MAgPIE 4.7
Open Source land-use modeling framework

Jan Philipp Dietrich
dixyzetrich@pik-xyzpotsdam.de


What is ?

Basics
Model of Agricultural Production and its Impact on the Environment
MAgPIE
cost minimization of consecutive time slices with a length of 5-20 years until 2100
dynamic recursive optimization
global | 5-20 world regions | 50-2000 spatial cluster
3 spatial layers
bringing together biophysical (plant growth, carbon, nutrients, water) and economic (costs, prices, demand, policies) aspects
balance biophysical and economic side

Basics
Flexible world regions



raw data
model input



ISO countries
Default world regions
Alternative aggregation
model input
powered by madrat » github.com/pik-piam/madrat
Spatial
Cluster
- Cluster build from 0.5° grid cells
- Clustering based on similarity of spatial explicit model inputs
- Outputs interpolated to 0.5° for visualization & analysis

Model Outputs





REMIND-MAgPIE Integrated Assessment Model
Model cascade
PIAM - Potsdam Integrated Assessment Modelling Framework



Model cascade
MESSAGEix
MAgPIE
code

Requirements
-
GAMS (non-free)|gams.com
including license for solver CONOPT and CPLEX - R (free) | r-project.org
-
R packages (free)
README.md in MAgPIE lists all packages currently required - GIT (free) | git-scm.com
Recommendations
-
Modern editor
e.g. VS Code| code.visualstudio.com -
Rstudio | rstudio.com
R editor -
PIK-CRAN | rse.pik-potsdam.de
PIK R package repository - GitHub account | github.com
Open Source


> 30 supporting R packages published under LGPL (copyleft), BSD-2 (non-copyleft), or other Open Source licenses
github.com/pik-piam

First steps / get the code
- Fork MAgPIE repository @ github.com/magpiemodel/magpie
make a copy of the repository in GitHub
- Clone forked repository
download the copy to your machine
- Follow instructions in README.md
- Run the model
Code Structure #1
GAMS Model Core
- model equations
- modules
- model simulation
R Pre- & Postprocessing Layer
- model configuration
- data download
- run management
- run compilation
- output processing
- visualization
- Model development in GAMS
- Model application in R
- R used to boost GAMS capabilities
Code Structure #2
GAMS Model Core
- core
- module 1
- realization A
- realization B
- module 2
- realization A
- realization B
- realization C
- module 1
...
- split a big system into many small components
- flexibility to switch between different implementations


