Terrell Russell, Ph.D
Executive Director
iRODS Consortium

DAViDD: Initial data management
solution for UNC's
READDI AViDD Center
May 28-31, 2024
iRODS User Group Meeting 2024
Amsterdam, Netherlands


The Rapidly Emerging Antiviral Drug Development Initiative AViDD Center (READDI-AC) is an NIH-funded public-private partnership focused on developing effective antiviral drugs to combat emerging viruses.
The READDI-AC at UNC-Chapel Hill is one of nine Antiviral Drug Discovery (AViDD) Centers funded by the US National Institute of Allergy and Infectious Disease (NIAID) at the National Institutes of Health.
$65M in 2022 - 40 Investigators, 23 Research Sites, 5 Countries
NIH Award 1U19AI171292-01
READDI-AC

The response to viral outbreaks has historically been reactive – vaccines and medications are developed only after a new virus emerges. Our mission is to proactively prepare for emerging viruses by developing antiviral drugs that are active against more than one virus in a family. These broad-spectrum antivirals will help safeguard the well-being of communities worldwide against existing viruses and will be more likely to be effective against future novel viruses in the same family.
Four families:
- Coronaviruses - causes SARS, MERS, COVID-19
- Filoviruses - includes Ebola, Marburg
- Flaviviruses - includes West Nile, Dengue, Zika
- Alphaviruses - includes Chikungunya, Equine Encephalitis

READDI-AC Mission

RENCI, as a subawardee, was tasked to assess, design, and develop the data management solution for the READDI-AC project.
Timeline
- Interviews - July-August 2022
-
Survey - August 2022
- Determination of existing lab workflows
- Document types, variety, size, volume
- Number and identity of humans in the loop
- Opportunities for automation
- Opportunities for cross-lab interactions
- Security considerations - Fall 2022
- Initial design of system - Fall 2022
- Paper evaluation - Fall 2022
- Initial implementation - Nov-Dec 2022
- Testing - Dec 2022
- Deployment - Jan 2023
- Evaluation - Q1 2023
- Iteration - through 2023

-
If you use instruments in your work, what is the format of files they produce?
-
Where do you currently record and store chemical or biological data? In what data format?
-
What keywords or other terms do you use to search for previously recorded data?
-
Do you use existing or public vocabularies, ontologies or other references?
-
Are you familiar with FAIR data sharing principles?
-
What are typical dataset sizes for each unit of work?
-
What is a typical data generation rate for your lab / unit?
-
What is a typical number of data files you generate per week?
-
Do you protect the stored data (do you require secure login/authentication to access your data?
-
How do you currently share your data with (i) your labmates, (ii) within UNC, (iii) with the rest of the world?
-
What software do you use to process or visualize your data?
-
Do you have a need to format your data for publication or presentation format?
-
What steps are manually processed today? What steps are automated today?
-
Where is manual processing required? Where can data processing be automated?
-
What limitations are your lab / group / team running into?
-
What is your highest priority or need for data capture or storage?
Discovery Questions

-
Not Big Data (yet)
-
1000s of files over the course of a year
-
Maybe 10s of GBs, but most much smaller
-
Human scale rate of ingest
-
-
A few formats, mostly open / convertible
-
doc, pdf, xls, ppt, csv, txt, prism, jpg
-
-
Some electronic lab notebooks
-
Mostly xls
-
Manually calculated / generated
-
-
Very little currently automated
-
Manual transcription from paper notebooks
-
Graphing done in Excel or (rarely) Jupyter notebooks
-
Discovery Findings
-
No shared naming conventions
-
For either data filenames or metadata
-
Sometimes consistent within a lab
-
Due to necessity
-
But not over time
-
-
-
Highest priority is access / sharing
-
This project's data needs a 'home'
-
-
No centralized data repository
-
No standardized data processing (raw to publication quality) and data upload protocols
-
No versioning protocols
-
Metadata currently non-existent
-
Negative results / Failed attempts not recorded
-
HEIGHTENED NEED for RDM due to new NIH reqs

There was very little process to automate - we were starting from scratch and these labs did not have much in common. Different instruments, different chemistry, different software, different processes, different formats.
Not their fault - they'd never been required to coordinate and collaborate in the past except via publications. This was a new mandate.
There would be two projects:
- People engineering
- hardest part, scientists do not want to change their processes
- requires many people to coordinate (expensive in time and effort)
- Software engineering
- a few puzzles to solve, but nothing too daunting
- security requirements demand working with other parties
Design and Preparation

