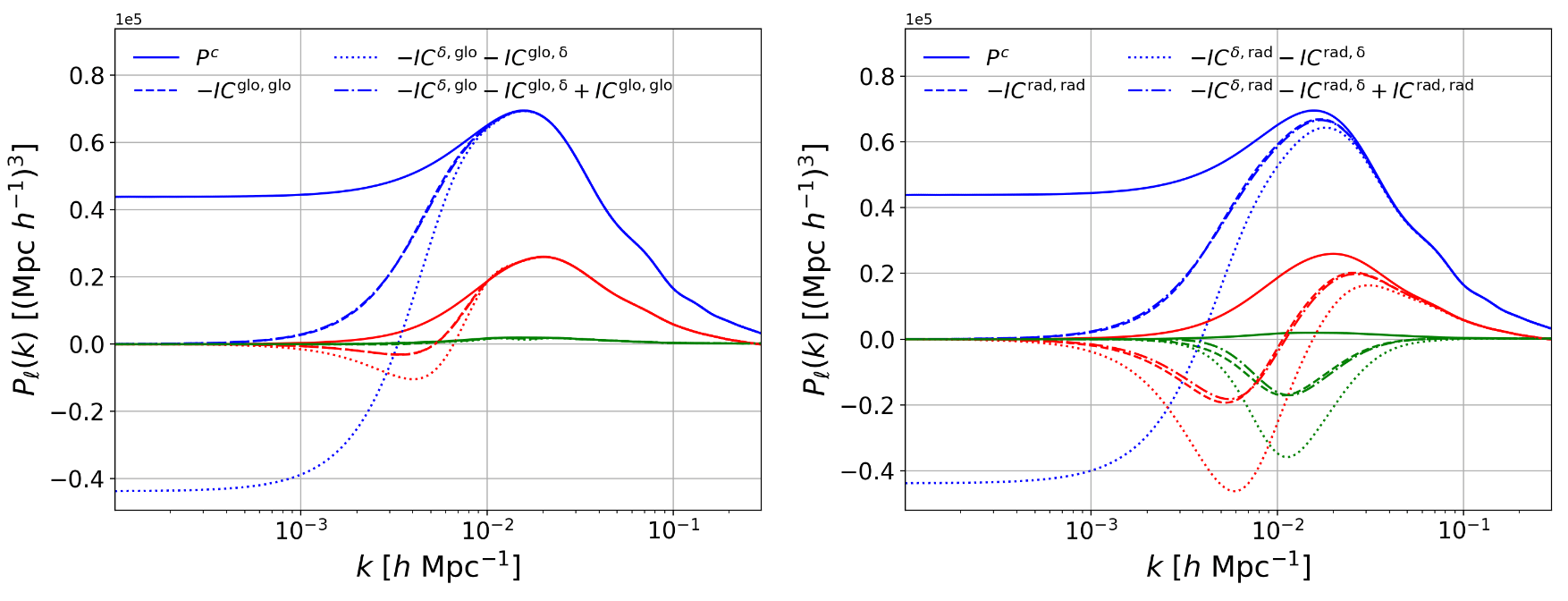

Radial Integral Constraint
\(\delta_g \propto n_g - \braket{n_g}\approx n_g - \bar n_g(r)\)
i.e. impose \(\bar \delta_g(r) = 0\)
Global Integral Constraint
\(\delta_g \propto n_g - \braket{n_g} \approx n_g - \bar n_g\)
i.e. impose \(\bar \delta_g = 0\)
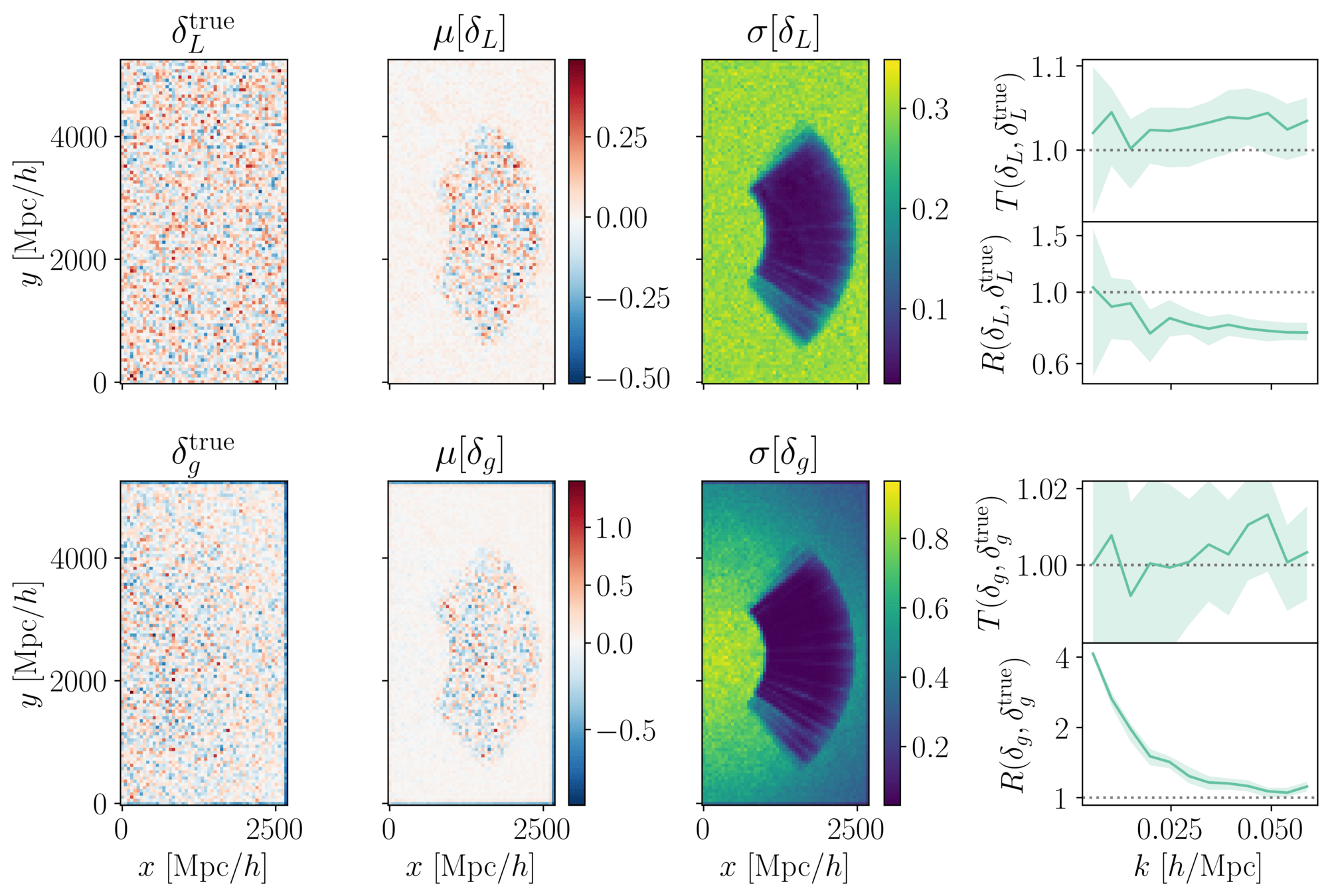