GAMS Modules
Code Structure #3
Naming conventions to..
- improve readability
- emulate local environments in GAMS
q13_cost_tc(i2) ..
v13_cost_tc(i2) =e= sum(ct, i13_land(i2) * i13_tc_factor(ct,i2)
* vm_tau(i2)**i13_tc_exponent(ct,i2)
* (1+pm_interest(i2))**15);1st Prefix q - eQuation v - Variable i - Input parameter p - Parameter
...
2nd Prefix
?m - module interface
?00 - module-internal
(2-digit module code)
more details:
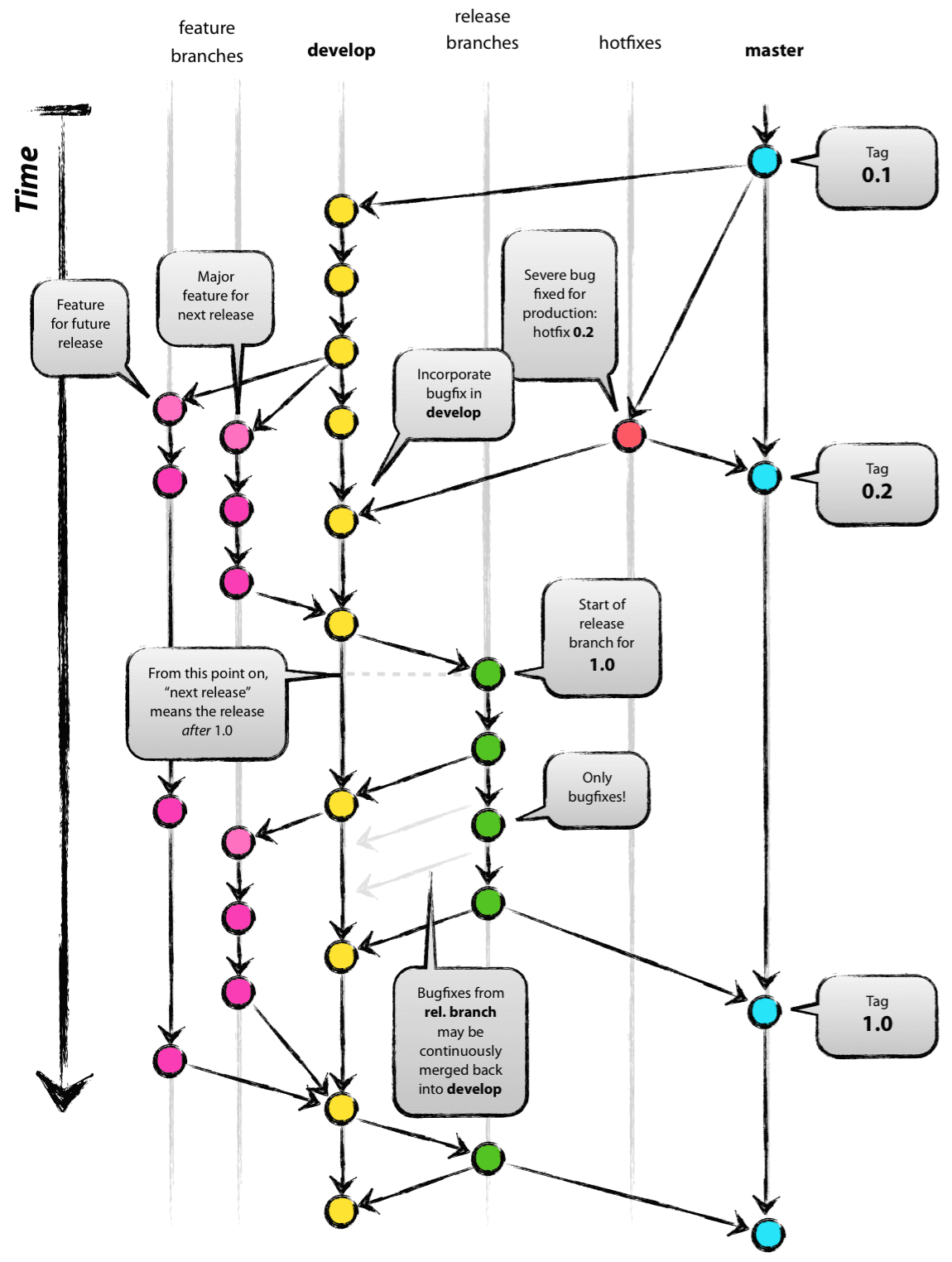
Repository Structure
- official releases in master branch
- most recent version in develop
- release candidates in release branch
- development in feature branches (f_<name>)
Configuration | default.cfg
# | (C) 2008-2024 Potsdam Institute for Climate Impact Research (PIK)
# | authors, and contributors see CITATION.cff file. This file is part
# | of MAgPIE and licensed under AGPL-3.0-or-later. Under Section 7 of
# | AGPL-3.0, you are granted additional permissions described in the
# | MAgPIE License Exception, version 1.0 (see LICENSE file).
# | Contact: magpie@pik-potsdam.de
##################
#### SETTINGS ####
##################
cfg <- list()
#### Main settings ####
# short description of the actual run
cfg$title <- "default"
# path to the submodel to be used relative to main model folder
cfg$model <- "main.gms" #def = "main.gms"
#### input settings ####
# which input data sets should be used?
cfg$input <- c(regional = "rev4.101_h12_magpie.tgz",
cellular = "rev4.101_h12_fd712c0b_cellularmagpie_c200_MRI-ESM2-0-ssp370_lpjml-8e6c5eb1.tgz",
validation = "rev4.101_h12_validation.tgz",
additional = "additional_data_rev4.48.tgz",
calibration = "calibration_H12_per_ton_fao_may22_glo_13Mar24.tgz")
# NOTE: It is recommended to recalibrate the model when changing cellular input data
# as well as for any other setting that would affect initial values in the model,
# e.g. changes in costs structure, NPI policies, etc.
#a list of repositories (please pay attention to the list format!) in which the
#files should be searched for. Files will be searched in all repositories until
#found, always starting with the first repository in the list. The argument must
#have the format of a named list with the url of the repository as name and a
#corresponding list of options such as username or password to access the
#repository as value. If no options are required the value has to be NULL. (e.g.
#list("ftp://my_pw_protected_server.de/data"=list(user="me",password=12345),
# "http://free_server.de/dat"=NULL))
#Please add system or user specific repositories (such as repos with limited
#access for) through the R option "magpie_repos". Through the append command
#below it will get merged into cfg$repositories
cfg$repositories <- append(list("https://rse.pik-potsdam.de/data/magpie/public"=NULL),
getOption("magpie_repos"))
# Should input data be downloaded from source even if cfg$input did not change?
cfg$force_download <- FALSE
# Should an existing output folder be replaced if a new run with the same name is started?
cfg$force_replace <- FALSE
# Settings for the yield calibration
# For this yield calibration (that uses results from a special MAgPIE calibration run
# triggered by the recalibrate switch) to be activated the switch s14_use_yield_calib needs
# to be turned on. (Default is off)
# This switch should only be activated for penalty_apr22 crop realization.
# For other realizations, it is recommended not to use this yield calibration.
#
# Switch to turn on/off recalibration of yields.
# * (TRUE): Yield calibration will be performed
# * (ifneeded): Yield calibration will only be executed if input data is
# * downloaded from repository
# * (FALSE): Yield calibration will not be performed
cfg$recalibrate <- FALSE # def = FALSE
# Up to which accuracy shall be recalibrated?
cfg$calib_accuracy <- 0.05 # def = 0.05
# What is the maximum number of iterations if the precision goal is not reached?
cfg$calib_maxiter <- 20 # def = 20
# Factor determining how much the divergence infuences the new
# calibration factor in each calibration iteration (0-1)
cfg$damping_factor <- 0.96 # def= 0.96
# switch on/of calibration of cropland (pasture will be left untouched)
cfg$calib_cropland <- TRUE # def= TRUE
# set upper limit for cropland calibration factor
cfg$crop_calib_max <- 1.5 # def= 1.5
# Selection type of calibration factors.
# If FALSE, calibration factors from the last iteration are used.
# If TRUE, calibration factors from the iteration with the lowest divergence are used.
cfg$best_calib <- TRUE # def = TRUE
# Settings for land conversion cost calibration (cropland)
# The calibration routine derives regional calibration factors for
# costs of cropland expansion and rewards for cropland reduction,
# with the goal to match historical regional cropland in 2015.
# * (TRUE): Land conversion cost calibration will be performed
# * (ifneeded): Land conversion cost calibration will only be executed if the input file "f39_calib.csv" is missing
# * (FALSE): Land conversion cost calibration will not be performed
cfg$recalibrate_landconversion_cost <- "ifneeded" #def "ifneeded"
# Up to which accuracy shall be recalibrated?
cfg$calib_accuracy_landconversion_cost <- 0.05 # def = 0.05
# What is the maximum number of iterations if the precision goal is not reached?
cfg$calib_maxiter_landconversion_cost <- 40 # def = 40
# Restart from existing calibration factors (TRUE or FALSE)
cfg$restart_landconversion_cost <- FALSE # def = FALSE
# Factor determining how much the divergence infuences the new
# calibration factor in each calibration iteration (0-1)
cfg$damping_factor_landconversion_cost <- 0.96 # def= 0.96
# Set upper limit for cropland calibration factor
cfg$cost_calib_max_landconversion_cost <- 3 # def= 3
# Set lower limit for cropland calibration factor
cfg$cost_calib_min_landconversion_cost <- 0.05 # def= 0.05
# Selection type of calibration factors.
# If FALSE, calibration factors from the last iteration are used.
# If TRUE, calibration factors from the iteration with the lowest divergence are used.
cfg$best_calib_landconversion_cost <- FALSE # def = FALSE
# Settings for NPI/NDC recalculation
# * (TRUE): NPI/NDC recalculation will be performed
# * (ifneeded): NPI/NDC recalculation will only be executed if current input files are zero
# * and policy switches (c32_aff_pol, c35_ad_pol) are set to "npi" or "ndc".
# * If policy switches are set to "none" (default) NPI/NDC recalculation will not be performed
# * (FALSE): NPI/NDC recalculation will not be performed
cfg$recalc_npi_ndc <- "ifneeded" # def = ifneeded
# * which national or subnational mappinng should be used
# * (iso): policies on the national levels
# * (bra): includes subnational policies for Brazil
cfg$policyregions <- "bra" # def = "bra"
#### magpie.gms settings ####
cfg$gms <- list()
# Set number of time steps (1-16) or type "less_TS" for remind time steps
cfg$gms$c_timesteps <- "coup2100"
# historic time steps
cfg$gms$c_past <- "till_2010"
# use of gdx files
cfg$gms$s_use_gdx <- 0 # def = 0
#* 0: gdx will not be loaded
#* 1: gdx is loaded in the first time step
#* 2: gdx is loaded in all time steps
# **----------------------------------------------------------------------------
# *** MODULES
# ***---------------------------------------------------------------------------
#### Useful shortcuts ####
# Vector of all iso countries (used for regional policy implementations)
all_iso_countries <- "ABW,AFG,AGO,AIA,ALA,ALB,AND,ARE,ARG,ARM,
ASM,ATA,ATF,ATG,AUS,AUT,AZE,BDI,BEL,BEN,
BES,BFA,BGD,BGR,BHR,BHS,BIH,BLM,BLR,BLZ,
BMU,BOL,BRA,BRB,BRN,BTN,BVT,BWA,CAF,CAN,
CCK,CHN,CHE,CHL,CIV,CMR,COD,COG,COK,COL,
COM,CPV,CRI,CUB,CUW,CXR,CYM,CYP,CZE,DEU,
DJI,DMA,DNK,DOM,DZA,ECU,EGY,ERI,ESH,ESP,
EST,ETH,FIN,FJI,FLK,FRA,FRO,FSM,GAB,GBR,
GEO,GGY,GHA,GIB,GIN,GLP,GMB,GNB,GNQ,GRC,
GRD,GRL,GTM,GUF,GUM,GUY,HKG,HMD,HND,HRV,
HTI,HUN,IDN,IMN,IND,IOT,IRL,IRN,IRQ,ISL,
ISR,ITA,JAM,JEY,JOR,JPN,KAZ,KEN,KGZ,KHM,
KIR,KNA,KOR,KWT,LAO,LBN,LBR,LBY,LCA,LIE,
LKA,LSO,LTU,LUX,LVA,MAC,MAF,MAR,MCO,MDA,
MDG,MDV,MEX,MHL,MKD,MLI,MLT,MMR,MNE,MNG,
MNP,MOZ,MRT,MSR,MTQ,MUS,MWI,MYS,MYT,NAM,
NCL,NER,NFK,NGA,NIC,NIU,NLD,NOR,NPL,NRU,
NZL,OMN,PAK,PAN,PCN,PER,PHL,PLW,PNG,POL,
PRI,PRK,PRT,PRY,PSE,PYF,QAT,REU,ROU,RUS,
RWA,SAU,SDN,SEN,SGP,SGS,SHN,SJM,SLB,SLE,
SLV,SMR,SOM,SPM,SRB,SSD,STP,SUR,SVK,SVN,
SWE,SWZ,SXM,SYC,SYR,TCA,TCD,TGO,THA,TJK,
TKL,TKM,TLS,TON,TTO,TUN,TUR,TUV,TWN,TZA,
UGA,UKR,UMI,URY,USA,UZB,VAT,VCT,VEN,VGB,
VIR,VNM,VUT,WLF,WSM,YEM,ZAF,ZMB,ZWE"
# * Based on https://tntcat.iiasa.ac.at/SspDb/dsd?Action=htmlpage&page=10#regiondefs
oecd90andEU <- "ALB,AUS,AUT,BEL,BIH,BGR,CAN,CYP,CZE,DNK,EST,FIN,FRA,
DEU,GRC,HUN,HRV,ISL,IRL,ITA,JPN,LUX,LVA,LTU,MLT,MNE,
NLD,NOR,NZL,POL,PRT,ROU,SRB,SVK,SVN,ESP,SWE,CHE,MKD,TUR,
GBR,USA"
# ***--------------------- 09_drivers ----------------------------------------
# * (aug17): default drivers
cfg$gms$drivers <- "aug17" # def = aug17
cfg$gms$c09_pop_scenario <- "SSP2" # def = SSP2
cfg$gms$c09_gdp_scenario <- "SSP2" # def = SSP2
# * options: SSP: "SSP1", "SSP2", "SSP2EU", "SSP3", "SSP4", "SSP5"
# * SDP: "SDP", "SDP_EI", "SDP_MC", "SDP_RC"
# * Note: SSP2EU a European Commission population/income scenario for
# * Ariadne project. SDP is same as SSP1 income, while SDP_EI, SDP_RC, SDP_MC
# * see different GDP trajectories as described in SHAPE project.
# * Physical activity level scenario:
cfg$gms$c09_pal_scenario <- "SSP2" # def = SSP2
# * options: SSP: "SSP1", "SSP2", "SSP2EU", "SSP3", "SSP4", "SSP5"
# * SDP: "SDP", "SDP_EI", "SDP_MC", "SDP_RC"
# Year until which all parameters are fixed to SSP2 values
cfg$gms$sm_fix_SSP2 <- 2020
# Year until which all parameters affected by climate change are fixed to historical values
cfg$gms$sm_fix_cc <- 2020
# ***--------------------- 10_land ----------------------------------------
# * (feb15): default land realization
# * (landmatrix_dec18): includes a land transition matrix
cfg$gms$land <- "landmatrix_dec18" # def = landmatrix_dec18
# * Artificial cost for balance variables (USD05MER per ha)
# * The balance variables in the land module avoid infeasibilities due to
# * differences in accuracy between parameters and variables in GAMS.
# * High costs make sure that the balance variables are only used as a last resort.
cfg$gms$s10_cost_balance <- 1000000 # def = 1000000
# ***--------------------- 11_costs ------ --------------------------------
# * (default): default cost realization
cfg$gms$costs <- "default" # def = default
# ***--------------------- 12_interest_rate ---------------------------------
# * (select_apr20): regional interest rates dependent on development state
cfg$gms$interest_rate <- "select_apr20" # def = select_apr20
# * Interest rate scenario:
# * Options: coupling (exogenous interest rate)
# * gdp_dependent (regional interest rate dependent on development state)
# * Note: Currently, coupling is included as an option of an exogenous interest
# * rate scenario. It is currently not part of coupled runs with REMIND.
# * Note: In the gdp_dependent scenario, the selected interest rate fully takes
# * effect in 2050. It slowly starts fading in by 2025.
cfg$gms$c12_interest_rate <- "gdp_dependent" # def = "gdp_dependent"
# * Interest rate coefficients:
# * The coefficients determine the regionally specific interest rate. They do
# * not represent the value of the interest rate.
# * The s12_interest_lic coefficient determines the intercept for future interest
# * rate determination. It represents the interest rate in low income countries.
cfg$gms$s12_interest_lic <- "0.1" # def = 0.1
# * The s12_interest_hic determines the speed of interest rate adjustment for
# * future interest rates. It represents the interest rate in high income countries.
cfg$gms$s12_interest_hic <- "0.04" # def = 0.04
# * Analog logic applies to historic interest rate coefficients
cfg$gms$s12_hist_interest_lic <- "0.1" # def = 0.1
cfg$gms$s12_hist_interest_hic <- "0.04" # def = 0.04
# * Note: It is also possible to choose a global interest rate by setting the
# * upper (s12_interest_lic) and the lower (s12_interest_hic) bound equal
# * This was formerly used for SSP runs and was set to SSP1, SSP5: 0.07 (hist),
# * 0.04 (future), SSP2: 0.07 (hist, future), SSP3: 0.07 (hist), 0.1 (future),
# * SSP4: s12_interest_lic=0.1, s12_interest_hic=0.04, s12_hist_interest_lic=0.07,
# * s12_hist_interest_hic=0.07
# * Regional interest rate policy scenario switch
# * Options: list of iso-codes of countries where above selected coefficients
# * should be in effect.
# * In all other countries, alternative coefficients (s12_interest_lic_noselect, s12_interest_hic_noselect,
# * s12_hist_interest_lic_noselect, s12_hist_interest_hic_noselect) that can be selected below
# * are in effect.
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: all iso countries
cfg$gms$select_countries12 <- all_iso_countries # def = all_iso_countries
# * Interest rate coefficients for non-selected countries:
cfg$gms$s12_interest_lic_noselect <- "0.1" # def = 0.1
cfg$gms$s12_interest_hic_noselect <- "0.04" # def = 0.04
cfg$gms$s12_hist_interest_lic_noselect <- "0.1" # def = 0.1
cfg$gms$s12_hist_interest_hic_noselect <- "0.04" # def = 0.04
# ***--------------------- 13_tc -----------------------------------------
# * (endo_jan22): endogenous technological change with full cost accounting and
# * stepwise updated crop and managed pastures area information
# * (exo): exogenous technological change (removes non-linearities from the model);
# * requires an existing model run with endo tc for generating the input file
# * f13_tau_scenario.csv
cfg$gms$tc <- "endo_jan22" # def = endo_jan22
# * tc cost scenario crops: low, medium or high
cfg$gms$c13_tccost <- "medium" # def = medium
# * ignore historical tau (1) or use it as lower bound (0)
cfg$gms$s13_ignore_tau_historical <- 1 # def = 1
# * Maximum regional tech cost expressed as share of regional GDP
# * A meaningful value would be 0.002. However, this bound causes infeasibilities in some cases.
# * Therefore, this bound is not used in the current model version.
cfg$gms$s13_max_gdp_shr <- Inf # def = Inf
# ***--------------------- 14_yield --------------------------------------
# * (managementcalib_aug19): calibrate potential LPJmL-yields to FAO regional numbers,
# * pasture yields increase based on exogenous demand-side proxy
# * for growth rate of cattle stocks
cfg$gms$yields <- "managementcalib_aug19" # def = managementcalib_aug19
# * yield scenario
# * options: cc (climate change)
# * nocc (no climate change)
# * nocc_hist (no climate change after year defined by sm_fix_cc)
cfg$gms$c14_yields_scenario <- "cc" # def = "cc"
# * switch determing the effectivity of translating crop tc into pasture yield
# * increase. Value has to be in the range of 0 (no pasture yield growth)
# * and 1 (pasture yields increase linearily with tau). Only used in the realizations:
# * biocorrect (tc of current time step) and managementcalib (tc of previous time step).
cfg$gms$s14_yld_past_switch <- 0.25 # def = 0.25
# * Switch that allows selecting how yield calibration factors will be calculated.
# * If 0, crop yields calibration will be calculated as multiplicative relative values.
# * If 1, calibration values will be limited to additive absolute values when modeled yields strongly
# * underestimate historical values. For more information, read the description on the 'preloop'
# * file of module 14_yield.
# * options: 1 (limit to absolute values)
# * 0 (pure relative calibration)
cfg$gms$s14_limit_calib <- 1 # def = 1
# * Switch on scaling of irrigated yields to AQUASTAT irrigated-to-rainfed yield
# * ratio on regional scale
# NOTE: It is recommended to recalibrate the model when changing this setting!
cfg$gms$s14_calib_ir2rf <- 1 # def = 1
# * Switch to include yield impacts of land degradation.
# * The state of yield-relevant nature's contributions to people (NCP)
# * is reported through ./modules/14_yields/input/f14_yld_ncp_report.cs3
# * based on estimated outcomes for soil loss or pollination sufficiency
# * per cluster from an existing model run.
# * options: 1 (yields are reduced based on yield reduction coefficients)
# * 0 (land degradation is switched off)
cfg$gms$s14_degradation <- 0 # def = 0
# Switch to toggle the use of yield calibration factors (that resulted from a calibration run).
# If 0, no yield calibration factors are used, meaning all calibration factors are set to 1.
# If 1, yield calibration factors are used. (For this option to function, either an existing calibration file
# must be supplied in the input directory, or yields must be recalibrated during preprocessing.)
# This switch should only be activated for penalty_apr22 crop realization.
# For other realizations, it is recommended not to use this yield calibration.
cfg$gms$s14_use_yield_calib <- 0 # def = 0
# ***--------------------- 15_food ---------------------------------------
# * (anthropometrics_jan18): estimates food using scenario dependent regression
# * and demography drivers
# * (anthro_iso_jun22): estimates food using scenario dependent regression
# * and demography drivers on iso level
cfg$gms$food <- "anthro_iso_jun22" # def = anthro_iso_jun22
# * switch between exogenous and endogenous food demand
# * options: 0 (exogenous food demand) and 1 (endogenous food demand)
cfg$gms$s15_elastic_demand <- 0 # def =0
# * maximal number of iterations between food and magpie model before
# * simulation proceeds to next time step
cfg$gms$s15_maxiter <- 5 # def = 5
# * convergence criteria: maximal allowed country-wise deviation in calculated
# * real income between iterations
cfg$gms$s15_convergence <- 0.005 # def = 0.005
# * food scenario for selected (and respectively not selected) countries
# * in scen_countries15
# * options: SSP: "SSP1", "SSP2", "SSP3", "SSP4", "SSP5"
# * SRES: "A1", "A2", "B1", "B2"
# * OTHER: "PB" (planetary boundaries)
cfg$gms$c15_food_scenario <- "SSP2" # def = SSP2
cfg$gms$c15_food_scenario_noselect <- "SSP2" # def = SSP2
# * Temporal development of ruminant meat share within the livestock food product
# * group (applied before food demand model is executed)
# * options: constant, halving2050, mixed
cfg$gms$c15_rum_share <- "mixed" # def = mixed