Software Engineering Requirements
- federated login for otherwise unaffiliated researchers
- secure enclave
- just files, mostly spreadsheets
- some annotation
- automation where possible
- search
- available for analysis with existing tooling
- probably via download

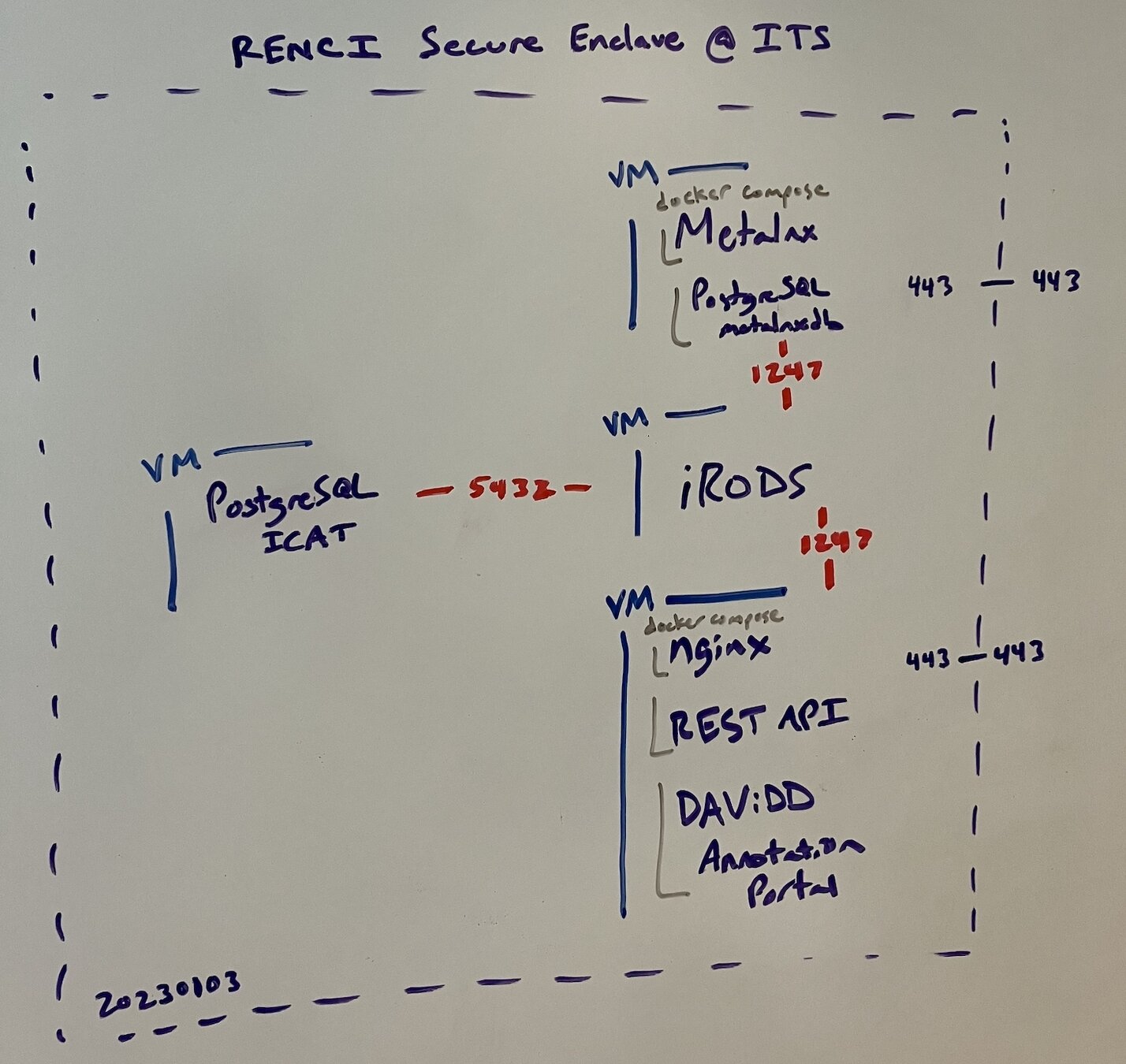
- 4 VMs
- RENCI Secure Enclave
- docker compose
- originally REST API
- later HTTP API
- CILogon providing identity
January 2023 - Initial Deployment