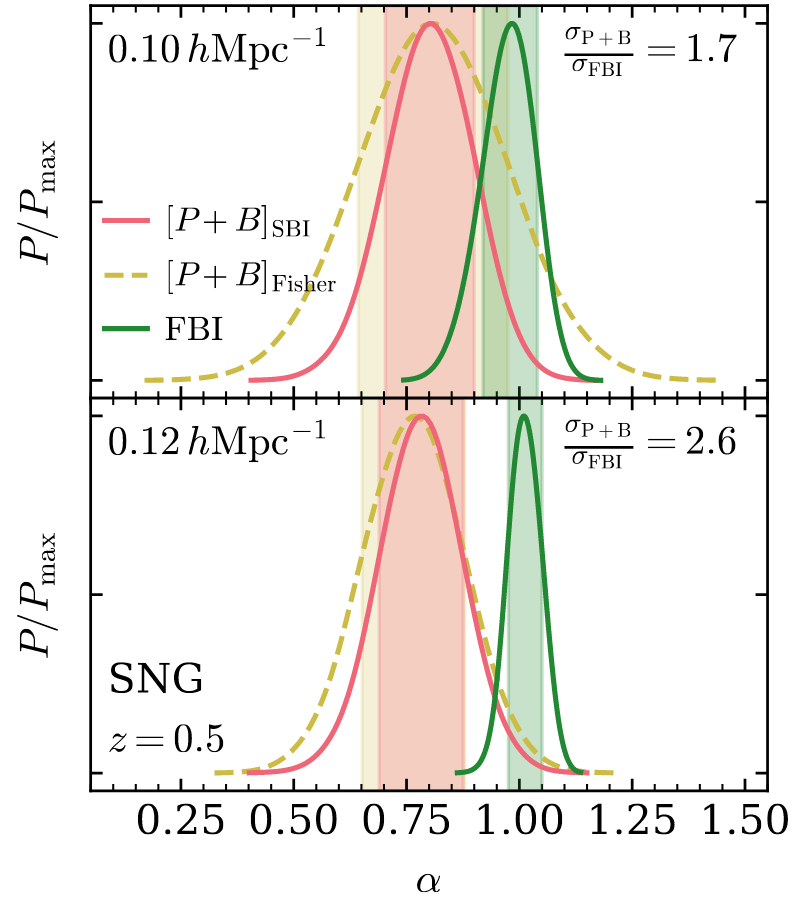
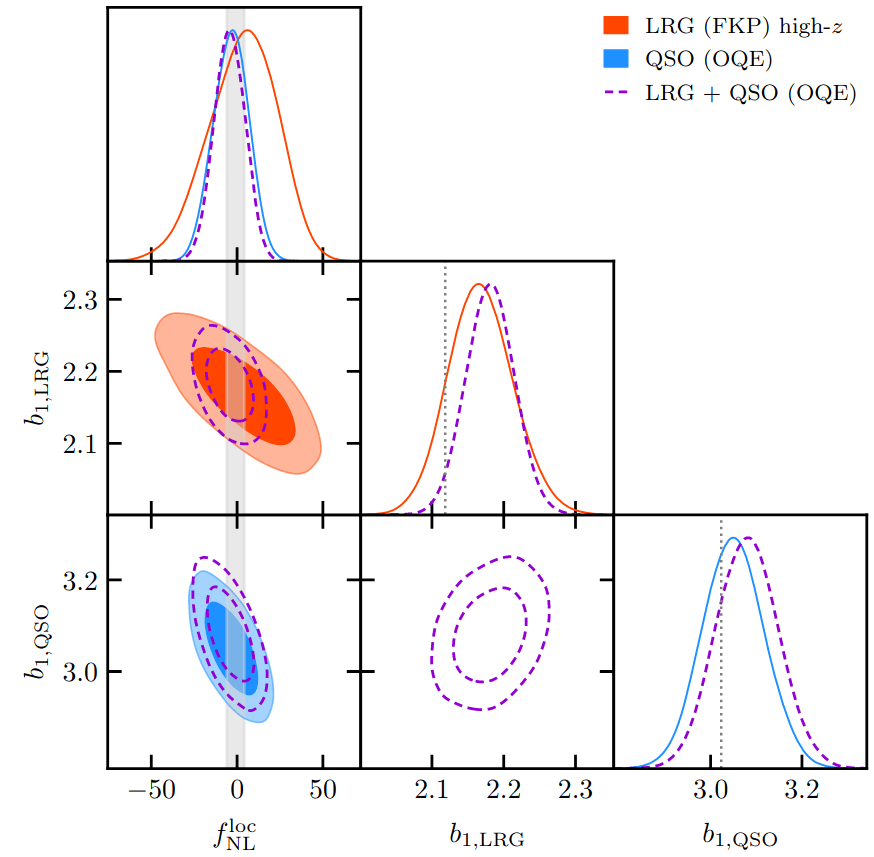
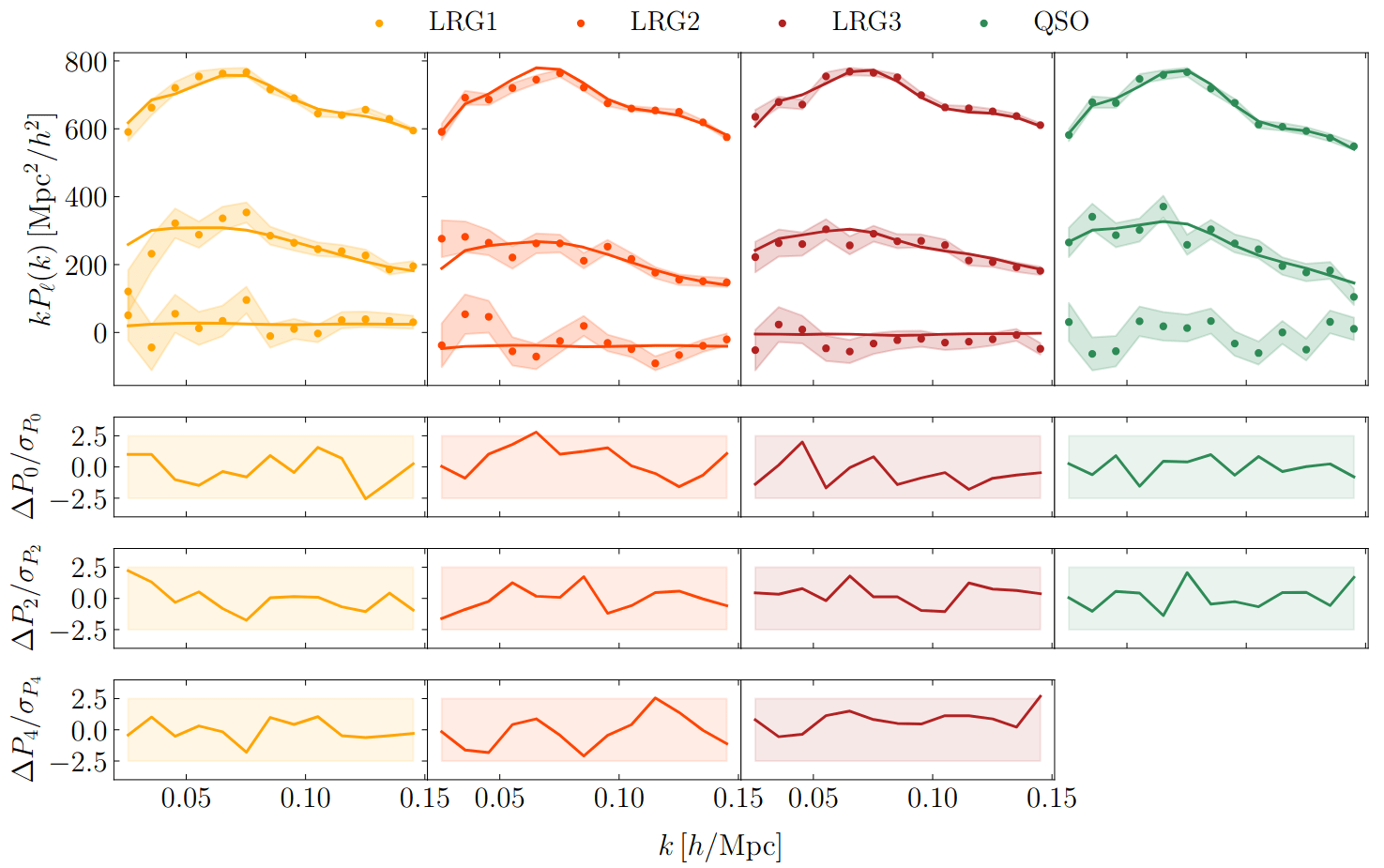
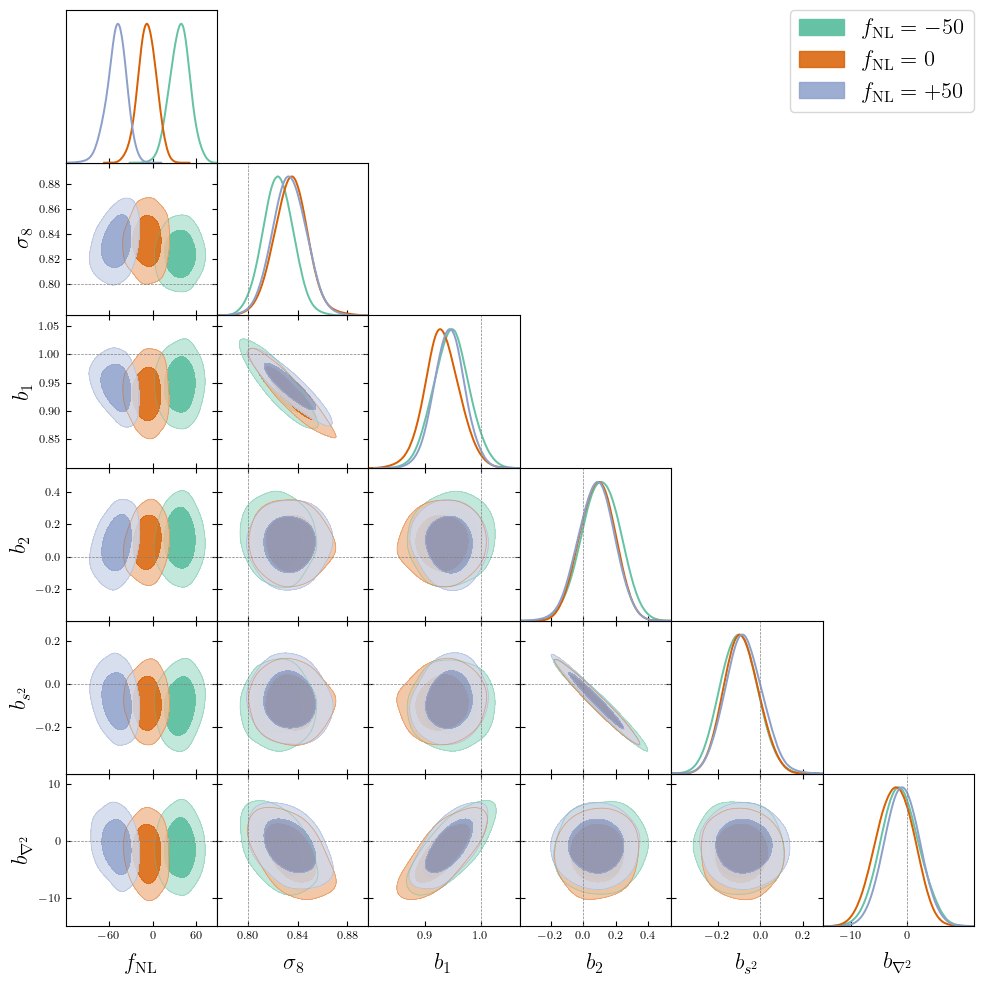
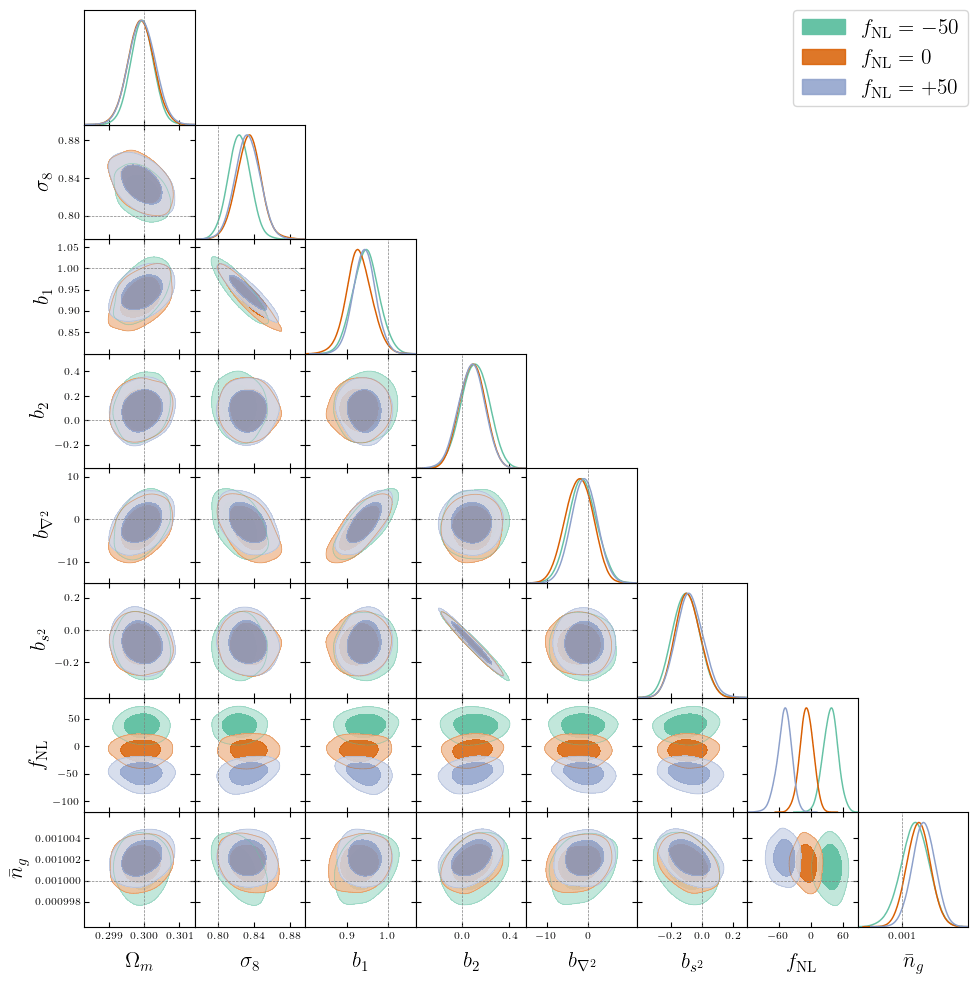
PRELIMINARYPreliminary results
on LRG SGC footprint
\(k_\mathrm{max} \approx 0.04\, [h/\mathrm{Mpc}]\)
\(\sigma[f_\mathrm{NL}] \approx 20\), consistent with power spectrum analysis (Chaussidon+2024)
Next steps:
- EFT model validation (w/o PNG) on AbacusSummit
- EFT model validation (w/ PNG) on PNG-Unitsims
- Systematics model validation on contaminated mocks
- Application to DESI DR1 LRG and QSO
To be answered
-
3 \(f_\mathrm{NL}\) "values" to infer:
- \(f_\mathrm{NL}\) in init field, \(b_\phi f_\mathrm{NL}\) and \(b_{\phi\delta} f_\mathrm{NL}\) in galaxy bias
-
"universality" relations robust at field-level?
\(b_\phi = 2 \delta_c (1 + b_1 - p)\) and \(b_{\phi \delta} = b_\phi - b_1 + \delta_c b_2\)
-
Redshift varying biases? templates?
- \(b_1(z) = a_1 (1+z)^2 + c_1\)? \(b_2(z)\), \(b_s^2(z)\),...?
- Redshift bins? Interpolation?
- Max resolution we can robustly + computationally push to
- \(k_\mathrm{max} \leq 0.14\, h/\mathrm{Mpc}\)?