# * Stronger ruminant fadeout in India
# * options: 0 (=off), 1 (=on)
cfg$gms$s15_rum_share_fadeout_india_strong <- 1 # def = 1
# * Milk share fadeout in India within the livestock food product
# * group (applied before food demand model is executed)
# * options: 0 (=off), 1 (=on)
cfg$gms$s15_milk_share_fadeout_india <- 1 # def = 1
# * Food substitution scenarios including functional forms, targets
# * and transition periods (applied after food demand model is executed)
# * options consist of 3 parts: functional form (lin,sigmoid), target (zero, 20pc, 50pc, 80pc, 90pc) and transition period (10_50: from 2010 to 2050, 20_50: from 2020 to 2050)
# * Example for sigmoid_50pc_20_50:
# * Functional form: sigmoid (S-shaped)
# * Target: 50percent reduction at end of transition period (2050 in this case)
# * Transition period: start in 2020, end in 2050
# * options: constant,
# * lin_zero_10_50, lin_zero_20_50, lin_zero_20_30, lin_zero_20_70, lin_50pc_20_50, lin_50pc_20_50_extend65, lin_50pc_20_50_extend80,
# * lin_50pc_10_50_extend90, lin_75pc_10_50_extend90, lin_80pc_20_50, lin_80pc_20_50_extend95, lin_90pc_20_50_extend95,
# * lin_99-98-90pc_20_50-60-100, sigmoid_20pc_20_50, sigmoid_50pc_20_50, sigmoid_80pc_20_50
cfg$gms$c15_rumscen <- "constant" # def = constant
cfg$gms$c15_fishscen <- "constant" # def = constant
cfg$gms$c15_alcscen <- "constant" # def = constant
cfg$gms$c15_livescen <- "constant" # def = constant
cfg$gms$c15_rumdairy_scp_scen <- "constant" # def = constant
cfg$gms$c15_rumdairyscen <- "constant" # def = constant
# * Set items of kfo_rd. This option allows for sensitivity scenarios.
# * kfo_rd is used in the food substitution scenarios c15_rumdairy_scp_scen and c15_rumdairyscen (see above)
# * options: "livst_rum,livst_milk", "livst_rum" or "livst_milk"
cfg$gms$kfo_rd <- "livst_rum" #def = livst_rum
# * Reduction of livestock products towards a kcal/cap/day intake target with or w/o substitution
# * maximum kcal/cap/day intake target used for downwards convergence of livestock products
cfg$gms$s15_kcal_pc_livestock_intake_target <- "430" # def = 430
# * choice temporal fade-out towards s15_kcal_pc_livestock_intake_target
# * options: constant, lin_zero_10_50, lin_zero_20_50, lin_zero_20_30, lin_zero_20_70
cfg$gms$c15_livescen_target <- "constant" # def = constant
# * fade-out of livestock products (0) or substitution of livestock products with plant-based products (1)
# * options: 0 (=fade-out), 1 (=substitution)
cfg$gms$s15_livescen_target_subst <- 1 # def = 1
# ***** Start configuration of exogenous food intake and waste scenarios *****
#
#
# * switch for transition to food waste scenarios
# * (1): transition towards exogenous food waste target
# * (0): regression-based estimation of food waste
cfg$gms$s15_exo_waste <- 0 # def = 0
#
# * scenario target for the ratio between food demand and intake
# * only activated if s15_exo_waste is set to 1
# * options: scalars >1
# * (1.1): corresponds to 10% food waste ~ quarter waste of HIC
# * (1.2): corresponds to 20% food waste ~ half waste of HIC
cfg$gms$s15_waste_scen <- 1.2 # def = 1.2
# * Switch for transition to exogenous diet scenarios
# * (EAT Lancet and National Institute of Nutrition (NIN))
# * (3): MAgPIE-specific realization of the EAT-Lancet diet, where model-internal
# * (regression-based) food demand projections are constrained by ranges for
# * intake targets of food groups to ensure healthy and sustainable diets as
# * recommended by the EAT-Lancet Commission (following Springmann et al. 2018 and Willett et al. 2019)
# * (2): transition towards exogenous diets (NIN for India and EAT for other regions)
# * Please use this switch in combination with cfg$gms$c15_EAT_scen <- "FLX_hmilk"
# * If only India's diet needs to be changed, then set cfg$gms$scen_countries15 <- "IND"
# * (1): transition towards exogenous diets and food demand parametrized
# * to a food-specific data set published by the EAT-Lancet Commission
# * (Willett et al., 2019). This EATLancet implementation is deprecated.
# * Please use MAgPIE-specific realization of the EAT-Lancet diet (3)
# * for EATLancet diet.
# * (0): regression-based estimation of diets and food demand
cfg$gms$s15_exo_diet <- 0 # def = 0
# * exogenous calorie scenario (EAT Lancet diet scenarios)
# * only activated if s15_exo_diet is > 0
# * options: healthy_BMI, 2100kcal, 2500kcal,
# * endo, no_underweight, no_overweight
# * half_overweight, no_underweight_half_overweight
# * healthy_BMI: all people have a BMI of 20-25
# * no_underweight: all people with BMI<20 have a BMI 20-25
# * no_overweight: all people with BMI>25 have a BMI 20-25
# * half_overweight: number of people with BMI>25 is halved compared to baseline (have BMI 20-25 instead)
# * no_underweight_half_overweight: combination of no_underweight and half_overweight
# * endo: no changes with respect to endogenous BMI
# * 2100kcal, 2500kcal: an exogenous target of 2100 or 2500 kcal, all
# * people have a BMI 20-25.
cfg$gms$c15_kcal_scen <- "healthy_BMI" # def = healthy_BMI
#
# * exogenous food-specific diet scenario (EAT Lancet diet scenarios)
# * only activated if s15_exo_diet is 1 or 2
# * options: BMK, FLX, PSC, VEG, VGN, FLX_hmilk, FLX_hredmeat
cfg$gms$c15_EAT_scen <- "FLX" # def = FLX
# * Sub-specifications: Which commodities shall be included in the diet shift
# * towards the selected scenario diet? Only selected (1) commodities will be
# * included, while the others will remain to be endogenous.
# * Any shift will be compensated to reach the calorie target by adjusting
# * staple calories.
cfg$gms$s15_exo_monogastric <- 1 # def = 1, options: 0,1
cfg$gms$s15_exo_ruminant <- 1 # def = 1, options: 0,1
cfg$gms$s15_exo_fish <- 1 # def = 1, options: 0,1
cfg$gms$s15_exo_fruitvegnut <- 1 # def = 1, options: 0,1
cfg$gms$s15_exo_roots <- 1 # def = 1, options: 0,1 (only relevant for new EATLancet realization s15_exo_diet=3)
cfg$gms$s15_exo_pulses <- 1 # def = 1, options: 0,1
cfg$gms$s15_exo_sugar <- 1 # def = 1, options: 0,1
cfg$gms$s15_exo_oils <- 1 # def = 1, options: 0,1
cfg$gms$s15_exo_brans <- 0 # def = 0 (as no target defined in EATLancet, they are kept at their projected level, unless switch activated (1), then brans are set to 0), options: 0,1
cfg$gms$s15_exo_scp <- 1 # def = 1, options: 0,1
# * Scenario target for the inclusion of alcohol in the EAT-Lancet diet
# * The EAT-Lancet diet only allows for added sugars,
# * but does not include processed food or alcohol.
# * Only activated if s15_exo_diet is set to 1 or 3
cfg$gms$s15_exo_alcohol <- 1 # def = 1, options: 0,1
# * Via 's15_alc_scen' a maximum target for alcohol consumption can be defined.
# * Only activated if s15_exo_alcohol = 1
# * (0): no alcohol consumption, as in the original version of the EAT-Lancet diet
# * (0.014): maximum target for alcohol consumption is 1.4% of total calorie consumption
# * (see Lassen et al., 2020)
cfg$gms$s15_alc_scen <- 0 # def = 0
#
#
# * Switch and specification of countries for which exogenous food scenarios
# * (EAT Lancet diet and food waste scenarios), food substitution scenarios and
# * c15_food_scenario shall be applied
# * Options: list of iso-codes of countries where exogenous scenario should be
# * in effect
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: all iso countries
cfg$gms$scen_countries15 <- all_iso_countries
#
# * Transition to exogenous food intake and waste scenarios including functional forms, targets
# * and transition periods (applied after the food demand model is executed). The resulting scenario
# * parametrisation is a linear combination of the default MAgPIE parametrisation and the exogenous
# * scenario data input.
# * Options consist of 3 parts: functional form (lin, sigmoid), target (zero, 20pc, 50pc, 80pc, 90pc;
# * pc: percent) and transition period (10_50: from 2010 to 2050, 20_50: from 2020 to 2050).
# * The target defines the relative contribution of the default MAgPIE parametrisation, e.g. zero
# * defines a full transition to the exogenous scenario data input.
# * Examples with target year y2030: lin_zero_20_30
# * Examples with target year y2050: lin_zero_20_50
# * Examples with target year y2070: lin_zero_20_70
# * For the full list of available options, see information on food substitution scenarios above.
# * Only active if at least one of the exogenous food intake and waste scenario
# * switches (s15_exo_diet and s15_exo_waste) is set to 1.
cfg$gms$c15_exo_foodscen <- "lin_zero_20_50" # def = lin_zero_20_50
# ***** End configuration of exogenous food intake and waste scenarios *****
# * calibration to historical values
# * options: 0, 1
cfg$gms$s15_calibrate <- 1 # def = 1
# ***--------------------- 16_demand -------------------------------------
# * (sector_may15): default for flexible regions
cfg$gms$demand <- "sector_may15" # def = sector_may15
# ***--------------------- 17_production ---------------------------------
# * (flexreg_apr16): default production aggregation
cfg$gms$production <- "flexreg_apr16" # def = flexreg_apr16
#* Additional switch to initialize cellular production
# * (on) : it initializes cellular production
# * (off) : it does not initialize cellular production
cfg$gms$c17_prod_init <- "on" # default = on
# ***--------------------- 18_residues -----------------------------------
# * (flexreg_apr16): detailed residue calculations
# * (off): off
cfg$gms$residues <- "flexreg_apr16" # def = flexreg_apr16
# * residue on field burning
# * options: phaseout - phaseout of residue burning to minimum burn share (0-10%)
# * constant - constant shares of on field burning (15-25%)
cfg$gms$c18_burn_scen <- "phaseout" # def = phaseout
# ***--------------------- 20_processing ---------------------------------
# * (substitution_may21) : processing with couple products allowing for substitution
# * (off): off
cfg$gms$processing <- "substitution_may21" # def = substitution_may21
# * SCP route
# * single-cell microbial protein production route
# * options: mixed, methane, sugar, cellulose, hydrogen
# * Mapping of scp route to feedstock: methane:foddr, sugar:sugar_cane, cellulose:begr
# * Note that hydrogen does not require land-based inputs
# * Mixed assumes equal shares of methane, sugar, cellulose and hydrogen
cfg$gms$c20_scp_type <- "sugar" # def = sugar
# ***--------------------- 21_trade --------------------------------------
# * (free_apr16): free trade without restrictions
# * (off): no trade at all
# * (exo): exogenously prescribed trade
# * (selfsuff_reduced): self-sufficiency based trade with trade costs related to exports
# * (selfsuff_reduced_bilateral22): same as `selfsuff_reduced` but with bilateral trade flows
cfg$gms$trade <- "selfsuff_reduced" # def = selfsuff_reduced
# * option for `exo` realization only:
# * fix `v21_manna_from_heaven` to zero (0) or not (1)
# Note: Without fixing to zero, v21_manna_from_heaven might be used unnecessarily in runs started with highres.R
cfg$gms$s21_manna_from_heaven <- 0
# * options for `selfsuff_reduced` and `selfsuff_reduced_bilateral22` realizations:
# * Commodities that can have additional imports to maintain feasibility
cfg$gms$k_import21 <- "wood, woodfuel"
# * Cost for additional imports to maintain feasibility in USD05MER per tDM
cfg$gms$s21_cost_import <- 10000 # def = 10000
# * trade balance reduction scenario
# * (l909090r808080): 10 percent trade liberalisation for secondary and
# * livestock products in 2030,2050,2100 and 20 percent for
# * crops
# * (l908080r807070): livestock/secondary: 10% in 2030, 20% in 2050,2100
# * crops: 20% in 2030, 30% in 2050,2100
# * (l909595r809090): livestock/secondary: 10% in 2030, 5% in 2050,2100
# * crops: 20% in 2030, 10% in 2050,2100
# * Initial values for the trade balance reduction in 1995 are 1, i.e. all trade in
# * 1995 happens through the self-sufficiency pool. For later time steps, additional
# * scenarios of trade liberalization (i.e. of allocation to the comparative advantage pool)
# * are implemented based on Schmitz et al. 2012 (also described in Popp et al. 2017)
cfg$gms$c21_trade_liberalization <- "l909090r808080" # def = l909090r808080
# * Fraction to ease self sufficiency pool trade for roundwood
cfg$gms$s21_trade_bal_damper <- 0.65 # def 0.65
# * whether trade tariff should be considered at all
# * (0) without trade tariff
# * (1) with trade tariff
cfg$gms$s21_trade_tariff <- 1 # def =1
# * whether to fade out trade tariffs (bilateral trade realization) # def = 0
cfg$gms$s21_trade_tariff_fadeout <- 0
# * start year of fadeout if s21_trade_tariff_fadeout = 1 # def = 2020
cfg$gms$s21_trade_tariff_startyear <- 2020
# * end year of fadeout if s21_trade_tariff_fadeout = 1 # def = 2050
cfg$gms$s21_trade_tariff_targetyear <- 2050
# ***--------------------- 22_land_conservation --------------------------------------
# * (area_based_apr22): Area-based conservation (baseline and future)
# * based on WDPA and conservation priority areas
cfg$gms$land_conservation <- "area_based_apr22" # def = area_based_apr22
# * Additional land conservation target based on conservation priority areas
# * during future time steps (after `cfg$gms$sm_fix_SSP2`).
# * ("none") No additional land conservation target (WDPA only)
# * ("30by30") 30 % of global land surface in Key Biodiversity Areas (KBA),
# * GSN Distinct Species Assemblages & Critical Connectivity Areas + WDPA
# * ("KBA") Key Biodiversity Areas + WDPA
# * ("GSN_DSA") Global Safety Net: Distinct Species Assemblages + WDPA
# * ("GSN_RarePhen") Global Safety Net: Rare Phenomena + WDPA
# * ("GSN_AreaIntct") Global Safety Net: Areas of Intactness + WDPA
# * ("GSN_ClimTier1") Global Safety Net: Climate Stabilisation Tier 1 + WDPA
# * ("GSN_ClimTier2") Global Safety Net: Climate Stabilisation Tier 2 + WDPA
# * ("CCA") Critical Connectivity Areas (Brennan et al. 2022) + WDPA
# * ("IrrC_XXpc") Land area that covers XX percent of total global irrecoverable carbon
# * as defined by Noon et al. (2022), where XX correponds to either
# * 50, 75, 95, or 99 percent + WDPA
# * ("IrrC_XXpc_30by30") 30by30 + Land area that covers XX percent of total global irrecoverable
# * carbon as defined by Noon et al. (2022), where XX correponds to either
# * 50, 75, 95, or 99 percent + WDPA
# * ("BH") Biodiversity Hotspots + WDPA
# * ("IFL") Intact Forest Landscapes + WDPA
# * ("BH_IFL") Biodiversity Hotspots + Intact Forest Landscapes + WDPA
# * ("GSN_HalfEarth") Full protection of areas within the Global Safety Net, which
# * roughly corresponds to 50 percent of the global land surface
# * ("PBL_HalfEarth") Ecoregion-based approach to protecting half of the global land surface
# * Note: c22_protect_scenario applies to countries selected in policy_countries22
# * c22_protect_scenario_noselect applies to all other countries.
cfg$gms$c22_protect_scenario <- "none" # def = None
cfg$gms$c22_protect_scenario_noselect <- "none" # def = None
# * Switch and specification of countries for which land conservation policies in
# * c22_protect_scenario apply.
# * Options: list of iso-codes of countries where land conservation should be applied
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: all iso countries
cfg$gms$policy_countries22 <- all_iso_countries
# * Whether land restoration is carried out in areas targeted by land conservation
# * (1) Land is fully restored in conservation priority areas
# * (0) Only remaining forest and other land areas are conserved
cfg$gms$s22_restore_land <- 1 # def = 1
# * Start and target year of land conservation option ('c22_protect_scenario').
# * Defines when full protection based on 'future options' is reached, using a
# * sigmoidal trajectory. NOTE: This switch is only relevant
# * if one of the 'future options' (e.g. BH) is chosen.
# * Start year:
cfg$gms$s22_conservation_start <- 2020 # def = 2020
# * Target year (year when full protection is reached):
cfg$gms$s22_conservation_target <- 2030 # def = 2030
# ***--------------------- 29_ageclass -----------------------------------
# * (feb21): Distribution of age-classes according to Poulter et al 2018
cfg$gms$ageclass <- "feb21" # def = feb21
# ***--------------------- 30_crop ---------------------------------------
# * (endo_apr21): Hard rotational constraints. Dynamic cropland and detailed cropland availability data at grid cell level.
# * (rotation_apr22): hard rotational constraints and fallow constraints with the option of different future scenarios. Dynamic cropland and detailed cropland availability data at grid cell level.
# * (penalty_apr22): rotational and fallow constraints are incentivized via penalty payments. Dynamic cropland and detailed cropland availability data at grid cell level.
# NOTE: It is recommended to recalibrate the model when changing this setting!
cfg$gms$crop <- "endo_apr21" # def = endo_apr21
# * (c30_bioen_type): switch for type of bioenergy crops; options: begr, betr, all
cfg$gms$c30_bioen_type <- "all" # def = "all"
# * (c30_bioen_water): switch for irrigation of bioenergy crops; options: rainfed, irrigated, all
cfg$gms$c30_bioen_water <- "rainfed" # def = "rainfed"
# * (c30_rotation_constraints): switch for rotational constraints: on, off
cfg$gms$c30_rotation_constraints <- "on" # def = "on"
# * (c30_rotation_scenario): scenarios for constraints.
# * In realization rotation_apr22: min (minimal constraints), default (best guess), good (good practice),
# * good_20div (good practice - 20% for other crops), setaside (default plus fallow),
# * legumes (minimum share of legumes), agroforestry (minimum share of perennials), sixfoldrotation
# * (crops can only repeat after 6 years), agroecology (mix of previous scenarios)
# * In realization penalty_apr22: none (no incentives), default (best guess), fallow (increased fallow incentives),
# * legumes (increased incentives for legumes), agroforestry (increased incentives for perennials), agroecology (mix)
# * In realization endo_apr21: no scenarios exist.
cfg$gms$c30_rotation_scenario <- "default" # def = "default"
# * Year by which rotation scenario is fully implemented:
# * Start year:
cfg$gms$s30_rotation_scenario_start <- 2020 # def = 2020
# * Target year (year when full implementation is reached):
cfg$gms$s30_rotation_scenario_target <- 2050 # def = 2050