Angular Application
- upload
- assays
- search
- compound profile
- FAQ
- profile information
iRODS Policy - Four recurring rules
- irule davidd_add_sweeper_to_queue
- irule davidd_add_compound_profile_sweeper_to_queue
- irule davidd_add_compound_profile_remover_to_queue
- irule davidd_add_assays_sweeper_to_queue
DAViDD - Application and Policy

irule davidd_add_sweeper_to_queue
- davidd_find_and_parse_uploaded_files
- davidd_parse_and_place_jsonfile
- parse python dict
- prepare avus_to_add
- decode file data, write it
- associate avus
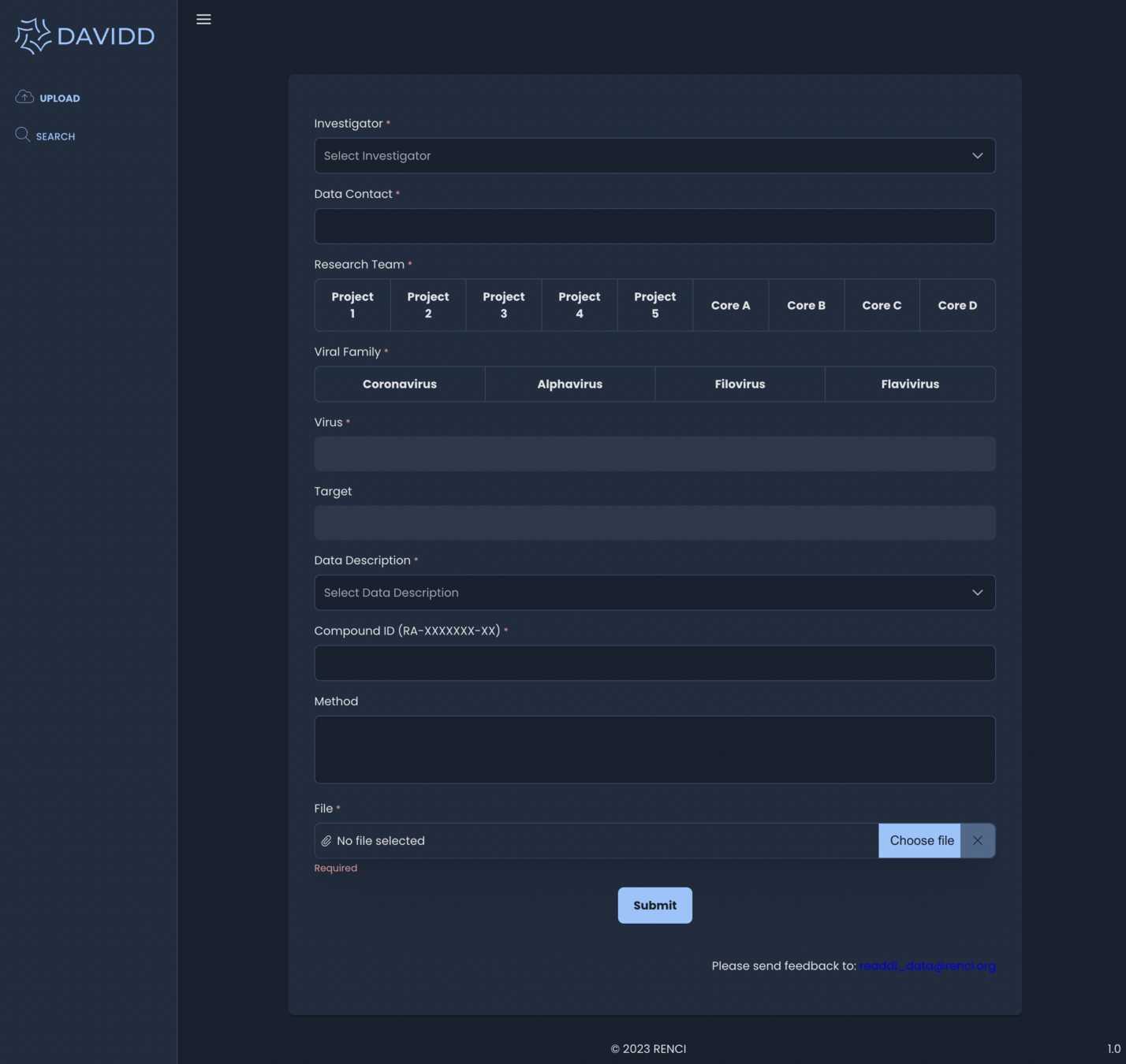
February 2023 - Upload