Where we are
Tally
- Currently visiting Montréal in Laurence Perreault-Levasseur team
- Talks:
- Optimal cosmo information extraction at Sesto (for Euclid people)
- DESI meeting at Berkeley (for colab)
- CoBALt at Institut Pascal (for inflation theorists)
- Bayesian Deep Learning 3 at APC (for deep learners)
- ED Festival (for particle physicists)
- GDR Cophy 2h tutorial
- Papers:
- stat paper at NeurIPS2024 (from master internship)
- Benchmarking Field-Level in review on JCAP
- PNG measurement at the field level in prep
- Teaching: Bachelor 2 Biostats (20h) and Master 1 Maths (15) courses at UPsaclay
- Formation:
- VSS, Science ethics, Sustainable dev. (Open Science left)
- Euclid summer school

Next steps
- Scientific:
- Validation for PNG inference and application to DESI
- Alternative sampling method for field-level inference
- Going Multi-GPU
- Manuscript:
- Detailed plan at the end of December, first chapter in January
- Looking and applying for postdocs this Autumn,
hence the meetings and visits...

Where we are
- Primordial PNG and linear matter field ✔️
- Structure formation (2LPT) ✔️
- Matter-galaxy connection (EFT) (to validate)
- Light-cone, curved-sky, RSD ✔️
- Alcock-Paczynski (to correct)
- Window, relaxing radial integral constraint ✔️
- Systematics (to implement)
-
Simple likelihood (to correct)
(to replace by EFT likelihood + systematics modeling)
Alcock-Paczynski Problems
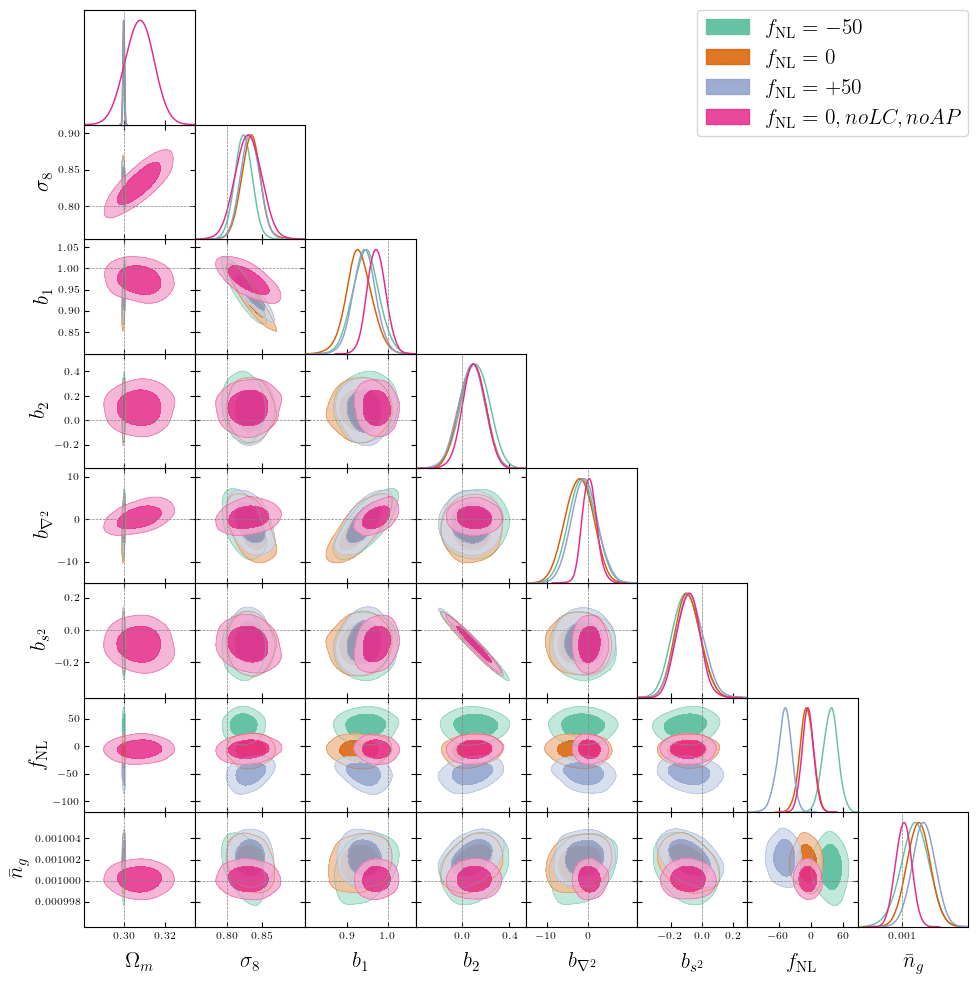
PRELIMINARY RESULTS\(\alpha_\mathrm{iso}\)/\(\Omega_m\) to well-constrained with AP
Alcock-Paczynski Problems
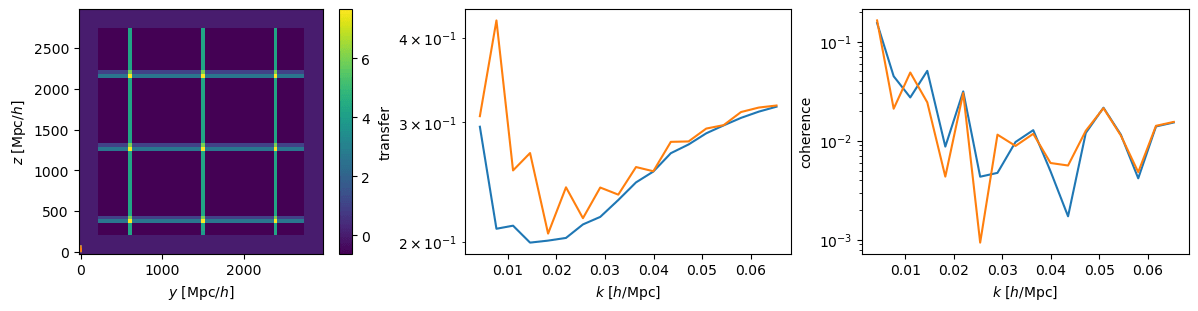
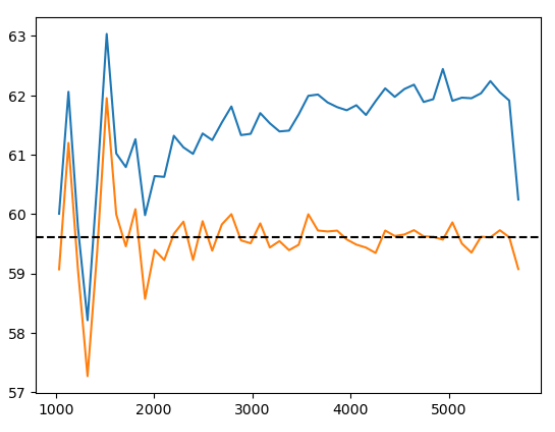
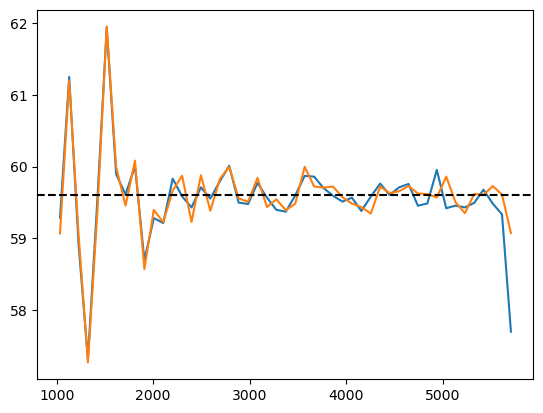
AP modifies simulated tracer density, that we in practice don't know
w/o Jacobian correction w/ Jacobian correction
r [Mpc/h]
r [Mpc/h]
n(r)
n(r)
Did not solved \(\alpha_\mathrm{iso}\) constraints. Probably discretization artificats:
Where we are
Next steps:
- Correct AP modeling
- EFT model validation (w/o PNG) on AbacusSummit
- EFT model validation (w/ PNG) on PNG-Unitsims
- Systematics modeling
- Primordial PNG and linear matter field ✔️
- Structure formation (2LPT) ✔️
- Matter-galaxy connection (EFT) (to validate)
- Light-cone, curved-sky, RSD ✔️
- Alcock-Paczynski (to correct)
- Window, relaxing radial integral constraint ✔️
- Systematics (to implement)
- Simple likelihood (to correct)