# * Switch to determine whether marginal land (suitability index below 0.33) should be included
# * in the total available cropland. Options are:
# * ("all_marginal"): All marginal land can be used for crop cultivation
# * ("q33_marginal"): The bottom tertile of marginal land is excluded
# * ("no_marginal"): Marginal land is completely excluded from crop cultivation
# * Note: Option "q33_marginal" produces the highest spatial correlation
# * with observed cropland patterns and is recommended for productive runs.
cfg$gms$c30_marginal_land <- "q33_marginal" # def = "q33_marginal"
# * Share of available cropland that is withheld for maintaining semi-natural vegetation (SNV)
# * in each square km in cropland areas, including grassland, forest and other land. For example,
# * a share of 0.2 corresponds to 20 % of SNV in terms of the available cropland.
# * The amount of cropland relocation is estimated based on external high resolution
# * land cover information from the Copernicus Global Land Service for the year 2019.
# * Accepted sensible values are between 0 and 0.5
# Note: s30_snv_shr applies to countries selected in policy_countries30
# s30_snv_shr_noselect applies to all other countries.
cfg$gms$s30_snv_shr <- 0 # def = 0
cfg$gms$s30_snv_shr_noselect <- 0 # def = 0
# * Year by which SNV policy ('s30_snv_shr') is fully implemented.
# * Start year (should be close to 2019):
cfg$gms$s30_snv_scenario_start <- 2020 # def = 2020
# * Target year (year when full implementation is reached):
cfg$gms$s30_snv_scenario_target <- 2030 # def = 2030
# * Switch and specification of countries for which SNV policy in
# * s30_snv_shr apply.
# * Options: list of iso-codes of countries where SNV policy should be applied
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: all iso countries
cfg$gms$policy_countries30 <- all_iso_countries
# * Land types included in the SNV policy. This option allows for sensitivity analyses.
# * plausible options: "secdforest, forestry, past, other",
# * "secdforest, other",
# * "secdforest, past, other" etc.
cfg$gms$land_snv <- "secdforest, forestry, past, other" #def = "secdforest, forestry, past, other"
# ***--------------------- 31_past ---------------------------------------
# * (static): static pasture
# * (endo_jun13): dynamic pasture
# * (grasslands_apr22): Grassland management separating managed pastures from rangelands
# NOTE: It is recommended to recalibrate the model when changing this setting!
cfg$gms$past <- "endo_jun13" # def = endo_jun13
# * Factor requirements (USD04 per ton DM)
cfg$gms$s31_fac_req_past <- 1 # def = 1
# * switch for unequal (1) or equal (0) sign in pasture production constraint q31_prod.
# * unequal means that pasture production can be lower than pasture area x pasture yield,
# * reflecting that not all pasture area is grazed.
cfg$gms$s31_unequal <- 1 # def = 1
# * Scenario for grassland, pasture suitability areas and pasture yields (only take effect for realization grasslands_apr22)
# * options: cc (climate change)
# * nocc (no climate change)
# * nocc_hist (no climate change after year defined by sm_fix_cc)
cfg$gms$c31_grassl_yld_scenario <- "cc" # def = "cc"
# * Switch that allows selecting how yield calibration factors will be calculated.
# * If 0, grassland yield calibration will be calculated as multiplicative relative values.
# * If 1, calibration values will be limited to additive absolute values when modeled yields strongly
# * underestimate historical values. For more information, read the description on the 'preloop'
# * file of module 31_past.
# * options: 1 (limit to absolute values)
# * 0 (pure relative calibration)
cfg$gms$s31_limit_calib <- 1 # def = 1
# ***--------------------- 32_forestry -----------------------------------
# * (dynamic_feb21): Dynamic forestry sector including afforestation with detailed age-classes
cfg$gms$forestry <- "dynamic_feb21" # def = dynamic_feb21
# * afforestation planing horizon (years)
cfg$gms$s32_planing_horizon <- 50 # def = 50
# * Settings for CO2 price driven afforestation (Growth curve and BII)
# * Growth curve for CO2 price driven afforestation
# * Switch for using natural vegetation (0) or plantation (1) growth curves towards LPJmL natural
# * vegetation carbon density.
# * Afforestation following plantation growth curves reflects managed or assistent regrowth,
# * and might also include non-native species.
cfg$gms$s32_aff_plantation <- 0 # def = 0
# * BII coefficient for CO2 price driven afforestation
# * Switch for using secondary vegetation (0) or timber (1) BII coefficients for CO2 price driven afforestation
# * The recommend setting is to map the BII coefficient to the choice of the growth curve.
# * natural vegetation growth curve (0): secondary vegetation BII coefficient (0)
# * plantation growth curve (1): timber BII coefficient (1)
# * However, afforestation with plantations (1) could possibly be done in a biodiversity friendly way (0)
cfg$gms$s32_aff_bii_coeff <- 0 # def = 0
# Afforestation policy
# * ("none"): no prescribed afforestation
# * ("npi"): prescribed afforestation based on NPI policies
# * ("ndc"): prescribed afforestation based on NPI+NDC policies
cfg$gms$c32_aff_policy <- "npi" # def = "npi"
# Maximum total global afforestation in Mha (Inf = no constraint)
# Note 1: the limit applies to the sum of endogenous CO2 price driven and exogenous NPI/NDC afforestation
# Note 2: a limit of 0 or below the level of exogenous NPI/NDC afforestation will be a automatically raised to the level of exogenous NPI/NDC afforestation
cfg$gms$s32_max_aff_area <- Inf # def = Inf
# Protection of afforested areas (endogenous CO2 price driven)
# 0=until end of planning horizon (see s32_planing_horizon)
# 1=forever
# Note: Without a price on CO2 emissions from land-use change in module 32_forestry,
# which is the current default (c56_emis_policy <- "reddnatveg_nosoil"),
# the recommended setting for s32_aff_prot is 1.
# Otherwise, the model can do re/afforestation on existing re/afforestation areas
# after the end of planning horizon without penalty for CO2 emissions.
cfg$gms$s32_aff_prot <- 1 # def = 1
# Type of afforestation constraint
cfg$gms$s32_max_aff_area_glo <- 1 # def = 1
# * (1): global limit based on s32_max_aff_area
# * (0): regional limit based on the input file "f32_max_aff_area.cs4"
# Switch to determine whether afforestation should be limited to
# certain latitudinal zones
# * ("unrestricted"): No regions excluded
# * ("noboreal"): Exclude boreal regions > 50deg N
# * ("onlytropical"): Afforestation only in tropical areas 20deg S-20deg N
cfg$gms$c32_aff_mask <- "noboreal" # def = "noboreal"
# Switch to determine whether biogeophysical responses to
# afforestation should be considered
# If this endogenous bgp effect is considered it is adviced to use the
# unrestricted afforestation mask
# to avoid superimposing the additional exogeneous bgp implementation
# * ("nobgp"): No biogeophysical influences
# * ("ann"): Annual BGP
# * ("djf"): Boreal winter BGP December January February
# * ("jja"): Boreal summer BGP June July August
cfg$gms$c32_aff_bgp <- "nobgp" # def = "nobgp"
# Switch for the different TCRE estimates which are used to
# translate the BGP temperature effect into their carbon equivalent.
# The estimates encompass the ensemble mean +/- SD of 20 CMIP5 models.
# Ceq ~ BGP/TCRE - Hence, high TCRE decreases the BGP effect.
# * ("ann_TCREmean"): Ensemble mean
# * ("ann_TCREhigh"): Ensemble mean + SD
# * ("ann_TCRElow"): Ensemble mean - SD
cfg$gms$c32_tcre_ctrl <- "ann_TCREmean"
# Switch for dynamic timber plantations
# "off" means that timber plantations are initialized in the highest
# age class and that there is no regrowth seen in such plantations.
# In addition, timber plantations are fixed to 1995 levels in the "off" case.
# In other cases, age-classes in timber plantations are initialized based on
# rotation length and can change dynamically over time.
# Note that this switch has no effect in the "static_sep16" realization,
# in which forestry area is assumed static.
# * 0= off
# * 1= Equal distribution
# * 2= FAO distribution based on planted forest estimates 2006
# * 3= Poulter distribution
# * 4= Manual distribution - Similar to equal distribution but higher weight on
# * younger age-classes as plantation areas increase strongly
# * in recent history (ca. >1990).
cfg$gms$s32_initial_distribution <- 0 # def = 0
# Switch fore regional or global interest rate for rotation length calculations.
# Using the global setting would mean that the timber plantation decisions are
# decoupled from other decisions in the model which are based on regional interest
# rates.
# * ("regional") = Regionally differentiated interest rates
# * ("global") = One global interest rate
cfg$gms$c32_interest_rate <- "regional" # def = "regional"
# Global interest rate for plantations in case c32_interest_rate switch is global
# Accepted values between 0 and 1.
cfg$gms$s32_forestry_int_rate <- 0.05 # def = 0.05
# Scenarios for using FAO scenarios of future proportion of production coming from
# Timber plantations used for deciding how much establishment should be made in
# simulation time step. This setting is only used when s32_hvarea is set to 2 ("Endogenous")
# * ("abare") = Global outlook for plantations. Australian Bureau of Agriculture and Resource Economics (ABARE) and Jaakko Pöyry Consulting 1999
# * ("brown") = The global outlook for future wood supply from forest plantations. Working paper GFPOS/WP/03 prepared for the 1999 Global Forest Products Outlook Study 1999
cfg$gms$c32_dev_scen <- "abare" # def = "abare"
# Scenarios for future development of timber production contribution to roundwood demand
# based on the settings from c32_dev_scen.
# * ("constant") = Constant share over time (1995-2250)
# * ("h5s5l5") = In every time step 5% increase from 1995 till 2020, 5% increase from 2025 till 2050, 5% increase from 2055 till 2250
# * ("h5s2l2") = In every time step 5% increase from 1995 till 2020, 2% increase from 2025 till 2050, 2% increase from 2055 till 2250
# * ("h5s2l1") = In every time step 5% increase from 1995 till 2020, 2% increase from 2025 till 2050, 1% increase from 2055 till 2250
# * ("h5s1l1") = In every time step 5% increase from 1995 till 2020, 1% increase from 2025 till 2050, 1% increase from 2055 till 2250
# * ("h5s1l05") = In every time step 5% increase from 1995 till 2020, 1% increase from 2025 till 2050, 0.5% increase from 2055 till 2250
# * ("h2s1l05") = In every time step 2% increase from 1995 till 2020, 1% increase from 2025 till 2050, 0.5% increase from 2055 till 2250
cfg$gms$c32_incr_rate <- "h5s2l1" # def = "h5s2l1"
# Harvesting switch for timber production
# * 0 = No harvested area from plantations, no age-class shifting (area held constant at 1995 levels)
# * This also means that no harvesting or establishment of new plantations takes place.
# * 1 = "Exogenous" scenario. Harvested area from plantations but with age-class shifting
# * All timber plantations are harvested at rotation age and are re-established
# * such that the total plantation area remains constant.
# * 2 = "Endogenous" scenario. Harvest from plantations including age-class shifting
# * All plantations are harvested at rotation age. Plantation establishment is endogenous.
cfg$gms$s32_hvarea = 0 # def = 0
# Type of rotation length selection criteria
# * ("mean_annual_increment") = Harvesting when the average annual increment is maximum
# * ("current_annual_increment") = Harvesting when the marginal increment is maximum
# * ("instantaneous_growth_rate") = Harvest when instantaneous growth rate is equal to interest rate
cfg$gms$c32_rot_calc_type <- "current_annual_increment"
# Generic forest shock scenarios. Disturbance sets percentage of all age classes back to 0
# * ("none") No disturbance applied
# * ("002lin2030") Disturbance applied to 0p2 percent per year linear phase-in over 20yrs from 0 in 2030
# * ("004lin2030") Disturbance applied to 0p4 percent per year linear phase-in over 20yrs from 0 in 2030
# * ("008lin2030") Disturbance applied to 0p8 percent per year linear phase-in over 20yrs from 0 in 2030
# * ("016lin2030") Disturbance applied to 1p6 percent per year linear phase-in over 20yrs from 0 in 2030
cfg$gms$c32_shock_scenario = "none" # def = "none"
# ***--------------------- 34_urban ---------------------------------------
# * 34_urban includes urban land
# * (static) static urban land fixed on 1995 patterns from LUH2v2
# * (exo_nov21) has exogenous future urban land patterns based on LUH2v2
cfg$gms$urban <- "exo_nov21" # def = exo_nov21
# * Additional settings for exo_nov21 realization only:
cfg$gms$c34_urban_scenario <- "SSP2" # def = SSP2
# * options: SSP: "SSP1", "SSP2", "SSP3", "SSP4", "SSP5"
# ***--------------------- 35_natveg --------------------------------------
# * 35_natveg includes primforest, secdforest and other land
# * (dynamic_feb21): Dynamic natural vegetation land with detailed age-classes
cfg$gms$natveg <- "dynamic_feb21" # def = dynamic_feb21
# Avoided Deforestation policy
# * ("none"): no avoided deforestation
# * ("npi"): avoided deforestation based on NPI policies
# * ("ndc"): avoided deforestation based on NPI+NDC policies
cfg$gms$c35_ad_policy <- "npi" # def = "npi"
# Avoided Other Land Conversion policy
# * ("none"): no avoided other land conversion
# * ("npi"): avoided other land conversion based on NPI policies
# * ("ndc"): avoided other land conversion based on NPI+NDC policies
cfg$gms$c35_aolc_policy <- "npi" # def = "npi"
# Distribution of age-classes during secondary forest initialization
# * (0): All secondary forest belongs to highest age class
# * (1): Equal distribution of secondary forest in all age-classes
# * (2): Distribution of secondary forest according to Poulter et al 2018 based on MODIS satellite data
cfg$gms$s35_secdf_distribution <- 0 # def = 0
# Damages in natural forests
# * (0): No damage simulated
# * (1): Damage from shifting agriculture (constant)
# * (2): Damage from shifting agriculture is faded out by the year defined in 'cfg$gms$c35_forest_damage_end'
# * (4): Generic forest disturbance scenarios defined in `f35_forest_shock.csv`
cfg$gms$s35_forest_damage <- 2 # def = 2
# * If option (2) above: Year by which damage from shifting agriculture has petered out
cfg$gms$s35_forest_damage_end <- 2050 #def = 2050
# Harvesting switch for timber production
# * 0 = No timber production from natveg, no age-class shifting
# * 1 = No timber production from natveg but with age-class shifting
# * 2 = Timber production from natveg including age-class shifting
cfg$gms$s35_hvarea <- 0 # def = 0
# Generic forest shock scenarios. Disturbance sets percentage of all age classes back to 0
# * ("none") No disturbance applied
# * ("002lin2030") Disturbance applied to 0p2 percent per year linear phase-in over 20yrs from 0 in 2030
# * ("004lin2030") Disturbance applied to 0p4 percent per year linear phase-in over 20yrs from 0 in 2030
# * ("008lin2030") Disturbance applied to 0p8 percent per year linear phase-in over 20yrs from 0 in 2030
# * ("016lin2030") Disturbance applied to 1p6 percent per year linear phase-in over 20yrs from 0 in 2030
cfg$gms$c35_shock_scenario = "none" # def = "none"
# ***--------------------- 36_employment -------------------------------
# * This module calculates agricultural employment.
# * (exo_may22): employment is calculated based on labor costs and not affecting the
# * optimization
cfg$gms$employment <- "exo_may22" # default = "exo_may22"
# * global minimum wage in USDMER05 per hour that needs to be reached in all countries by 2050
cfg$gms$s36_minimum_wage <- 0 # default = 0 (no minimum wage)
# * A scenario that increases wages can either be fully related to productivity increase
# * (leading to lower employment for the same total labor costs), or keep productivity
# * constant (leading to the same employment for higher total labor costs), or to a
# * mixture between productivity increase an total labor cost increase.
# * The scalar `s36_scale_productivity_with_wage` describes how high the labor productivity
# * gain should be relative to the increase in hourly labor costs. If set to 0,
# * productivity stays constant, if set to 1 productivity scales proportional to hourly
# * labor costs. Other positive values lead to a mixture of productivity gain and increase
# * in total costs.
cfg$gms$s36_scale_productivity_with_wage <- 0 # default = 0
# ***--------------------- 37_labor_prod ---------------------------------
# * This module provides a labour productivity factor (0-1),
# * which reflects the efficiency of labour under changing environmental conditions (1 = no change).
# * The labour productivity factor is currently only considered in the sticky_labor
# * realization of the [38_factor_costs] module.
# * (off): labor productivity factor fixed to 1
# * (exo): labor productivity factor is affected by climate change impacts
cfg$gms$labor_prod <- "off" # default = off
# * Additional setting for exo realization only:
# * Climate change impact scenario. Options: "rcp119", "rcp585"
cfg$gms$c37_labor_rcp <- "rcp119" # default = "rcp119"
# * Heat assessment metric. Options: "ISO", "HOTHAPS"
cfg$gms$c37_labor_metric <- "ISO" # default = "ISO"
# * Labour intensity. Options: "300W", "400W"
cfg$gms$c37_labor_intensity <- "400W" # default = "400W"
# * Uncertainty over ESMs. Options: "enslower", "ensmean", "ensupper"
cfg$gms$c37_labor_uncertainty <- "ensmean" # default = "ensmean"
# ***--------------------- 38_factor_costs -------------------------------
# * Make sure you use the corresponding calibration file to the selected realization
# * Please, check details in input settings.
# * (per_ton_fao_may22) factor costs fixed per ton
# * (sticky_feb18) factor costs including investments in capital
# * (sticky_labor) based on sticky_feb18 + labor productivity factor included
# NOTE: It is recommended to recalibrate the model when changing this setting!
# NOTE: In case the maximum number of iterations is reached without convergence
# in the calibration, please consider to use the best_calib setting for the calibration run
cfg$gms$factor_costs <- "per_ton_fao_may22" # default = per_ton_fao_may22