Associated metadata from upload form available to search and browse
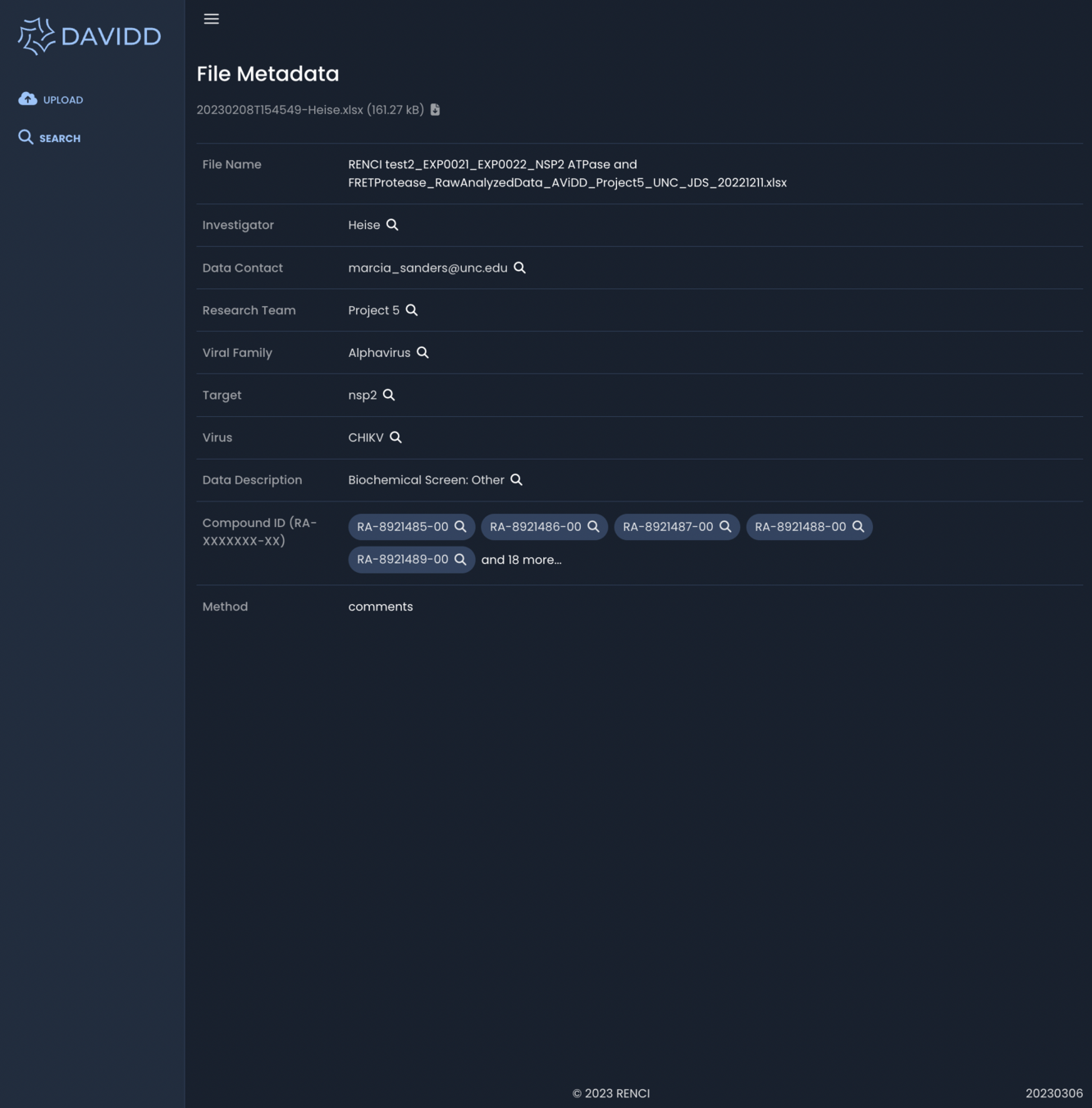
March 2023 - File Metadata