Some useful programming tools
-
NumPyro
- Probabilistic Programming Language (PPL)
- Powered by JAX
- Integrated samplers
-
JAX
-
GPU acceleration
- Just-In-Time (JIT) compilation acceleration
- Automatic vectorization/parallelization
- Automatic differentiation
-
GPU acceleration
JAX in practice?
-
GPU accelerate
- JIT compile
- Vectorize/Parallelize
- Auto-diff
import jax.numpy as np
# then enjoyfunction = jax.jit(function)
# function is so fast now!gradient = jax.grad(function)
# too bad if you love chain ruling by handvfunction = jax.vmap(function)
pfunction = jax.pmap(function)
# for-loops are for-loosers
Probabilistic Modeling and Compilation
def simulate(seed):
rng = np.random.RandomState(seed)
x = rng.randn()
y = rng.randn() + x**2
return x, yfrom scipy.stats import norm
log_prior = lambda x: norm.logpdf(x, 0, 1)
log_lik = lambda x, y: norm.logpdf(y, x**2, 1)
log_joint= lambda x, y: log_lik(y, x) + log_prior(x)grad_log_prior = lambda x: x**2
grad_log_lik = lambda x, y: stack([2 * x * (y - x**2), (y - x**2)])
grad_log_joint = lambda x, y: grad_log_lik(y, x) + stack([grad_log_prior(x), zeros_like(y)])
# Hessian left as an exercise...To fully define a probabilistic model, we need
- its simulator
- its associated joint log prob
- possibly its gradients, Hessian... (useful for inference)
We should only care about the simulator!
\(Y\)
\(X\)
$$\begin{cases}X \sim \mathcal N(0,1)\\Y\mid X \sim \mathcal N(X^2, 1)\end{cases}$$
my model on paper
my model on code
What if we modify to fit variances? non-Gaussian dist? Recompute everything? :'(


NumPyro in practice?
def model():
x = sample('x', dist.Normal(0, 1))
y = sample('y', dist.Normal(x**3, 1))
return y
render_model(model, render_distributions=True)
y0 = dict(y=seed(model, 42)())
obs_model = condition(model, y0)
logd_fn = lambda x: log_density(obs_model,(),{},{'x':x})[0]from jax import jit, vmap, grad
force_vfn = jit(vmap(grad(logd_fn)))kernel = infer.NUTS(obs_model)
mcmc = infer.MCMC(kernel, n_warmup, n_samples)
mcmc.run(jr.key(43))
samples = mcmc.get_samples()- Probabilistic Programming
- JAX machinery
- Integrated samplers