# * Regional (reg) or global (glo) factor requirements (applicable for per_ton_fao_may22,
# * sticky_feb18, sticky_labor). The regional version keeps factor requirements constant
# * after 2010, the global version uses values from 2005 for all years. Typically, the
# * same version (glo or reg) should be chosen for c70_fac_req_regr for consistency.
# NOTE: It is recommended to recalibrate the model when changing this setting!
cfg$gms$c38_fac_req <- "glo" # default "glo"
# * Only relevant for sticky_labor implementation:
# * Year until which the capital share should be fixed to the historical values, not allowing for endogenous
# * substitution between labor and capital.
cfg$gms$s38_startyear_labor_substitution <- 2020 # default 2020
# * The following two settings are to create exogenous scenarios in which capital is substituted by labor to increase
# * employment:
# * The taget labor share defins a global target for labor share out of labor + capital needed per unit
# * of output. If 0, no target is set.
cfg$gms$s38_target_labor_share <- 0 # default 0
# * The target year defines the year for which the global target labor share should be set.
cfg$gms$s38_targetyear_labor_share <- 2050
# * The target fulfillment share indicates the share by which a region moves from its baseline
# * labor share towards the target value
cfg$gms$s38_target_fulfillment <- 0.5 # default 0.5
# ***--------------------- 39_landconversion -----------------------------
# * (calib): Costs for cropland expansion are scaled with a regional calibration factor
# * Costs for pasture and forestry expansion are global static
cfg$gms$landconversion <- "calib" # def = calib
# * Cost for cropland expansion before calibration (USD05MER per hectare)
cfg$gms$s39_cost_establish_crop <- 10000 #def = 10000
# * Reward for cropland reduction before calibration (USD05MER per hectare)
cfg$gms$s39_reward_crop_reduction <- 6000
# * Cost for pasture land expansion (USD05MER per hectare)
cfg$gms$s39_cost_establish_past <- 8000 #def = 8000
# * Cost for foresty land expansion (USD05MER per hectare)
cfg$gms$s39_cost_establish_forestry <- 1000 #def = 1000
# * Cost for urban land expansion (USD05MER per hectare)
cfg$gms$s39_cost_establish_urban <- 10000 #def = 10000
# * Switch for ignoring land conversion cost calibration factors
# * Options: 1 (ignore calibration factors)
# * 0 (use calibration factors)
cfg$gms$s39_ignore_calib <- 0 #def = 0
# ***--------------------- 40_transport ----------------------------------
# * (off): no transport costs
# * (gtap_nov12): transport costs applied to crops and ruminant livestock
# * products calibrated based on magpie-output (dynamic pasture)
# * to match GTAP data
cfg$gms$transport <- "gtap_nov12" # def = gtap_nov12
# * scalar to introduce pasture transport costs
cfg$gms$s40_pasture_transport_costs <- 0 # def = 0
# ***--------------------- 41_area_equipped_for_irrigation ---------------
# * (static): no expansion
# * (endo_apr13): endogenous, cost driven expansion
cfg$gms$area_equipped_for_irrigation <- "endo_apr13" # def = endo_apr13
# * switch for initialization area
# * (LUH2v2): area equipped for irrigation based on LUH2v2 irrigated areas
# * (Mehta2022): area equipped for irrigation from Mehta et al. (2022)
cfg$gms$c41_initial_irrigation_area <- "LUH2v2" # def = LUH2v2
# * Sets the rate of depreciation of irrigation infrastructure in every timestep.
# * Only applicable when (endo_apr13) realization is selected
cfg$gms$s41_AEI_depreciation <- 0 # def = 0
# ***--------------------- 42_water_demand --------------------------------
# * (agr_sector_aug13): fixed fraction of water available is reserved
# * for other uses
# * (all_sectors_aug13): manufacturing, electricity and domestic demand are
# * retrieved from WATERGAP data.
cfg$gms$water_demand<- "all_sectors_aug13" # def = all_sectors_aug13
# * water demand scenario
# * options: cc (climate change)
# * nocc (no climate change)
# * nocc_hist (no climate change after year defined by sm_fix_cc)
cfg$gms$c42_watdem_scenario <- "cc" # def = "cc"
# * Choice of fraction of available water that is not
# * available for agriculture (only affects agr_sector_aug13 realization)
cfg$gms$s42_reserved_fraction <- 0.5 # def = 0.5
# * Scenario for non agricultural water demand from WATERGAP
# * (only affects all_sector_aug13 realization)
# * (1): ssp1
# * (2): ssp2
# * (3): ssp3
cfg$gms$s42_watdem_nonagr_scenario <- 2 # def = 2
# * Switch to determine the irrigation efficiency scenario
# * (1): global static value
# * (2): regional static values from gdp regression
# * (3): gdp driven increase
cfg$gms$s42_irrig_eff_scenario <- 2 # def = 2
# * Irrigation efficiency
# * (Only used if global static value is requested)
cfg$gms$s42_irrigation_efficiency <- 0.66 # def=0.66
# * Environmental flow protection policy
# * (off): no EFP policy
# * (on): global EFP policy
# * (mixed): EFP policy only in hic regions
cfg$gms$c42_env_flow_policy <- "off" # def = "off"
# * Start and target year of environmental flow protection policy ('c42_env_flow_policy').
# * Defines when linear fading in of policy starts and when the target of
# * full protection based on 's42_env_flow_scenario' is reached
# * Start year:
cfg$gms$s42_efp_startyear <- 2020 # def = 2020
# * Target year (year when full protection is reached):
cfg$gms$s42_efp_targetyear <- 2040 # def = 2040
# * Switch and specification of countries for which environmental flow policy
# * shall apply.
# * Options: list of iso-codes of countries where EFP should be in effect.
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: all iso countries
cfg$gms$EFP_countries <- all_iso_countries # def = all_iso_countries
# * Environmental flow protection scenario
# * (0): do not consider environmental flows.
# * s42_env_flow_base_fraction and
# * s42_env_flow_fraction have no effect.
# * (1): Reserve a certain fraction of available water
# * specified by s42_env_flow_fraction for
# * environmental flows
# * (2): Each grid cell receives its own value for
# * environmental flow protection based on LPJ
# * results and a calculation algorithm by Smakhtin 2004.
# * s42_env_flow_fraction has no effect.
cfg$gms$s42_env_flow_scenario <- 2 # def = 2
# * Fraction of available water that is reserved for the environment
# * in case of a protection policy
# * In which regions and timesteps a protection policy is in place is determined
# * in the file EFR_protection_policy.csv in the input folder
# * of the 42_water_demand module
cfg$gms$s42_env_flow_fraction <- 0.2 # def = 0.2
# * Fraction of available water that is reserved for the environment in case of
# * missing protection policy
cfg$gms$s42_env_flow_base_fraction <- 0.05 # def = 0.05
# * Switch to activate pumping costs (only available for India currently), Set to 1 if want to use for India
cfg$gms$s42_pumping <- 0 # def = 0
# * Year in which pumping costs will be implemented
cfg$gms$s42_multiplier_startyear <- 1995 # def = 1995
# * Scalar to change pumping costs for sensitivity analysis, takes numeric values
# * Default value is 0 for all regions, set value to 1 for India default analysis
# * Only applicable when s42_pumping is equal to 1
cfg$gms$s42_multiplier <- 0 # def = 0
# ***--------------------- 43_water_availability --------------------------
# * (total_water_aug13): surface and ground water resources available
cfg$gms$water_availability <- "total_water_aug13" # def = total_water_aug13
# * water availability scenario
# * options: cc (climate change)
# * nocc (no climate change)
# * nocc_hist (no climate change after year defined by sm_fix_cc)
cfg$gms$c43_watavail_scenario <- "cc" # def = "cc"
# ***------------------------- 44_biodiversity ------------------------------
# * (bii_target): Biodiversity Intactness Index (BII) for 71 biomes with the option to define a lower bound for BII at biome level
# * (bv_btc_mar21): Global optimization of range-rarity weighted biodiversity stocks based on a price for losses and gains
cfg$gms$biodiversity <- "bii_target" # def = bii_target
# * Start year for linear increase towards BII target value (in bii_target) or biodiversity target price (in bv_btc_mar21)
cfg$gms$s44_start_year <- 2025 #def = 2025
# * Year in which the BII target value (in bii_target) or biodiversity target price (in bv_btc_mar21) is reached
cfg$gms$s44_target_year <- 2100 #def = 2100
# ** Options for realization `bii_target`
# * This realisation allows to define a target value and year for the Biodiversity Intactness Index (BII)
# * that is applied in each biome type of each biogeographic realm or at the global level.
# * The lower bound increases in each biome linearly from the BII level in the start year (s44_start_year) to the value defined in `s44_bii_lower_bound` in the target year (s44_target_year).
# * The BII has been proposed as a control variable for changes in ecosystem functioning at the global scale
# * and has shown to capture increasing levels of ecosystem degradation. Steffen et al. (2015, 10.1126/science.1259855) have proposed
# * a premliminary boundary of 0.9 at the biome level (safe space) but with an uncertainty range of 0.9-0.3,
# * in which ecosystem degradation may undermine stable ecosystem functioning and the supply of nature's
# * contributions to people (NCP). With BII values below 0.3, the area's biodiversity has been depleted,
# * causing high risk of ecosystem collapse. Estimates for the global BII range between 0.7975 in 1970 and 0.7754
# * in 2000. https://www.nhm.ac.uk/our-science/data/biodiversity-indicators/biodiversity-intactness-index-data
# * In MAgPIE, the global BII value is about 0.8 in 1995 and declines to 0.795 by 2020.
# * Lower bound for BII in target year; plausible range: 0-1
cfg$gms$s44_bii_lower_bound <- 0 #def = 0
# * Indicative outcomes for `s44_bii_lower_bound` based on c44_bii_decrease = 1, s44_start_year = 2025 and s44_target_year = 2100.
# * < 0.7: continued decrease of global BII
# * 0.74: no further decrease of global BII
# * 0.76: moderate increase of global BII
# * >= 0.78: (very) strong increase of global BII accomplished by high conversion of pasture to non-forest natural land
# * BII lower bound implementation (binary)
# * options: 0 (no decrease allowed) or 1 (decrease allowed)
# * 0 (no decrease allowed): If the biome-level BII in `s44_start_year` exceeds the lower bound defined in `s44_bii_lower_bound, the BII value in the start year is used as lower bound for BII in all future time steps.
# * 1 (decrease allowed): The lower bound for BII can decrease over time, depending on the biome-level BII in the start year and the lower bound defined in `s44_bii_lower_bound.
# * Note: The BII constraint is defined as a minimum constraint. Therefore, the actual BII can be higher than the lower bound.
cfg$gms$c44_bii_decrease <- 1 #def = 1
# ** Options for realization `bv_btc_mar21`
# * Price for biodiversity stock loss/gain in target year (USD per ha)
# * Plausible range: 0-10000
# * Indicative outcomes
# * 0: decrease of BII
# * 3000: rather constant BII
# * 6000: moderate increase of BII
# * 10000: stronger increase of BII
cfg$gms$s44_target_price <- 0 #def = 0
# * Price for biodiversity stock loss/gain in start year (USD per ha)
cfg$gms$s44_start_price <- 0 #def = 0
# ***------------------------- 45_climate ---------------------------------
# * (static): static koeppengeiger climate classification data
cfg$gms$climate <- "static" # def = static
# ***------------------------- 50_nr_soil_budget --------------------------
# * (off): off
# * (macceff_aug22): nr efficiency dependent on MACC curves
cfg$gms$nr_soil_budget <- "macceff_aug22" # def = macceff_aug22
# * Scenario for baseline nr efficiency (excluding additional improvements based
# * on marginal abatmeent cost curves) on croplands for selected (and
# * respectively non-selected) countries in cropneff_countries and pastneff_countries
# *
# * Options:
# * "constant",
# * Scenarios keep baseline NUE constant.
# * "baseeff_add3_add10_add20_max75", (SSP1)
# * "baseeff_add3_add5_add10_max65", (SSP2)
# * "baseeff_add3_add0_add0_max55", (SSP3)
# * "baseeff_add3_add10_add15_max75", (SSP4)
# * "baseeff_add3_add5_add15_max75", (SSP5)
# * "baseeff_add3_add15_add25_max65", (lowTech)
# * "baseeff_add3_add15_add25_max75", (MaxMitigation)
# * In the baseeff_add scenarios, the baseline NUE starts from historical
# * values in 2010. The three numbers signify how many percentage points
# * the efficiency increases until 2020,2050 and 2100, always relative to
# * 2010. The max efficiency is the maximum base efficiency in 2050
# * and 2100. "baseeff_add3_add15_add25_max65" means that China with a
# * current NUE of 41 increases to 44% by 2010, 56% by 2050 and 65% by 2100,
# * while the USA, with a current NUE of 71% has a falling base NUE to 65%
# * by 2050 and 2100. These base NUEs can however be improved by the MACCs,
# * leading to a max NUE of 85% for both China and USA in 2100.
# * The recommmended mapping to storylines is shown above in brackets.
# * "maxeff_add3_glo75_glo85", "maxeff_add3_glo75_glo80",
# * "maxeff_add3_glo60_glo65", "maxeff_add3_glo65_glo75",
# * Scenarios with a globaly uniform target NUE. This target NUE is achieved
# * by the combination of full MACCs and baseline NUE. First number is the
# * again additive efficiency increase until 2020, second number is
# * max NUE in 2050, third in 2100. Example: maxeff_add3_glo75_glo85
# * means efficiency increases a regional specific 3 percentag point
# * improvement until 2020, and then it achieves a globally uniform 75%
# * efficiency in y2050; the baseline NUE value is therefore about 63%,
# * while the MACCs can increase this to 75%. In 2100, the baseline NUE
# * increases to 74%, which can achieve 85% if combined with
# * the MACCs.
# * "maxeff_ZhangBy2030", "maxeff_ZhangBy2050"
# * These two scenarios reach the max NUE of Zhang et al under application
# * of the full MACCs by 2030 or 2050, and then rise a bit further to
# * achieve a maximum NUE of 80% by 2100.
cfg$gms$c50_scen_neff <- "baseeff_add3_add5_add10_max65" # def = baseeff_add3_add5_add10_max65
cfg$gms$c50_scen_neff_noselect <- "baseeff_add3_add5_add10_max65" # def = baseeff_add3_add5_add10_max65
# * For pastures, only two scenarios exist: a constant scenario, and one where
# * the minimum NUE increases to 55,60 and 65% for 2020,2050 and 2100, to avoid
# * that pastures which do not have a nutrient deficit are fertilized
# * (historical NUE may be low despite no fertilization)
# * Options:
# * constant, constant_min55_min60_min65
cfg$gms$c50_scen_neff_pasture <- "constant_min55_min60_min65" # def = constant_min55_min60_min65
cfg$gms$c50_scen_neff_pasture_noselect <- "constant_min55_min60_min65" # def = constant_min55_min60_min65
# * Switch and specification of countries for which above defined (c50_scen_neff,
# * c50_scen_neff_pasture) apply, respectively.
# * For all other countries c50_scen_neff_noselect / c50_scen_neff_pasture_noselect
# * apply.
# * Options: list of iso-codes of countries where selected neff scenario should
# * be in effect.
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: above defined neff scenario applies to all iso countries
cfg$gms$cropneff_countries <- all_iso_countries
cfg$gms$pastneff_countries <- all_iso_countries
# ***--------------------- 51_nitrogen ------------------------------------
# * (rescaled_jan21): IPCC emissions factors rescaled with efficiency
# * (off): no nitrogen calculations
cfg$gms$nitrogen <- "rescaled_jan21" # def = rescaled_jan21
# ***--------------------- 52_carbon --------------------------------------
# * (normal_dec17): regrowth of carbon stocks in all c pools starts from pasture levels
# * (off): carbon calculations deactivated
cfg$gms$carbon <- "normal_dec17" # def = normal_dec17
# * RCP scenario of land carbon sink adjustment factor, from Grassi et al 2021
# * options: RCP19, RCP26, RCP34, RCP45, RCP60, RCPBU,
# * nocc (fixed to values from 1995),
# * nocc_hist (RCPBU until year defined in sm_fix_cc, fixed thereafter)
cfg$gms$c52_land_carbon_sink_rcp <- "RCPBU"
# * carbon scenario
# * options: cc (climate change)
# * nocc (no climate change)
# * nocc_hist (no climate change after year defined by sm_fix_cc)
cfg$gms$c52_carbon_scenario <- "cc" # def = "cc"
# * Minimum threshold of carbon density (tC/ha) in timber plantations
cfg$gms$s52_plantation_threshold <- 8 # def = 8
# ***--------------------- 53_methane -------------------------------------
# * (ipcc2006_aug22): IPCC 2006 methodology
# * (off): methane calculations deactivated
cfg$gms$methane <- "ipcc2006_aug22" # def = ipcc2006_aug22
# ***--------------------- 54_phosphorus ----------------------------------
# * (off): calculations deactivated
cfg$gms$phosphorus <- "off" # def = off
# ***--------------------- 55_awms ---------------------------------------
# * (ipcc2006_aug16): animal waste management systems
# * (based on IPCC 2006 Guidelines)
# * (off): deactivated animal waste management
cfg$gms$awms <- "ipcc2006_aug16" # def = ipcc2006_aug16
# * scenario for animal waste management.
# * option: "ssp1", "ssp2", "ssp3", "ssp4", "ssp5", "constant", "a1", "a2", "b1","b2",
#* "GoodPractice"
# * Note: c55_scen_conf applies to countries selected in scen_countries55
# * c55_scen_conf_noselect applies to all other countries.
# * Available scenarios for c55_scen_conf_noselect are identical to c55_scen_conf
cfg$gms$c55_scen_conf <- "ssp2" # def = ssp2
cfg$gms$c55_scen_conf_noselect <- "ssp2" # def = ssp2
# * Switch and specification of countries for which awm scenario in
# * c55_scen_conf applies.
# * Options: list of iso-codes of countries where awm scneario should be applied
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: all iso countries
cfg$gms$scen_countries55 <- all_iso_countries
# ***--------------------- 56_ghg_policy ----------------------------------
# * (price_aug22): applies pollutant prices to different emission types and calculates the reward for CDR from afforestation
cfg$gms$ghg_policy <- "price_aug22" # def = price_aug22