GenQuery
- matching on file name and metadata
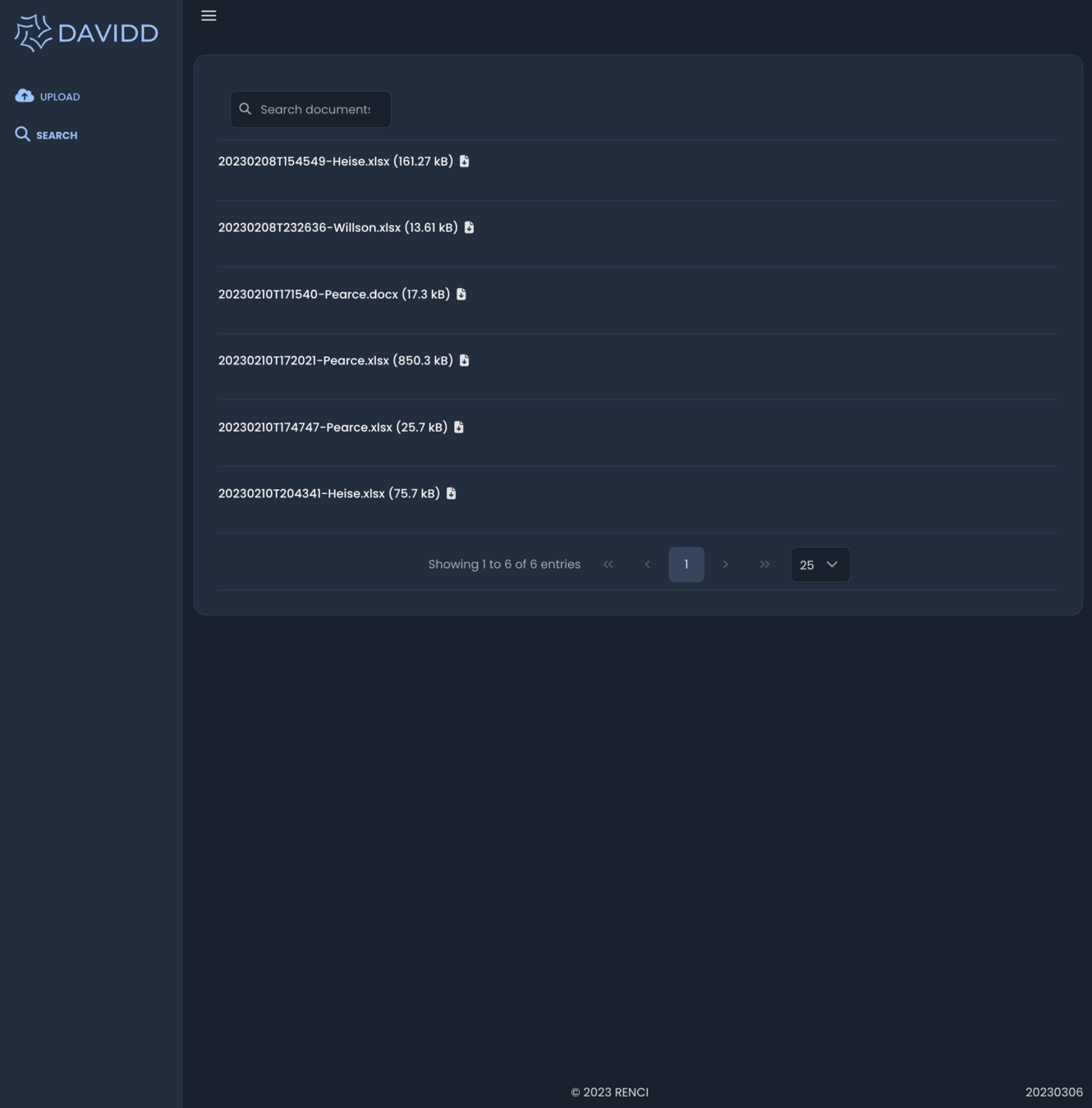
March 2023 - Search