Modeling with Numpyro



y_sample = seed(model, 42)()
log_joint = lambda x, y: log_density(model,(),{},{'x':x, 'y':y})[0]obs_model = condition(model, y_sample)
logp_fn = lambda x: log_density(obs_model,(),{},{'x':x})[0]def model():
x = sample('x', dist.Normal(0, 1))
y = sample('y', dist.Normal(x**2, 1))
return yfrom jax import jit, vmap, grad
force_vfn = jit(vmap(grad(logp_fn)))- Define model as simulator... and that's it
- Simulate, get log prob
- +JAX machinery
- Condition model
- And more!
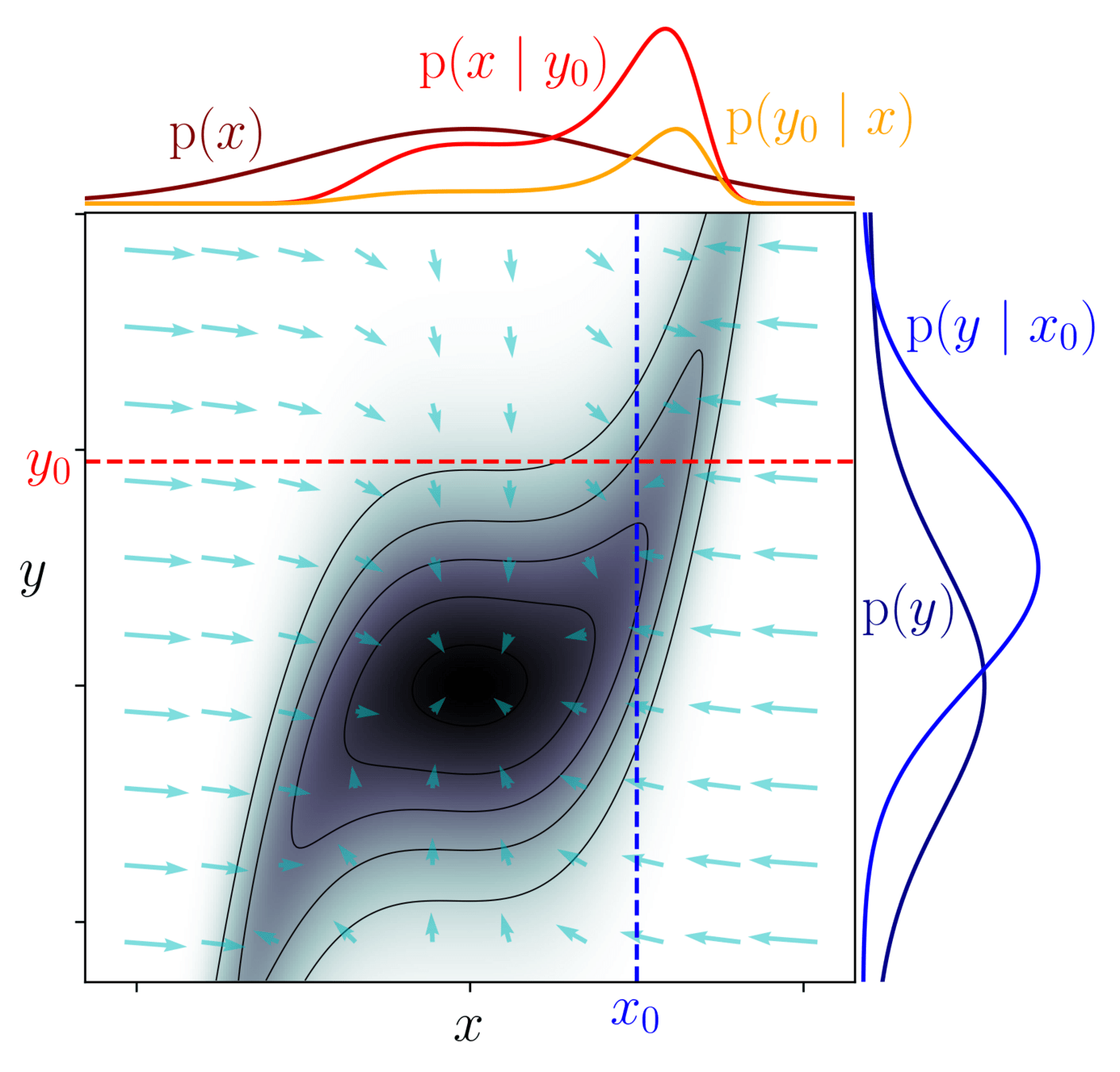
Why care about differentiable model?
-
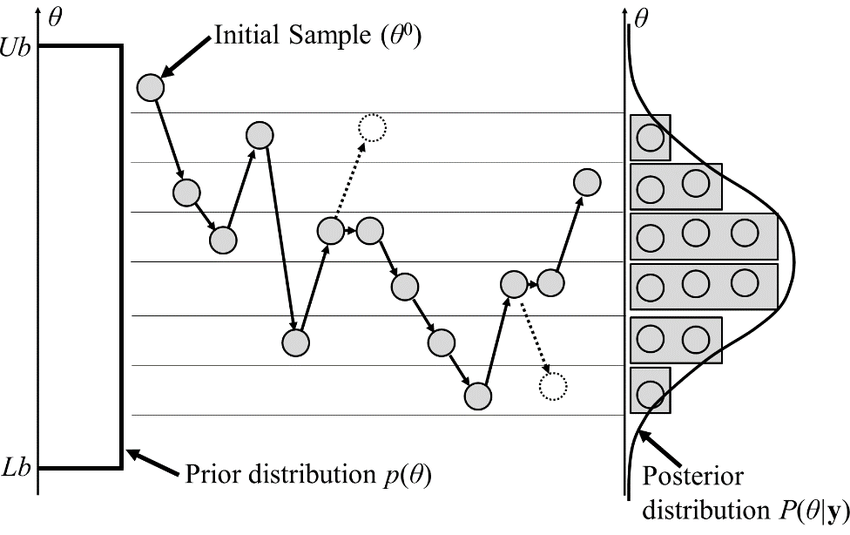
Classical MCMCs
-
agnostic random moves
+ MH acceptance step
= blinded natural selection
- small moves yield correlated samples
-
agnostic random moves
- SOTA MCMCs rely on the gradient of the model log-prob, to drive dynamic towards highest density regions

gradient descent
posterior mode
Brownian
exploding Gaussian
Langevin
posterior
+
=