# * Switch for scaling GHG price with development state (1=on 0=off)
cfg$gms$s56_ghgprice_devstate_scaling <- 0 # def = 0
# * reduction factor for CO2 price (only used in price_jan19)
# * lowers the economic incentive for CO2 emission reduction (avoided deforestation) and afforestation
cfg$gms$s56_cprice_red_factor <- 1 # def = 1
# * GHG emission price scenario
# * Note: For best consistency it is recommended to use trajectories from the most recent
# * coupled REMIND-MAgPIE runs. Currently, this is R32M46.
# * Available options:
# * none: no 2nd generation bioenergy demand
# * R32M46: Coupled REMIND-MAgPIE runs with REMIND 3.2.1.dev412 and MAgPIE 4.6.11
# * NPi: Current policies; limit peak warming to 3.0°C
# * NDC: Nationally determined contributions; limit peak warming to 2.5°C
# * PkBudg650: Peak Budget with 650 GtCO2 until net-zero CO2 emissions; well-below 1.5°C in 2100
# * PkBudg1050: Peak Budget with 1050 GtCO2 until net-zero CO2 emissions; well-below 2.0°C in 2100
# * R32M46-SDP_MC-NPi, R32M46-SDP_MC-NDC, R32M46-SDP_MC-PkBudg650,
# * R32M46-SSP1-NPi, R32M46-SSP1-NDC, R32M46-SSP1-PkBudg1050, R32M46-SSP1-PkBudg650,
# * R32M46-SSP2EU-NPi, R32M46-SSP2EU-NDC, R32M46-SSP2EU-PkBudg1050, R32M46-SSP2EU-PkBudg650,
# * R32M46-SSP5-NPi, R32M46-SSP5-NDC, R32M46-SSP5-PkBudg1050, R32M46-SSP5-PkBudg650,
# * R21M42: Coupled REMIND-MAgPIE runs with REMIND 2.1 and MAgPIE 4.2
# * NPi: Current policies; above 3.0°C in 2100
# * PkBudg900: Budget with 900 GtCO2; well-below 1.5°C in 2100 (PkBudg1000 for SDP)
# * PkBudg1300: Budget with 1300 GtCO2; well-below 2.0°C in 2100
# * R21M42-SDP-NPi, R21M42-SDP-PkBudg1000, R21M42-SDP-PkBudg1100, R21M42-SDP-PkBudg900,
# * R21M42-SSP1-NPi, R21M42-SSP1-PkBudg1100, R21M42-SSP1-PkBudg1300, R21M42-SSP1-PkBudg900,
# * R21M42-SSP2-NPi, R21M42-SSP2-PkBudg1100, R21M42-SSP2-PkBudg1300, R21M42-SSP2-PkBudg900,
# * R21M42-SSP5-NPi, R21M42-SSP5-PkBudg1100, R21M42-SSP5-PkBudg1300, R21M42-SSP5-PkBudg900,
# * R2M41: Coupled REMIND-MAgPIE runs with REMIND 2.0 and MAgPIE 4.1
# * R2M41-SSP2-NPi, R2M41-SSP2-NDC, R2M41-SSP2-Budg1300, R2M41-SSP2-Budg600, R2M41-SSP2-Budg950
# * Standalone REMIND runs from Strefler et al 2021; well-below 2.0°C in 2100
# * https://www.nature.com/articles/s41467-021-22211-2
# * PIK_GDP, PIK_H2C, PIK_HBL, PIK_HOS, PIK_LIN, PIK_NPI, PIK_OPT
# * SSP Database 2018, various SSP, RCP and Model combinations
# * https://tntcat.iiasa.ac.at/SspDb
# * Caution: using trajectories from other models or older MAgPIE versions might be
# * inconsistent with the current MAgPIE version.
# * SSPDB-SSP1-20-IMAGE,SSPDB-SSP1-20-REMIND-MAGPIE,
# * SSPDB-SSP1-26-IMAGE,SSPDB-SSP1-26-REMIND-MAGPIE,
# * SSPDB-SSP1-34-IMAGE,SSPDB-SSP1-34-REMIND-MAGPIE,
# * SSPDB-SSP1-37-REMIND-MAGPIE,
# * SSPDB-SSP1-45-IMAGE,SSPDB-SSP1-45-REMIND-MAGPIE,
# * SSPDB-SSP1-Ref-IMAGE,SSPDB-SSP1-Ref-REMIND-MAGPIE,
# * SSPDB-SSP2-18-MESSAGE-GLOBIOM,SSPDB-SSP2-19-MESSAGE-GLOBIOM,
# * SSPDB-SSP2-20-MESSAGE-GLOBIOM,SSPDB-SSP2-20-REMIND-MAGPIE,
# * SSPDB-SSP2-26-MESSAGE-GLOBIOM,SSPDB-SSP2-26-REMIND-MAGPIE,
# * SSPDB-SSP2-34-MESSAGE-GLOBIOM,SSPDB-SSP2-34-REMIND-MAGPIE,
# * SSPDB-SSP2-37-REMIND-MAGPIE,
# * SSPDB-SSP2-45-MESSAGE-GLOBIOM,SSPDB-SSP2-45-REMIND-MAGPIE,
# * SSPDB-SSP2-60-MESSAGE-GLOBIOM,SSPDB-SSP2-60-REMIND-MAGPIE,
# * SSPDB-SSP2-Ref-MESSAGE-GLOBIOM,SSPDB-SSP2-Ref-REMIND-MAGPIE,
# * SSPDB-SSP3-34-AIM-CGE,SSPDB-SSP3-45-AIM-CGE,
# * SSPDB-SSP3-60-AIM-CGE,SSPDB-SSP4-26-GCAM4,
# * SSPDB-SSP4-34-GCAM4,SSPDB-SSP4-45-GCAM4,
# * SSPDB-SSP4-60-GCAM4,SSPDB-SSP4-Ref-GCAM4,
# * SSPDB-SSP5-20-REMIND-MAGPIE,SSPDB-SSP5-26-REMIND-MAGPIE,
# * SSPDB-SSP5-34-REMIND-MAGPIE,SSPDB-SSP5-37-REMIND-MAGPIE,
# * SSPDB-SSP5-45-REMIND-MAGPIE,SSPDB-SSP5-60-REMIND-MAGPIE,
# * SSPDB-SSP5-Ref-REMIND-MAGPIE,
# * Used for producing coupled runs with REMIND-MAgPIE or for exogenous input (see below)
# * coupling
# * Note: c56_pollutant_prices applies to countries selected in policy_countries56
# * c56_pollutant_prices_noselect applies to all other countries.
# * Available scenarios for c56_pollutant_prices_noselect are identical to c56_pollutant_prices
# * (see above) except for emulator and coupling (which can only be chosen for c56_pollutant_prices)
cfg$gms$c56_pollutant_prices <- "R32M46-SSP2EU-NPi" # def = R32M46-SSP2EU-NPi
cfg$gms$c56_pollutant_prices_noselect <- "R32M46-SSP2EU-NPi" # def = R32M46-SSP2EU-NPi
# * The following two settings can be used to provide exogenous ghg prices
# * via a file that is not part of the input data. This is currently used
# * in the REMIND-MAgPIE coupling to read in the REMIND data.
# * Takes effect only if cfg$gms$c56_pollutant_prices is set to "coupling"
# * Use ghg prices from the mif file specified here
cfg$path_to_report_ghgprices <- NA
# * Mute GHG prices until (and including) the year given here
# * Note: minimum C price (see below) will be set regardless of this setting
cfg$gms$c56_mute_ghgprices_until <- "y2030" # def = y2030
# * Minium C price (USD per tC) for future time steps until (and including) the year defined in `c56_mute_ghgprices_until`
# * Note: in case of NDC policy a C price of 18 USD per tC (5 USD per tCO2) is used to guide land-use decisions (set via scenario_config.csv)
cfg$gms$s56_minimum_cprice <- 0 # def = 0
# * Switch and specification of countries for which pollutant pricing in
# * c56_pollutant_prices applies.
# * Options: list of iso-codes of countries where ghg policy should be applied
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: all iso countries
# * Note: Only for exogenous scenarios. Does not take any effect when "coupling" /
# * "emulator" is selected above.
cfg$gms$policy_countries56 <- all_iso_countries # def = all_iso_countries
# * Switch for C price driven afforestation (1=on 0=off)
cfg$gms$s56_c_price_induced_aff <- 1 # def = on
# * C price used as incentive for afforestation
# * Note: This setting should only be changed by experienced users
# * Options: forestry_vegc, primforest_vegc, secdforest_vegc
# * This setting interacts with `c56_emis_policy`.
# * If "forestry" is not included in `c56_emis_policy` (e.g. reddnatveg_nosoil), there will be no C-price driven afforestation unless `c56_cprice_aff` is changed to primforest_vegc or secdforest_vegc.
cfg$gms$c56_cprice_aff <- "secdforest_vegc" # def = secdforest_vegc
# * C price expectation for afforestation decision-making in years
# * 0 is myopic behaviour (only C price of current time step)
# * reflects perfect-foresight if equal to s32_planing_horizon
# * should not be higher than s32_planing_horizon
cfg$gms$s56_c_price_exp_aff <- 50 # def = 50
# * Share of carbon credits for re/afforestation projects set aside in a buffer to
# * account for potential failure of permanent carbon storage.
# * Alternatively, this parameter can be interpreted as a reduction factor
# * of the carbon price for non-permanent CDR.
# * Values > 0 will reduce the incentive for c-price-induced re/afforestation
cfg$gms$s56_buffer_aff <- 0.5 # def = 0.5
# * Upper limit for CH4 and N2O GHG price (USD05MER per tC)
# * Limits GHG prices selected in c56_pollutant_prices to the chosen value.
# * CH4 and N2O GHG prices are limited by default to 4000 USD05MER per tC equivalent,
# * which induces the maximum abatement possible with c57_macc_version = "PBL_2022"
# * Beyond 4000 USD05MER per tC equivalent no further technical mitigation is possible
# * but would increase agricultural prices.
# * Suggested limits for different c57_macc_version settings
# * PBL_2007: 1000 (200 steps, 5 USD each)
# * PBL_2019: 4000 (200 steps, 20 USD each)
# * PBL_2022: 4000 (200 steps, 20 USD each)
cfg$gms$s56_limit_ch4_n2o_price <- 4000 # def = 4000
# * NOTE: The following 2 options for emission pricing have strong interactions in runs with CO2 prices and should only be changed by experienced users.
# * The main purpose of these options is to regulate the pricing of positive emissions. Note that re/afforestation is covered by a separate mechanism.
# * GHG emission pricing policies
# * options:
# * none (not including any GHG sources),
# * redd_nosoil (Above ground CO2 emis from LUC in forest; all CH4 and N2O emissions),
# * redd_nosoil_nopeat (same as redd_nosoil but without GHG emissions from peatlands),
# * reddnatveg_nosoil (Above ground CO2 emis from LUC in forest and natveg; all CH4 and N2O emissions),
# * reddnatveg_nosoil_nopeat (same as reddnatveg_nosoil but without GHG emissions from peatlands),
# * redd+_nosoil (Above ground CO2 emis from LUC in forest and forestry; all CH4 and N2O emissions),
# * redd+_nosoil_nopeat (same as redd+_nosoil but without GHG emissions from peatlands),
# * redd+natveg_nosoil (Above ground CO2 emis from LUC in forest, forestry and natveg; all CH4 and N2O emissions),
# * redd+natveg_nosoil_nopeat (same as redd+natveg_nosoil but without GHG emissions from peatlands),
# * all (CO2 emis from LUC in all LUs; all CH4 and N2O emissions), all_nosoil (Above ground CO2 emis from LUC for all LUs; all CH4 and N2O emissions)
# * all_nosoil: Above ground CO2 emis from LUC for all LUs; all CH4 and N2O emissions
# * sdp_cropeff (only ch4 from rice), sdp_livestock (only ch4 and n2o from livestock), sdp_peatland (co2, ch4 and n2o from peatland),
# * sdp_redd (only co2 from vegc), sdp_soil (only co2 from litc and soilc), sdp_redd_soil (only co2 from vegc, litc, soilc),
# * sdp_redd_soil_peat (only co2 from vegc, litc, soilc and co2, ch4 and n2o from peatland), sdp_all (all individual sdp scenarios combined)
# * gcs_lbs (land-based mitigation: above-ground CO2 emis from LUC in forest, forestry and natveg; co2, ch4 and n2o from peatland),
# * gcs_res (mitigation in production systems: all agricultural non-CO2 emissions)
# * ecoSysProtAll: (Above ground CO2 emis from LUC in forest, forestry, natveg; All types of emis from peatland; All CH4 and N2O emis),
# * ecoSysProtForest: (Above ground CO2 emis from LUC in forest, forestry; All CH4 and N2O emis except peatland),
# * ecoSysProtPrimForest: (Above ground CO2 emis from LUC in primary forest; All CH4 and N2O emis except peatland),
# * ecoSysProtOff: (All CH4 and N2O emis except peatland),
# * ecoSysProtAll_agMgmtExclN2O: (Above ground CO2 emis from LUC in forest, forestry, natveg; All types of emis from peatland; All CH4 emis, no further N2O emis)
# * ecoSysProtAll_agMgmtExclCH4: (Above ground CO2 emis from LUC in forest, forestry, natveg; All types of emis from peatland; All N2O emis, no further CH4 emis)
# * ecoSysProtAll_agMgmtOff: (Above ground CO2 emis from LUC in forest, forestry, natveg; All types of emis from peatland; No further CH4/N2O/other emis related to ag. management)
cfg$gms$c56_emis_policy <- "reddnatveg_nosoil" # def = reddnatveg_nosoil
# * CO2 emissions subject to carbon pricing
# * options: actual, actualNoAcEst
# * actual: CO2 emissions for pricing are based on the difference of actual carbon stocks between time steps
# * actualNoAcEst: CO2 emissions for pricing are based on actual carbon stocks but
# * without newly established forest and non-forest areas. Note that re/afforestation is covered by a separate mechanism.
c56_carbon_stock_pricing <- "actualNoAcEst" # def = actualNoAcEst
# ***--------------------- 57_maccs ---------------------------------------
# * (on_aug22): maccs for non-CO2 emission mitigation activated
cfg$gms$maccs <- "on_aug22" # def = on_aug22
# * Version of MACCs
# * options: PBL_2007, PBL_2019, PBL_2022
cfg$gms$c57_macc_version <- "PBL_2022" # def = PBL_2022
# * MACC scenario (only for PBL_2022)
# * options: Default, Optimistic, Pessimistic
cfg$gms$c57_macc_scenario <- "Default" # def = Default
# * Exogenous forced mitigation, independent of pollutant prices
# * Overwrites signal of mitigation price by choosing maximum mitigation levels
# * This avoids that emission tax is rolled over to food prices, only the
# * the mitigaton costs are rolled over to prices.
# * options: -1, 1 to 201 for each MACC mitigation step
# * -1: pollutant-price driven maccs
# * 0: inactive MACCs (price level zero)
# * 201: maximum level of mitigation
cfg$gms$s57_maxmac_n_soil <- -1 # def = -1
cfg$gms$s57_maxmac_n_awms <- -1 # def = -1
cfg$gms$s57_maxmac_ch4_rice <- -1 # def = -1
cfg$gms$s57_maxmac_ch4_entferm <- -1 # def = -1
cfg$gms$s57_maxmac_ch4_awms <- -1 # def = -1
# ***--------------------- 58_peatland ------------------------------------
# * (off): Peatland area and associated GHG emissions are assumed zero
# * (v2): Peatland area is initialized with degraded and intact peatland area for the year 2022,
# * and projected into the future based on changes in managed land.
# * For boreal and tropical climates, GHG emissions are calculated using emission
# * factors from the IPCC 2013 Wetland supplement. For temperate climates, more recent
# * estimates from Tiemeyer et al 2020 are used.
cfg$gms$peatland <- "v2" # def = v2
# * peatland rewetting
# * options: 0 (off)
# * Inf (on)
cfg$gms$s58_rewetting_switch <- Inf # def = Inf
# * One-time and recurring costs for peatland rewetting (USD05MER per ha)
cfg$gms$s58_cost_rewet_onetime <- 7000 # def = 7000
cfg$gms$s58_cost_rewet_recur <- 200 # def = 200
# * One-time and recurring costs for peatland drainage (USD05MER per ha)
# * One-time costs apply on the drainage of intact and rewetted peatland
# * Recurring costs apply on the level of drained and managed peatland
cfg$gms$s58_cost_drain_intact_onetime <- 0 # def = 0
cfg$gms$s58_cost_drain_rewet_onetime <- 0 # def = 0
cfg$gms$s58_cost_drain_recur <- 0 # def = 0
# * Switch for fixing peatland area until the year given by s58_fix_peatland to historic levels (not available in `off`).
# * Note: In the realisation `v2`, historic peatland area is only available for the year 2022.
# * Due to the 5-year time step limitation in MAgPIE, the suggested default is 2020.
# * Fixing peatland area in previous time steps provides a better
# * proxy for GHG emissions from peatlands than assuming no peatland area.
cfg$gms$s58_fix_peatland <- 2020 # def = 2020
# ***------------------------- 59_som -------------------------------------
# * (static_jan19): static soil carbon loss for cropland
# * (cellpool_aug16): dynamic soil organic matter pool on cellular level
# * (cellpool_jan23): dynamic soil organic matter pool on cellular level
# * with updated, regionalized stock change factors (IPCC guidelines 2019)
cfg$gms$som <- "static_jan19" # def = static_jan19
# * static realization switch
# * options: cellular (use preprocessed cellular stock change factors)
# * cluster (use cshare_released on cluster within gams)
cfg$gms$c59_static_spatial_level <- "cellular"
# * cellpool realization switches
# * som climate impact scenario
# * options: cc (climate change)
# * nocc (no climate change)
# * nocc_hist (no climate change after year defined by sm_fix_cc)
cfg$gms$c59_som_scenario <- "cc" # def = "cc"
# * irrigation feedback
# * options: on (higher carbon sequestration under irrigation)
# * off (no carbon sequestration under irrigation)
cfg$gms$c59_irrigation_scenario <- "on" # def = "on"
# * Exogenous nr release through som loss (only in static realization)
# * options: constant (constant from 2020)
# * fadeout_2050 (fading out till 2050)
cfg$gms$c59_exo_scen <- "constant"
# ***--------------------- 60_bioenergy -----------------------------------
# * (1stgen_priced_dec18): exogenous and price-based 1st generation bioenergy
# * demand, 2nd generation residues exogeneous,
# * 2nd generation betr and begr coupled with REMIND.
cfg$gms$bioenergy <- "1stgen_priced_dec18" # def = 1stgen_priced_dec18
# * 1st generation bioenergy demand scenarios based on Lotze Campen (2014)
# * (phaseout2020): increase until 2020, followed by phaseout until 2050
# * (const2020): increase until 2020, constant thereafter
# * (const2030): increase until 2030, constant thereafter
cfg$gms$c60_1stgen_biodem <- "const2020" # def = const2020
# * 2nd generation bioenergy demand scenario
# * Note: For best consistency it is recommended to use trajectories from the most recent
# * coupled REMIND-MAgPIE runs. Currently, this is R32M46.
# * Available options:
# * none: no 2nd generation bioenergy demand
# * R32M46: Coupled REMIND-MAgPIE runs with REMIND 3.2.1.dev412 and MAgPIE 4.6.11
# * NPi: Current policies; limit peak warming to 3.0°C
# * NDC: Nationally determined contributions; limit peak warming to 2.5°C
# * PkBudg650: Peak Budget with 650 GtCO2 until net-zero CO2 emissions; well-below 1.5°C in 2100
# * PkBudg1050: Peak Budget with 1050 GtCO2 until net-zero CO2 emissions; well-below 2.0°C in 2100
# * R32M46-SDP_MC-NPi, R32M46-SDP_MC-NDC, R32M46-SDP_MC-PkBudg650,
# * R32M46-SSP1-NPi, R32M46-SSP1-NDC, R32M46-SSP1-PkBudg1050, R32M46-SSP1-PkBudg650,
# * R32M46-SSP2EU-NPi, R32M46-SSP2EU-NDC, R32M46-SSP2EU-PkBudg1050, R32M46-SSP2EU-PkBudg650,
# * R32M46-SSP5-NPi, R32M46-SSP5-NDC, R32M46-SSP5-PkBudg1050, R32M46-SSP5-PkBudg650,
# * R21M42: Coupled REMIND-MAgPIE runs with REMIND 2.1 and MAgPIE 4.2
# * NPi: Current policies; above 3.0°C in 2100
# * PkBudg900: Budget with 900 GtCO2; well-below 1.5°C in 2100 (PkBudg1000 for SDP)
# * PkBudg1300: Budget with 1300 GtCO2; well-below 2.0°C in 2100
# * R21M42-SDP-NPi, R21M42-SDP-PkBudg1000, R21M42-SDP-PkBudg1100, R21M42-SDP-PkBudg900,
# * R21M42-SSP1-NPi, R21M42-SSP1-PkBudg1100, R21M42-SSP1-PkBudg1300, R21M42-SSP1-PkBudg900,
# * R21M42-SSP2-NPi, R21M42-SSP2-PkBudg1100, R21M42-SSP2-PkBudg1300, R21M42-SSP2-PkBudg900,
# * R21M42-SSP5-NPi, R21M42-SSP5-PkBudg1100, R21M42-SSP5-PkBudg1300, R21M42-SSP5-PkBudg900,
# * R2M41: Coupled REMIND-MAgPIE runs with REMIND 2.0 and MAgPIE 4.1
# * R2M41-SSP2-NPi, R2M41-SSP2-NDC, R2M41-SSP2-Budg1300, R2M41-SSP2-Budg600, R2M41-SSP2-Budg950
# * Standalone REMIND runs from Strefler et al 2021; well-below 2.0°C in 2100
# * https://www.nature.com/articles/s41467-021-22211-2
# * PIK_GDP, PIK_H2C, PIK_HBL, PIK_HOS, PIK_LIN, PIK_NPI, PIK_OPT
# * SSP Database 2018, various SSP, RCP and Model combinations
# * https://tntcat.iiasa.ac.at/SspDb
# * Caution: using trajectories from other models or older MAgPIE versions might be
# * inconsistent with the current MAgPIE version.
# * SSPDB-SSP1-20-IMAGE,SSPDB-SSP1-20-REMIND-MAGPIE,
# * SSPDB-SSP1-26-IMAGE,SSPDB-SSP1-26-REMIND-MAGPIE,
# * SSPDB-SSP1-34-IMAGE,SSPDB-SSP1-34-REMIND-MAGPIE,
# * SSPDB-SSP1-37-REMIND-MAGPIE,
# * SSPDB-SSP1-45-IMAGE,SSPDB-SSP1-45-REMIND-MAGPIE,
# * SSPDB-SSP1-Ref-IMAGE,SSPDB-SSP1-Ref-REMIND-MAGPIE,
# * SSPDB-SSP2-18-MESSAGE-GLOBIOM,SSPDB-SSP2-19-MESSAGE-GLOBIOM,
# * SSPDB-SSP2-20-MESSAGE-GLOBIOM,SSPDB-SSP2-20-REMIND-MAGPIE,
# * SSPDB-SSP2-26-MESSAGE-GLOBIOM,SSPDB-SSP2-26-REMIND-MAGPIE,
# * SSPDB-SSP2-34-MESSAGE-GLOBIOM,SSPDB-SSP2-34-REMIND-MAGPIE,
# * SSPDB-SSP2-37-REMIND-MAGPIE,
# * SSPDB-SSP2-45-MESSAGE-GLOBIOM,SSPDB-SSP2-45-REMIND-MAGPIE,
# * SSPDB-SSP2-60-MESSAGE-GLOBIOM,SSPDB-SSP2-60-REMIND-MAGPIE,
# * SSPDB-SSP2-Ref-MESSAGE-GLOBIOM,SSPDB-SSP2-Ref-REMIND-MAGPIE,
# * SSPDB-SSP3-34-AIM-CGE,SSPDB-SSP3-45-AIM-CGE,
# * SSPDB-SSP3-60-AIM-CGE,SSPDB-SSP4-26-GCAM4,
# * SSPDB-SSP4-34-GCAM4,SSPDB-SSP4-45-GCAM4,
# * SSPDB-SSP4-60-GCAM4,SSPDB-SSP4-Ref-GCAM4,
# * SSPDB-SSP5-20-REMIND-MAGPIE,SSPDB-SSP5-26-REMIND-MAGPIE,
# * SSPDB-SSP5-34-REMIND-MAGPIE,SSPDB-SSP5-37-REMIND-MAGPIE,
# * SSPDB-SSP5-45-REMIND-MAGPIE,SSPDB-SSP5-60-REMIND-MAGPIE,
# * SSPDB-SSP5-Ref-REMIND-MAGPIE,
# * Used for producing coupled runs with REMIND-MAgPIE or for exogenous input (see below)
# * coupling
# * Note: c60_2ndgen_biodem applies to countries selected in scen_countries60
# * c60_2ndgen_biodem_noselect applies to all other countries.
cfg$gms$c60_2ndgen_biodem <- "R32M46-SSP2EU-NPi" # def = R32M46-SSP2EU-NPi