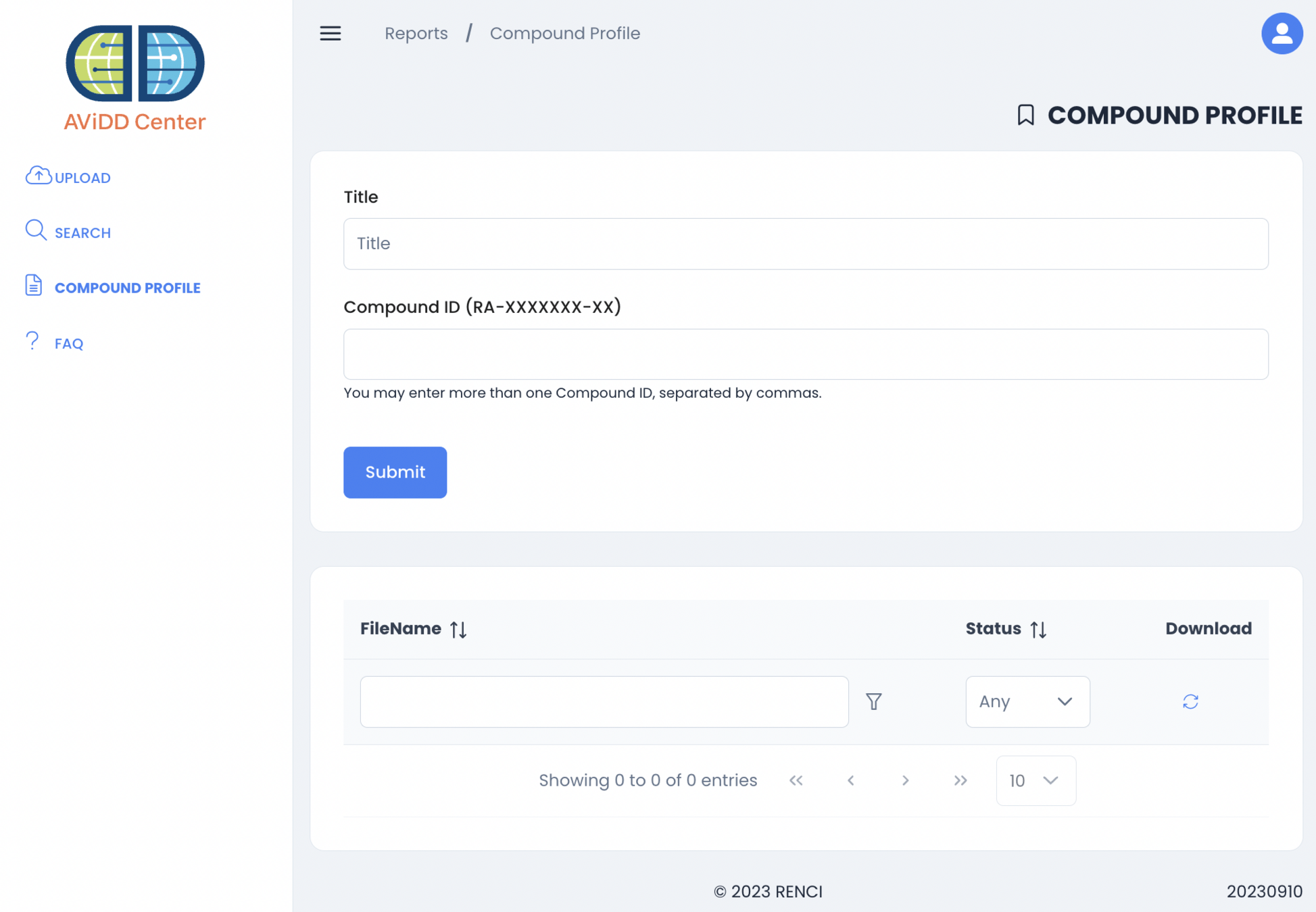
irule davidd_add_compound_profile_sweeper_to_queue
- davidd_process_requested_profile
- davidd_process_queued_file
- davidd_walk_collection_for_compound_info
- use openpyxl, read spreadsheets, populate new one
irule davidd_add_compound_profile_remover_to_queue
- davidd_remove_old_compound_profiles
- defined by compound_profile_removal_age_in_minutes
September 2023 - Compound Profile