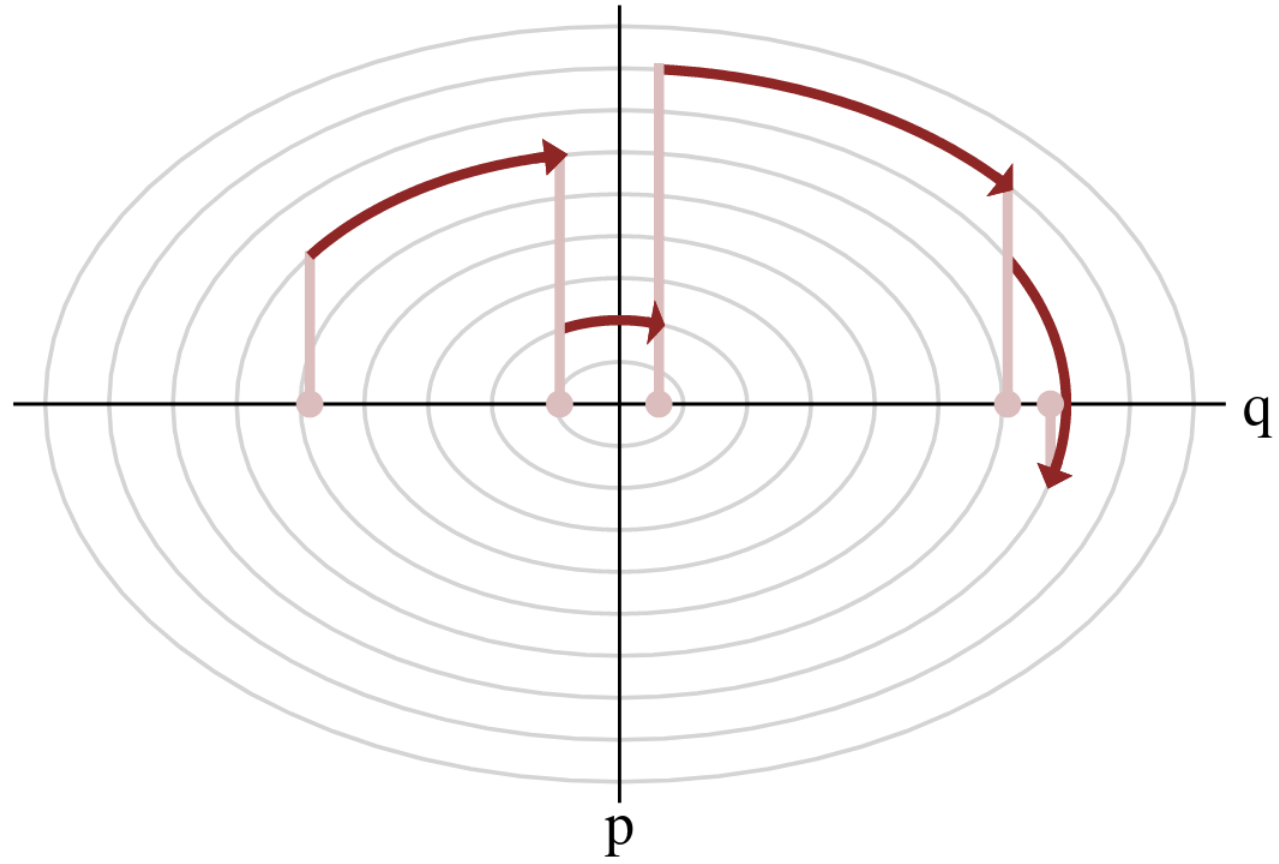
Hamiltonian Monte Carlo (HMC)
- To travel farther, add inertia.
- sample particle at position \(q\) now have momentum \(p\) and mass matrix \(M\)
- target \(\boldsymbol{p}(q)\) becomes \(\boldsymbol{p}(q , p) := e^{-\mathcal H(q,p)}\), with Hamiltonian $$\mathcal H(q,p) := -\log \boldsymbol{p}(q) + \frac 1 2 p^\top M^{-1} p$$
- at each step, resample momentum \(p \sim \mathcal N(0,M)\)
- let \((q,p)\) follow the Hamiltonian dynamic during time length \(L\), then arrival becomes new MH proposal.

Variations around HMC
-
No U-Turn Sampler (NUTS)
- trajectory length \(L\) auto-tuned
- samples drawn along trajectory
- NUTSGibbs i.e. alternating sampling over parameter subsets.
Why care about differentiable model?

Variations around HMC
-
No U-Turn Sampler (NUTS)
- trajectory length auto-tuned
- samples drawn along trajectory
- NUTSGibbs i.e. alternating sampling over parameter subsets
-
Model gradient drives sample particles towards high density regions
-
Hamiltonian Monte Carlo (HMC):
to travel farther, add inertia
- Yields less correlated chains

3) Hamiltonian dynamic
1) mass \(M\) particle at \(q\)
2) random kick \(p\)
2) random kick \(p\)
1) mass \(M\) particle at \(q\)
3) Hamiltonian dynamic

Hugo SIMON
PhD student supervised by
Arnaud DE MATTIA and François LANUSSE


- Field-level Bayesian inference
- High-dimensional sampling
- Differentiable N-body simulations
- Primordial non-Gaussianity from DESI


Hugo SIMON
PhD student at CEA Paris-Saclay, supervised by
Arnaud DE MATTIA and François LANUSSE


- Field-level Bayesian inference
- High-dimensional sampling
- Differentiable N-body simulations
- Primordial non-Gaussianity field-level inference from DESI






Thank you!