cfg$gms$c60_2ndgen_biodem_noselect <- "R32M46-SSP2EU-NPi" # def = R32M46-SSP2EU-NPi
# * The following setting can be used to provide exogenous bioenergy demand
# * via a .mif file that is not part of the input data. This is currently used
# * in the REMIND-MAgPIE coupling to read in the REMIND data.
# * Takes effect only if cfg$gms$c60_2ndgen_biodem is set to "coupling"
cfg$path_to_report_bioenergy <- NA
# * Switch and specification of countries for which 2nd gen bioenergy demand
# * scenario applies.
# * Options: list of iso-codes of countries where scenario should be applied
# * Note: must be written in the format: "IND, BRA, DEU"
# * Default: all iso countries
# * Note: Only for exogenous scenarios. Does not take any effect when "coupling" /
# * "emulator" selected.
cfg$gms$scen_countries60 <- all_iso_countries
# * residue demand for 2nd generation bioenergy scenarios
# * options: ssp1, ssp2, ssp3, ssp4, ssp5, off
cfg$gms$c60_res_2ndgenBE_dem <- "ssp2" # def = ssp2
# * bioenergy demand level
# * (1): regional
# * (0): global
cfg$gms$c60_biodem_level <- 1 # def = 1
# * Minimum dedicated 2nd generation bioenergy demand assumed in each region (mio. GJ per yr)
# * Without a minimum demand, there is the risk that some regions won't have a price for 2nd generation bioenergy.
# * Therefore, the minimum demand is of particular importance for the coupling with REMIND.
cfg$gms$s60_2ndgen_bioenergy_dem_min <- 1 # def = 1
# * first generation bioenergy subsidy (USD05MER per ton)
cfg$gms$c60_bioenergy_subsidy <- 300 # def = 300
# ***--------------------- 62_material ------------------------------------
# * (exo_flexreg_apr16): default
cfg$gms$material <- "exo_flexreg_apr16"
# * Biomass demand for bioplastics is based on a logistic curve projecting
# * bioplastic demand, which matches the historic demand in 2020, and is
# * defined by the maximum demand for bioplastics s62_max_dem_bioplastics
# * (in mio. tonnes) and the midpoint s62_midpoint_dem_bioplastics (i.e.
# * where bioplastic demand is half of the maximum demand).
# * If maximum demand is 0, biomass demand for bioplastic production is not included.
# * Projected total plastic use in the "Global Ambiton" scneario of the OECD is
# * about 600 mio. tonnes in 2050 -> E.g. 200 mio. tonnes in 2050 could be a
# * reasonable (but ambitious) bioplastic scenario. With midpoint 2050 this
# * would mean s62_max_dem_bioplastic = 400.
cfg$gms$s62_max_dem_bioplastic <- 0 # def = 0
cfg$gms$s62_midpoint_dem_bioplastic <- 2050 # def = 2050
# ***--------------------- 70_livestock -----------------------------------
# * (fbask_jan16): default feed basket realization
cfg$gms$livestock <- "fbask_jan16" # def = fbask_jan16
# * Year until the pasture management factor is fixed to 1
cfg$gms$s70_past_mngmnt_factor_fix <- "2005" # def = 2005
# * feed scenario
# * options: SSP: ssp1, ssp2, ssp3, ssp4, ssp5
# * SDP: SDP, SDP_EI, SDP_MC, SDP_RC
# * other: constant
cfg$gms$c70_feed_scen <- "ssp2" # def = ssp2
# * Feed substitution scenarios.
# * options consist of 3 parts: functional form (lin,sigmoid), target (zero, 20pc, 50pc, 80pc, 90pc) and transition period (10_50: from 2010 to 2050, 20_50: from 2020 to 2050)
# * Example for sigmoid_50pc_20_50:
# * Functional form: sigmoid (S-shaped)
# * Target: 50percent reduction at end of transition period (2050 in this case)
# * Transition period: start in 2020, end in 2050
# * options: constant,
# * lin_zero_10_50, lin_zero_20_50, lin_zero_20_30, lin_zero_20_70, lin_50pc_20_50, lin_50pc_20_50_extend65, lin_50pc_20_50_extend80,
# * lin_50pc_10_50_extend90, lin_75pc_10_50_extend90, lin_80pc_20_50, lin_80pc_20_50_extend95, lin_90pc_20_50_extend95,
# * lin_99-98-90pc_20_50-60-100, sigmoid_20pc_20_50, sigmoid_50pc_20_50, sigmoid_80pc_20_50
# * Cereal feed substituted by SCP
cfg$gms$c70_cereal_scp_scen <- "constant" # def = constant
# * Fodder feed substituted by SCP
cfg$gms$c70_foddr_scp_scen <- "constant" # def = constant
# * Regional (reg) or global (glo) regression for livestock factor requirements.
# * Typically, the same version (glo or reg) should be chosen for c38_fac_req
# * for consistency.
cfg$gms$c70_fac_req_regr <- "glo" # def = glo
# ***--------------------- 71_disagg_lvst -----------------------------------
# * (off): default
# * (foragebased_aug18): Disaggregation of livestock to cells
# * with high pasture and fodder availability
# * (foragebased_jul23): Disaggregation of livestock to cells
# * with high pasture and fodder availability
# * (improved and flexibitized)
cfg$gms$disagg_lvst <- "foragebased_jul23" # def = foragebased_jul23
# ***--------------------- 73_timber -----------------------------------
# * (default): Demand and production of timber products
cfg$gms$timber <- "default" # def = default
# Logical switch to turn on or off timber demand (wood and woodfuel).
# Note that "on" requires dynamic timber plantations (s32_hvarea=2 and s35_hvarea=2)
# for actual results.
# Vice-versa, in the "off" case, timber plantations should be static (s32_hvarea=0/1 and s35_hvarea=0/1)
# for a simplified representation of land demand for timber production from plantations
# (implicitly assuming the same rate for plantation harvest and establishment).
# * 1=on
# * 0=off
cfg$gms$s73_timber_demand_switch <- 0 # def = 0
# Setting to define if the model should be forward looking or not in terms of
# seeing the future demand for current timestep establishment of new plantations
# in the forward setting, the model sees only current demand for establishment
# decisions in the historical time period but then sees future demand for non
# historic time periods. In myopic setting, the model sees always the current
# timber demand for establishment decisions.
# * 1 = forward looking. Model sees future demand for establishment in current step
# * 0 = myopic. Model sees current demand for establishment in current step
cfg$gms$s73_foresight <- 0 # def = 0
# harvesting cost per ton of dry matter produced (USD/tDM)
s73_timber_prod_cost <- 2000 # def = 2000
# harvesting cost per ha of forests (USD/ha)
s73_timber_harvest_cost <- 2000 # def = 2000
# Cost multiplier for harvesting costs to make natural vegetation harvest expensive
# than timber plantation harvest. This provides a signal to the model to harvest
# timber plantations first.
s73_cost_multiplier <- 1.5 # def = 1.5
# Cost of production without using any land in case the model is running into infeasibilities.
# This is a last ditch effort for the model and the variable associated with this cost
# should not be used in a normally feasible model run (USD/tDM)
s73_free_prod_cost <- 1000000 # def = 1000000
# Switch for modifying woody biomass demand starting in 2035
# * ("default") = Default paper demand
# * ("nopaper") = Diminishing paper demand
# * ("construction") = Higher demand for construction wood in future
cfg$gms$c73_wood_scen <- "default" # def = "default"
# Building material demand
# * ("BAU") = Business as usual, only 0.5% of urban dwellers need timber buildings
# * ("10pc") = 10% of urban dwellers need timber buildings
# * ("50pc") = 50% of urban dwellers need timber buildings
# * ("90pc") = 90% of urban dwellers need timber buildings
cfg$gms$c73_build_demand <- "BAU" # def = "BAU"
# Linearized multiplier for scaling construction wood demand for easier demand
# scenario tests. The expansion factor is applied over next 80 years starting in
# 2020.
cfg$gms$s73_expansion <- 0 # def = 0
# ***------------------- 80_optimization ------------------------------------
# * (nlp_apr17): solve procedure solving the whole, nonlinear problem
# * at once
# * (lp_nlp_apr17): alternative approach in which the nonlinear terms are
# * fixed first so that the linear problem can be solved
# * before the full problem is solved
# * (nlp_par): Parallel optimization of regions. Allows to use higher
# * spatial resolution but works only with exogenous trade patterns
# * from a run with lower resolution solved with nlp_apr17.
# * Usage: add "extra/highres" as output script. This will start a
# * second model run with higher resolution.
# * Make sure that the cellular input file specified in
# * scripts/output/extra/highres.R exists.
# * See scripts/output/extra/highres.R for details.
cfg$gms$optimization <- "nlp_apr17" # def = nlp_apr17
# maximal number of solve iterations
cfg$gms$s80_maxiter <- 30
# * (conopt3): conopt3
# * (conopt4): conopt4
# * (conopt4+cplex): conopt4 followed by cplex with landdiff optimization
# * (conopt4+conopt3): conopt4 followed by conopt3
cfg$gms$c80_nlp_solver <- "conopt4" # def = conopt4
# * 1: using optfile for specified solver settings
# * 0: default settings (optfile will be ignored)
cfg$gms$s80_optfile <- 1
#*******************************END MODULE SETUP********************************
#### Other settings (e.g. clustering, gdx files, ...): ####
# Choosing scenario for coupling with MAGICC for emissions outside the food system
# (options) bjoernAR6_C_RemSDP-900-MagSSP1.mif, bjoernAR6_C_RemSSP1-1000-MagSDP.mif,
# bjoernAR6_C_SDP-NPi.mif, bjoernAR6_C_SDP-PkBudg1000.mif,
# bjoernAR6_C_SSP1-Base.mif, bjoernAR6_C_SSP1-NDC.mif,
# bjoernAR6_C_SSP1-NPi.mif, bjoernAR6_C_SSP1-PkBudg900.mif,
# bjoernAR6_C_SSP2-Base.mif, bjoernAR6_C_SSP2-NDC.mif,
# bjoernAR6_C_SSP2-NPi.mif, bjoernAR6_C_SSP2-PkBudg900.mif
cfg$magicc_emis_scen <- "bjoernAR6_C_SSP2-NDC.mif"
# Decide whether the runs should be run sequentially (TRUE),
# or in parallel (FALSE)
# NA means that this decision is taken automatically
# (typically on cluster = FALSE and locally = TRUE)
cfg$sequential <- NA
# Selection of QOS to be used for submitted runs on cluster.
# Will be ignored for all other runs.
# * options: short (24h max, no preemption)
# * short_maxMem (same as short but with 16 CPUs and max Memory)
# * medium (1 week max, no preemption)
# * priority (immediate start, but slots limited to 5 in parallel)
# * priority_maxMem (same as priority but with 16 CPUs and max Memory)
# * standby (1 week max, preemption possible)
# * standby_dayMax (24h max, preemption possible)
# * standby_maxMem_dayMax (same as standby_dayMax but with 16 CPUs and max Memory)
# * standby_maxMem (same as standby but with 16 CPUs and max Memory)
# * NULL (educated guess of best option based
# * available resources)
cfg$qos <- NULL # def = NULL
# How should log information be treated?
# (0:no output, 2:write to full.log 3:show in console)
cfg$logoption <- 2
# Should output.R generate output?
# List of output scripts that should be used
# Available scripts can be found in scripts/output/
cfg$output <- c("output_check", "extra/disaggregation", "rds_report")
# Set the format for the results folder
# :date: is a placeholder for the current time stamp (e.g. "results:date:")
# :title: is a placeholder for the current run title (cfg$title)
cfg$results_folder <- "output/:title::date:"
# Results folder used by the "highres.R" output script
# If NULL, the default set in "highres.R" is used (currently a subfolder in "output")
# To place highres runs directly in the "output" folder, change to "output"
# To place highres runs in a specific subfolder change to "output/HR"
cfg$results_folder_highres <- NULL
# Which files should be copied into the output folder?
cfg$files2export <- list()
# Files that should be copied before MAgPIE is started
cfg$files2export$start <- c("input/info.txt",
"input/avl_land_full_t_0.5.mz",
"input/luh2_side_layers_0.5.mz",
"modules/14_yields/input/lpj_yields_0.5.mz",
"modules/22_land_conservation/input/wdpa_baseline_0.5.mz",
"modules/22_land_conservation/input/consv_prio_areas_0.5.mz",
"modules/30_crop/endo_apr21/input/avl_cropland_0.5.mz",
"modules/50_nr_soil_budget/input/f50_NitrogenFixationRateNatural_0.5.mz",
"modules/50_nr_soil_budget/input/f50_AtmosphericDepositionRates_0.5.mz",
"input/f34_urbanland_0.5.mz",
"input/spatial_header.rda",
"scripts/run_submit/submit.R",
"scripts/run_submit/submit_*.sh",
".Rprofile",
"input/clustermap*.rds",
"input/lpj_envflow_total_*.mz",
"input/lpj_watavail_total_*.mz",
"input/validation.mif",
"calib_*.cs3",
"land_conversion_cost_calib_*.cs3",
"input/spamplot_*.pdf",
"input/regionmapping*.csv",
"input/kba_land_0.5.mz",
"input/f58_peatland_area_0.5.mz",
"input/MAPSPAM_croparea_0.5.mz",
"input/LUH2_croparea_0.5.mz")
# Files that should be copied after the MAgPIE run is finished
cfg$files2export$end <- NULL
# Expert use only, should not be set manually. Path to a renv.lock file
# defining the package environment for newly started runs.
cfg$renv_lock <- NULL
# Folder run statistics should be submitted to
cfg$runstatistics <- "/p/projects/rd3mod/models/statistics/magpie"
# name of the overall model (just used for reporting purposes)
# should usually not be changed
cfg$model_name <- "MAgPIE"
# a list of additional information characterizing this run. Can be used
# to save relevant information about the run and can, in contrast to all other
# setting, contain list elements which do not exist in the reference
# configuration
cfg$info <- list()
# Should the restart files of each iteration be kept in the output folder (TRUE)
# or deleted after the run finished (FALSE)
cfg$keep_restarts <- FALSE
# Should the model run in developer mode? This will loosen some restrictions,
# such as temporary toleration of coding etiquette violations
# Please make sure to set it to FALSE for production runs!
cfg$developer_mode <- FALSE
# Should the model run in debug mode?
# Download script will copy files from input to destination folder instead of
# moving it allowing to check whether something in the move/copy process goes
# wrong
cfg$debug <- FALSE
################################################################################Start & Output scripts
Rscript start.R
Main selection of MAgPIE start scripts
-----------------------------------------------------------------
-> Scripts in this selection are actively managed and work out <-
-> of the box <-
-----------------------------------------------------------------
1: default | start run with default.cfg settings
2: check code | Checking code for consistency issues
3: download data | just download default.cfg input data
4: test runs | Test routine for standardized test runs
5: forestry | start run with Forestry (Endogenous)
6: compilation check | download input and compile main.gms
Alternatively, choose a start script from another selection:
7: extra | Additional MAgPIE start scripts
8: projects | Project-specific MAgPIE start scripts
9: deprecated | Deprecated scripts
# ------------------------------------------------
# description: start run with default.cfg settings
# position: 1
# ------------------------------------------------
# Load start_run(cfg) function which is needed
# to start MAgPIE runs
source("scripts/start_functions.R")
#start MAgPIE run
start_run(cfg="default.cfg")
In-code documentation
powered by goxygen » github.com/pik-piam/goxygen
[...]
*' @equations
*' ![Investment-yield ratio in relation to $\tau$-factor
*' [@dietrich_forecasting_2014]](tcc_regression.png){ width=60% }
*'
*' Relative technological change costs `v13_cost_tc` are calculated as a
*' heuristically derived power function of the land use intensity `vm_tau` for
*' the investment-yield-ratio (see figure above) multiplied by the current
*' regional crop areas `pc13_land` (taken from previous time step) and shifted
*' 15 years into the future using the region specific interest
*' rate `pm_interest`:
q13_cost_tc(i2, tautype) ..
v13_cost_tc(i2, tautype) =e= sum(ct, pc13_land(i2, tautype) *
i13_tc_factor(ct) * sum(supreg(h2,i2),vm_tau(h2,tautype))**
i13_tc_exponent(ct) * (1+pm_interest(ct,i2))**15);
*' The shifting is performed because investments into technological change
*' require on average 15 years of research before a yield increase is achieved,
*' but the model has to see costs and benefits concurrently in order to take the
*' right investment decisions (see also @dietrich_forecasting_2014). Investment
*' costs are scaled in relation to crop area, since a wider areal coverage means
*' typically also higher variety in biophysical conditions, which would require
*' more research for the same overall intensity boost.
*'
*' In order to get the full investments required for the desired intensification
*' the relative technological change costs are multiplied with the given
*' intensification rate. These full costs are then distributed over an infinite
*' time horizon by multiplication with the interest rate `pm_interest(i)`
*' (annuity with infinite time horizon):
q13_tech_cost(i2, tautype) ..
v13_tech_cost(i2, tautype) =e= sum(supreg(h2,i2), vm_tau(h2,tautype)/pcm_tau(h2,tautype)-1)
* v13_cost_tc(i2,tautype) * sum(ct,pm_interest(ct,i2)/(1+pm_interest(ct,i2)));
q13_tech_cost_sum(i2) ..
vm_tech_cost(i2) =e= sum(tautype, v13_tech_cost(i2, tautype));
[...]Applications