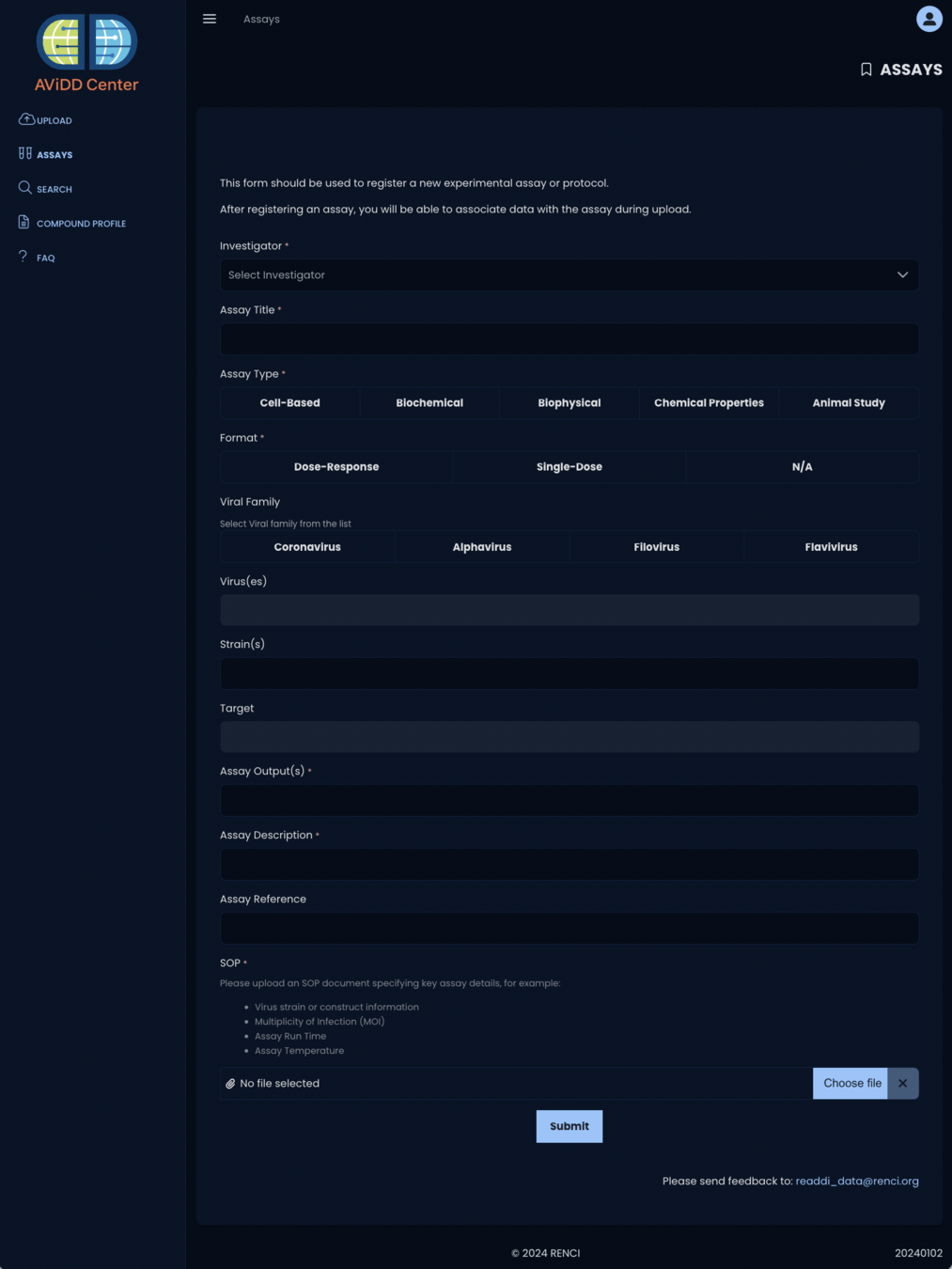
irule davidd_add_assays_sweeper_to_queue
- davidd_find_and_parse_assay_files
- davidd_parse_and_place_jsonfile
January 2024 - Assays

Discovery and prototyping were a success
- 4 labs interviewed
- Many challenges identified and lessons learned
- 3 federated login architectures attempted
- Selected CILogon.org
353 datafiles uploaded in the first year
- 105 Coronavirus
- 173 Alphavirus
- 27 Filovirus
- 48 Flavivirus
Summary

Having identified the main requirements and bench-to-data process, the project selected an existing commercial vendor for its extensive GUI and compound-specific analysis tooling.
RENCI continues to develop database-level tooling focused on chemical compound information and linkages with other tools in the ecosystem.
Acknowledgements
- NIH
- Ava Vargason, Nat Moorman, Ralph Baric, Tim Willson, Toni Baric
- Oleg Kapeljushnik, Kory Draughn, Alex Tropsha, Robert Hubal, Kelyne Kenmogne, Carrie Pasfield, Patrick Patton
The Future
Thank you!
Questions?