Evolution of the global forest plantation area
between 1995 and 2100 in an SSP2 world.






New features in 4.7

What's new?
Structurally, not much...
Look and feel quite comparable to 4.0
If you know how to use any 4.x version you (mostly) know how to use any other 4.x version
two notable exceptions:
renv integration (4.6.2|4.6.8)
MAgPIE runs in an isolated
R package environment
superregional layer (4.3.5)
it is now possible to have a
second regional layer
What's new?
# Changelog
All notable changes to this project will be documented in this file.
The format is based on [Keep a Changelog](https://keepachangelog.com/en/1.0.0/).
## [4.7.2] - 2024-04-02
### changed
- **21_trade** Revision of trade module. Replaced `cfg$gms$s21_trade_bal_damper` in favour of `cfg$gms$k_import21`, which allows for additional imports to maintain feasibility
- **58_peatland** Threshold in equations changed from 1e-10 to 1e-8 to avoid rare divisions by zero
- **70_livestock** if `c70_fac_req_regr` is set to `reg`: use of USDA/FAO values for historic factor requirements for livestock instead of using regression values and change of calibration year from 2005 to 2010 for regional factor requirements regression
- **config** updated FSEC scenario config for revision and included new calibration file (after cost fix in preprocessing)
- **scripts** modified output reporting for SEALS to account for forestry plantations
### added
- **30_crop** Improved representation of cropland requiring relocation in response to introducing semi-natural habitat at the 1 km level based on high-resolution satellite imagery.
- **config** added `.codeCheck` with additonal configuration when running `gms::codeCheck`
- **scripts** add additional BII reporting variables in FSDP_collect.R
- **scripts** added a new validation_cell.R output script that generates a pdf with the comparison of magpie land use and crop type outputs with LUH and MAPSPAM historical data at cellular resolution.
### removed
- **core** removed no longer needed set `si` Suitability classes
### fixed
- **52_carbon** i52_land_carbon_sink was not identical before 2020 for different RCPs. Fixed by setting to RCPBU until the year defined in sm_fix_cc.
- **inputdata** currency fixed in historic value of production for crops and livestock which affects e.g. total labor costs and in turn hourly labor costs, bugfix in aggregation weight of capital cost share out of factor costs
## [4.7.1] - 2024-02-28
### changed
- **15_food** Added improved EAT Lancet diet implementation (EAT 2p0)
- **21_trade** s21_trade_bal_damper for roundwood changed from 0.75 to 0.65
- **31_past** in grasslands_apr22 realization: changed structure of f31_pastr_suitability to align with ssp-rcp specific input data formulation. Changed input filename from cs3 to cs2. Added `cc`, `nocc` and `nocc_hist` options for `i31_manpast_suit` and changed input gams code from table to parameter. Climate scenario assignment moved from preloop.gms to input.gms. Removed pastSuit set in sets.gms as not needed anymore. Adjusted not_used.txt in both grasslands_apr22 and static realizations.
- **default.cfg and scenario_config.csv** Default for cfg$gms$c56_emis_policy changed from `redd+natveg_nosoil` to `reddnatveg_nosoil`,i.e. timber plantations are excluded from carbon pricing by default
- **default.cfg** changed default for `cfg$gms$s32_aff_prot` from 0 to 1
- **default.cfg** changed default for `cfg$gms$s56_buffer_aff` from 0.2 to 0.5
- **default.cfg** Default for cfg$gms$c56_cprice_aff changed from `forestry_vegc` to `secdforest_vegc`
- **default.cfg** Default for cfg$gms$peatland changed from `on` to `v2`
- **default.cfg** update default `cfg$gms$c56_pollutant_prices` and `cfg$gms$c60_2ndgen_biodem` to `R32M46-SSP2EU-NPi`
- **default.cfg** update input data to rev 4.99 (new validation data, new EATLancet recommendations, new semi-natural vegtation cropland data)
- **scenario_config.csv** removed erroneous setting `cc` from column `input`
- **scenario_config.csv** settings for cfg$gms$s35_secdf_distribution in `ForestryEndo` and `ForestryExo` changed from `2` to `0`
- **start_functions** Check if cfg$recalibrate is consistent with cfg$gms$s14_use_yield_calib
### added
- **32_forestry** new interfaces `vm_land_forestry`, `pcm_land_forestry` `vm_landexpansion_forestry` and `vm_landreduction_forestry`
- **56_ghg_policy_** added new trajectories for R32M46
- **60_bioenergy** added new trajectories for R32M46
- **scripts** added peatland to output/extra/disaggregation.R
### removed
- **58_peatland** removed realization "on"
- **default.cfg** Removed description of cfg$gms$c31_past_suit_scen since no longer needed due to changes in 31_past described below. Its function is now done by cfg$gms$c31_grassl_yld_scenario.
### fixed
- **21_trade** introduced s21_manna_from_heaven for fixing v21_manna_from_heaven to zero. Without fixing to zero, v21_manna_from_heaven was used unnecessarily in runs started with highres.R
- **32_forestry** bugfix unit p32_observed_gs_reg
- **32_forestry** keep c-density for timber plantations constant after rotation length to avoid unrealistic carbon sequestration in unharvested timber plantation
- **35_natveg** bugfixes ac_est
- **35_natveg** removed scaling of pm_carbon_density_ac
- **52_carbon** bugfix acx long-term carbon density
- **scripts** bugfix highres.R for bioenergy demand and GHG prices in coupled runs
- **scripts** disaggregation_LUH2.R which no longer relies on the old raster-based write.magpie for nc files
- **scripts** fixed disaggregation.R and disaggregation_LUH2.R to be used with 67k
- **scripts** fixed memory spike leading to crashes in disaggregation.R
- **scripts** fixed writing of NetCDF files in output/reportMAgPIE2SEALS.R
## [4.7.0] - 2023-12-11
### changed
- **14_yields_and_config** The new default is to not use yield calibration factors from a calibration run. The switch s14_use_yield_calib can optionally reenable the use of yield calibration factors.
- **36_employment** regression between hourly labor regression and GDP pc changed from linear to log-log
- **inputdata** Now using inputdata rev4.94 which is based on 67420 cells (67k, previously 59k)
- **scripts** For the emulator scripts select a different bioenergy demand variable that excludes bioenergy sources other than second generation bioenergy crops. Set the minimal bioenergy demand to zero. Both avoid artificial clustering of data points and allow for better fits.
- **scripts** LUH2_disaggregation output script was modified. Specifically, flooded area was made compatible with the LUH definition, cropland and grazing land were added to the states.nc file, and specific naming/details (datatype, zname, xname, and yname) were added when creating the .nc files.
### added
- **14_yields/config** Added option for considering impacts of land degradation on yields. If `s14_degradation` is switched to 1, MAgPIE will include cluster-specific information on the state of nature's contributions to people relevant for yields `./modules/14_yields/input/f14_yld_ncp_report.cs3`.
- **18_residues** Included cluster-level residue realization, for cluster-level production of residues (but balancing of recycling and burning budgets remains at region-level, for computational lightness)
- **32_forestry** new interface `vm_land_forestry`
- **58_peatland** added realization "v2" with updated peatland map and GHG emission factors
### fixed
- **inputdata** There was a major bug (related to proj/terra) in the rev4.91 inputdata that was fixed with rev4.92
- **inputdata** There was another bug (terra default na.rm changed) in the inputdata that was fixed with rev4.93
- **scripts** Fixed a bug in NPI/NDC calculations leading to missing AD policies when run with 67k
## [4.6.11] - 2023-09-05
### changed
- **scripts** All time steps between 2015 and 2050 are now reported to SEALS
### fixed
- **70_livestock** fixed division by zero that could occur depending on the scenario set-up
## [4.6.10] - 2023-08-16
### changed
- **config** update preprocessing to newest input data v4.88 with new transport costs
- **GitHub action** the github action is now faster, because it installs binary packages from Posit Package Manager
- **scenario_config.csv** update preprocessing to newest input data v4.88
- **scripts** output.R is now faster, because it no longer searches runfolder renvs for full.gms files
### fixed
- **scripts** check_config does not warn about c_input_gdx_path anymore while running empty model
- **scripts** fixed erronoeous if clause in output.R
- **scripts** fixed output/extra/disaggregation_LUH2.R. The script was not working any more because magpie4::protectedArea was changed to return protected area for all land types. Moreover, the script now also works for runs without dynamic forestry (default run) but with a warning message.
## [4.6.9] - 2023-07-27
### fixed
- **70_livestock** consideration of milk demand in the calculation of the pasture management factor
- **scripts** Fixed inaccuracies and inconsistent application of SNV policies during disaggregation in luscale::interpolateAvlCroplandWeighted(), which is called in extra/disaggregation.R
## [4.6.8] - 2023-07-17
### changed
- **41_area_equipped_for_irrigation** new AEI data (Mehta2022) replacing old Siebert data
- **80_optimization** printing of solprint when solver status is 7 re-activated
- **scripts** start_functions.R can now handle clusters per region flexibly
- **scripts** the REMIND-MAgPIE coupling now uses renv
### added
- **31_past** added `cc`, `nocc` and `nocc_hist` options for `c31_past_suit_scen` and `c31_grassl_yld_scenario`
- **32_carbon** added `nocc` and `nocc_hist` option for `c52_land_carbon_sink_rcp`
- **71_disagg_lvst** added new realisation `foragebased_jul23` which solves GAMS issues at higher spatial resolutions
- **config** added `cfg$results_folder_highres` which allows to modify the output folder used in the `highres.R` output script
- **config** new area equipped for irrigation (AEI) data in preprocessing (4.87)
- **scenario_config.csv** added a scenario for the NGFS project
- **scripts** New output script for reporting disaggregated land use patterns to the SEALS (Spatial Economic Allocation Landscape Simulator) downscaling model
### removed
- **config** `s80_num_nonopt_allowed`
- **scripts** removed .snapshot.Rprofile and the Rprofile.R script, renv now fully supersedes snapshots
### fixed
- **30_crop** corrected q30_cropland in module realization rotation_apr22, where fallow land was on the wrong side of the equation
- **71_disagg_lvst** reworked `foragebased_aug18` (including removal of linear version and correction of balance flow calculation)
- **80_optimization** resolve was not working in nlp_par realization due to `s80_num_nonopt_allowed`
- **config** corrected wrong names of parameters for peatland costs
- **config** updated scenario configs to newest preprocessing (4.87)
## [4.6.7] - 2023-05-10
### changed
- **09_drivers** Harmonization of sets for population, gdp, pal and demography
- **56_ghg_policy** added emission policies without GHG emissions from peatlands
- **config** added scenario `SSP2EU` in scenario_config.csv
- **config** modified `eat_lancet_diet` in scenario_config.csv
- **config** update of additional data to rev4.43
- **config** update of regional and cellular inputs to 4.85 in default.cfg and scenario_config.csv
- **scripts** added output script for forest area change at cluster level
- **scripts** NDC/NPI calculations can now handle 59k and 67k cell inputs
### added
- **15_food** added an option in `s15_exo_diet` to allow for exogenous diet scenario for India
## [4.6.6] - 2023-05-10
### changed
- **config** updated scenario_fsec.csv to reflect new GST validation
- **scripts** included new output indicator for water
- **scripts** updated global surface temperature maps to new RCPs per scenario
## [4.6.5] - 2023-03-29
### changed
- **22_land_conservation** Replaced old options for land conservation by new conservation priority areas. These include among others a new 30by30 template (based on Key Biodiversity Areas, unprotected habitat in Biodiversity Hotspots, Ecoregions with a high beta-diversity from the Global Safety Net (Dinerstein et al. 2020) and critical connectivity areas (Brennan et al. 2022), a new Half Earth template based on the Global Safety Net (Dinerstein et al. 2020) and land conservation of irrecoverable carbon (Noon et al. 2022).
- **56_ghg_policy** renamed `cfg$mute_ghgprices_until` to `cfg$gms$c56_mute_ghgprices_until` and changed the default to `y2030`, i.e. no GHG emission pricing in the AFOLU sector before (and including) 2030. This setting will be also used in coupled REMIND-MAgPIE runs.
- **config** input data revision to rev4.82 to include new conservation priority areas
- **config** new options for conservation priority areas (including new 30 by 30 protection)
- **scripts** calc_calib.R bug fix. If the calibration factor of a region is equal to the maximum allowed value, its divergence is set the maximum allowed divergence.
- **scripts** Disaggregation of BII merged into standard extra/disaggregation.R
- **scripts** Disaggregation of land use to 0.5° now takes land conservation into account - i.e. cropland expansion is not mapped to areas that are subject to land conservation
### added
- **56_ghg_policy** added switch `s56_minimum_cprice`
- **config** minimum CO2 price (`s56_minimum_cprice`) of 5 USD per tCO2 (18 USD per tC) for all future time steps in case of NDC policy to guide land-use decisions
- **scripts** added output script which writes landuse data on cluster resolution to a shapefile
### removed
- **56_ghg_policy** removed `s56_ghgprice_phase_in` and `s56_ghgprice_start`
- **scripts** removed argument `mute_ghgprices_until`, now handeld in GAMS code
### fixed
- **31_past** fixed pasture suitability to SSP2 before and including 2020 (only relevant for grassland implementation)
- **56_ghg_policy** the renamed switch `c56_mute_ghgprices_until` is now always used for coupled as well as standalone runs.
- **scripts** Fixed occasional memory failure in the disaggregation script
## [4.6.4] - 2023-02-22
### changed
- **15_food** Interpret EAT-Lancet guidelines not as target but as lower/upper limits
- **config** changed order of output scripts. Some functions in rds_report require gridded outputs.
- **config** input data revision to rev4.81 for trade margin bugfix
- **config** scenario_fsec.csv updated input data tgz
- **config** scenario_fsec.csv updated to new biodiversity scenario
- **scripts** fsec.R and project_FSEC_Scenarios.R include capitalSubst and landscapeElements scenarios
- **scripts** highres.R changed default resolution to c1000
- **scripts** recalibrate.R and recalibrate_realization were modified to always use best_calib for the yield calibration.
- **scripts** updated FSEC scenario start and output scripts
- **scripts** when manually running output scripts for multiple runs the lockfile is only created once
- **sticky_labor** changed labor cost share constraint from regional to cellular level
- **sticky_labor** renamed equation `q38_labor_capital_ratio` to `q38_labor_share_target`
### added
- **15_food** half_overweight scenario added
- **21_trade** New Bilateral trade realization selfsuff_reduced_bilat22 for bilateral trade within selffsuff constraints
- **32_forestry** added switch `s32_aff_prot` for protection of afforested areas (0=until end of planning horizon 1=forever)
- **56_ghg_policy** added two scenarios for GHG emission pricing and options for afforestation
- **config** added options for afforestation assumptions and updated additional data to 4.38
- **config** added setting cfg$keep_restarts which controls whether restart files should be kept after a run finished
- **config** changed default for `s_use_gdx` from 2 to 0
- **scripts** added restart points after each time step from which the model can now be restarted if the simulation aborts at some point
- **scripts** added SLURM dayMax submission type for standby QOS
- **sticky_labor** `nl_fix`, `nl_relax` and `nl_release` added
### removed
- **42_water_demand** removed fm_multicropping factor because of fallow inconsistency
### fixed
- **14_yields** nl_fix updated to current equation
- **32_forestry** pm_land_conservation(t,j,"secdforest","restore") now accounts for the rotation length in timber plantations to avoid infeasibilities
- **44_biodiversity** added regional layer `i` in `bii_target` realisation to make it compatible with the high-resolution parallel optimization output script
- **59_som** division by zero prevented by if condition
- **scripts** fixed a bug where renvs for high resolution runs were missing some packages
- **scripts** fixed in the calc_calib.R script the saving of calib_factors used in each iteration to ensure that they correspond to the divergence reported. Changed divergence from zero to NA for those iterations where calib_factors are above the limit. The best_calib selection criterion was changed from selecting the factors of the iteration with the lowest standard deviation to the selection, for each region, of the factor of the iteration with the lowest divergence. Also, factors from the first iteration are now not considered, and if two different factors had the same divergence for a region, the one of the latest iteration is picked.
## [4.6.3] - 2023-01-19
### changed
- **15_food** changed `anthro_iso_jun22` realisation such that results in case of `exo_diet = 1/0` and `exo_waste = 1/0` are identical until 2020
- **30_crop** identical assumptions for bioenergy until 2020
- **38_factor_costs** changed name of set `req` to `factors` (also used in 11_costs, 57_maccs, 70_livestock)
- **38_factos_costs** sticky_labor realization: included option to set a labor share target
- **62_material** Bioplastic demand identical in all scenarios until 2020
- **config** added `s38_target_labor_share`, `s38_targetyear_labor_share` and `s38_target_fulfillment` to define labor share target scnarios
- **config** and **38_factor_costs** changed name of `s38_fix_capital_need` to `s38_startyear_labor_substitution`
- **config** update input data to rev4.79
### added
- **31_past** added additional limitation (single climate scenario input) for **grasslands_apr22**
- **59_som** added new **cellpool_jan23** realization with updated 2019 IPCC guidelines values
- **scripts** added start script which starts an empty model just regenerating a previous run
### fixed
- **scripts** make sure that `c_title` in the GAMS code is not containing dots which otherwise could lead to compilation errors
## [4.6.2] - 2023-01-12
### changed
- **36_employment** include labor costs from MACCs in employment
- **38_factor_cots** renamed `p36_cost_shares` to `pm_cost_share_crops`
- **57_maccs** split costs into labor and capital
- **config** scenario_fsec.csv update
- **config** updated SHAPE SDP scenarios in scenario_config.csv
- **scripts** FSDP_collect FSDP_process fsec.R updates
### added
- **renv** MAgPIE now runs in an isolated, stable, reproducible R package environment
- **scripts** added output script that generates a short mif containing only variables relevant for the REMIND coupling
- **scripts** added output script that generates subnational validation outputs for fable_India projects
- **setup** required R packages are automatically installed
## [4.6.1] - 2022-12-13
### changed
- **42_water_demand** replaced `f42_env_flow_policy` with macro
- **30_crop** replaced `f30_scenario_fader` with macro
- **30_crop/config** changed switch `c30_rotation_scenario_speed` to `s30_rotation_scenario_target`
- **30_crop/config** changed switch `c30_snv_target` to `s30_snv_scenario_target`
- **config** changed default value for `c30_marginal_land` from `'all_marginal'` to `'q33_marginal'` for better spatial cropland patterns
### added
- **10_land** added `vm_lu_transitions` as interface
- **10_land** added interface `fm_land_iso` for consistency
- **30_crop/config** added switch `s30_rotation_scenario_start`
- **30_crop/config** added switch `s30_snv_scenario_start`
- **30_crop/config** added switch for set `land_snv`for defining which land cover types are allowed in the semi-natural vegetation policy in cropland scenarios
- **39_landconversion** scalar `s39_reward_crop_reduction` provides a cropland reduction reward
### removed
- **10_land** removed `feb15` realization
- **10_land** removed the interfaces `vm_croplandexpansion` and `vm_croplandreduction`
- **30_crop** removed `f30_scenario_fader.csv`input
- **39_landconversion** removed `s39_reward_shr`
- **config** validation_short.R output script is no longer run by default after each run
- **scripts** removed remind2::deletePlus in coupling interface of start_function
### fixed
- **59_som** fixed land use change tracking for non-cropland pools in the `cellpool_aug16` realization
- **config** changed default value for s56_limit_ch4_n2o_price from 1000 to 4000 for consistency with c57_macc_version = "PBL_2022"
- **scripts** rewrite of land conversion cost calibration script `landconversion_cost.R`
## [4.6.0] - 2022-11-09
### changed
- **18_residues** bugfix in `q18_cost_prod_res`
- **36_employment** included calculations for minimum wage scenario
- **38_factor_costs** included labor cost scaling in case of wage scenario
- **42_water_demand** account for multiple cropping in water requirements
- **51_nitrogen** moved maccs into emission modules. change of interface from vm_btm_reg to vm_emissions_reg
- **52_carbon** change of interface from vm_btm_reg to vm_emissions_reg
- **52_carbon** Soil C of urban areas set to soil C of natural other land
- **57_maccs** default changed from PBL2007 to PBL2022
- **58_peatland** moved maccs into emission modules. change of interface from vm_btm_reg to vm_emissions_reg
- **59_som** Now calculates soil C for fallow and urban areas
- **62_material** added biomass demand for bioplastic production
- **70_livestock** included labor cost scaling in case of wage scenario
- **config** added `s62_max_dem_bioplastic` and `s62_midpoint_dem_bioplastic` to define bioplastic scenario
- **config** adjusted PR template
- **config** best_calib set to FALSE in default
- **config** new switches `s36_minimum_wage`, `s36_scale_productivity_with_wage`, and `s38_fix_capital_need`
- **config** non-food system emission MAGICC switch
- **config** updated config to new module setup of MACCs
- **config** updated default realization of 15_food from anthropometrics_jan18 to anthro_iso_jun22
- **config** updated SHAPE SDP scenarios in scenario_config.csv
- **documentation** added literature
- **inputs** update of NPi for China (additional data 4.30)
- **inputs** updated f56_emis_policy (additional data 4.29)
- **inputs** updated non-food initial prices, MACCs curves, and removed suitability threshold of 0.1 in all_marginal setting
- **scripts** added single time step run to test runs
- **scripts** fix in start_functions for the calibration setting `ifneeded`
- **scripts** FSDP_collect handles the health impacts data provided by Marco Springmann, distributed it into the scenario's various reports. It performs a similar operation for global nutrient surplus (which must be calculated on the grid-level and then aggregated).- **13_tc** relaxed vm_tau upper limit
- **scripts** log files are now written in a subfolder "logs"
- **scripts** output/extra/disaggregation_BII.R adjusted BII output for primary and secondary other land
- **scripts** quit with exit code = gams status at the end of submit.R
- **scripts** update of rds_report to allow gridded intermediate outputs
- **scripts** updated FSEC start and output scripts
- **scripts** updated FSEC start scripts and related config files to introduce new scenarios
- **scripts/start** cleanup of old start scripts
### added
- **14_yields** added input file containing AQUASTAT yield calibration factors and switch `s14_calib_ir2rf` in default.cfg to activate this yield calibration
- **15_food** added new realization with country level exogenous diets, product-specific intake estimates, new scenarios for exogenous BMI and decomposition switches for EAT Lancet diets. Simplified code and improved iteration procedure.
- **50_nr_soil_budget** new module realization for more consistent MACCs implementation. change of interface from vm_btm_reg to vm_emissions_reg
- **53_methane** moved maccs into emission modules. change of interface from vm_btm_reg to vm_emissions_reg
- **56_ghg_policy** added new ecosystem protection scenarios
- **56_ghg_policy** new module realization for more consistent MACCs implementation
- **57_maccs** added new Marginal Abatement Cost Curve (MACCs) data set from PBL (PBL2022)
- **57_maccs** new more consistent maccs implementation. different mapping of emission sources to maccs.
- **f32_forest** added the option to run generic disturbance scenarios of secondary forest types determined in `f32_forest_shock.csv`
- **f35_natveg** added the option to run generic disturbance scenarios of primary forest determined in `f35_forest_shock.csv`
- **scripts** added output script creating a merged .csv for dietaryIndicators and caloricSupply outputs
- **scripts** added output script creating a merged .csv for dietaryIndicators and caloricSupply outputs
- **scripts** added output script creating a set of outputs for Alessandro Passaro in the FSEC context
- **scripts** added output script creating a set of outputs for Simon Dietz in the FSEC context
- **scripts** added output script for gridded crop diversity indices
- **scripts** added output script running MAGICC7 on a MAgPIE scenario
- **scripts** added output script, FSDP_process creating a merged .csv and .gdx for dietaryIndicators and caloricSupply outputs
- **scripts** added output scripts for FSEC FSDP runs
- **scripts** added water output script for FSEC model runs
### removed
- **15_food** removed read-in of non-needed input file "f15_calib_factor_FAOfsupply_iso"
- **38_factor_costs** removed `mixed_reg_feb17` realization
- **50_nr_soil_budget** old inconsistent module realizations
- **53_methane** old inconsistent module realizations
- **56_ghg_policy** old inconsistent module realizations
- **57_maccs** old inconsistent module realizations
### fixed
- **38_factor_costs** fixed calibration of share parameter in `sticky_labor` realization
- **43_water_availability** added missing years after 2100 in "f43_wat_avail" to avoid infeasibilities in coupled runs with less_ts timesteps
- **59_som** corrected the som pool due to the carbon transfer from other and primary forest to secondary forest before optimization (presolve)
- **scripts** fixed some bugs related to background execution of start/output scripts
## [4.5.0] - 2022-07-07
### changed
- **09_drivers** separation of GDP and population scenarios
- **09_drivers** changed `i09_gdp_pc_mer_iso` to `im_gdp_pc_mer_iso`
- **11_costs** Split of production costs per sector, addded new separated costs to the costs function.
- **13_tc** changed vm_tech_cost upper bound to share of regional GDP PPP (s13_max_gdp_shr)
- **13_tc** Replace endo_jan18 realization by endo_jan22. The new realization adds a new dimension to vm_tau separating crop from managed pastures tau.
- **15_food** added more options to define convergence towards exogenous food intake and waste scenarios accounting for different transition periods
- **18_residues** The variable that include production costs vm_cost_prod for residues changed to a new independent variable called vm_cost_prod_kres
(specific to residues).
- **30_crop** renamed switch `s30_set_aside_shr`, `s30_set_aside_shr_noselect` and `c30_set_aside_target` to `s30_snv_shr`, `s30_snv_shr_noselect` and `c30_snv_target`.
- **31_past** The variable that include production costs vm_cost_prod for pasture changed to a new independent variable called vm_cost_prod_past
(specific to past).
- **31_past** added new realization implementing the separation of rangelands and managed pastures for the production of grass biomass.
- **32_forestry** simplification and bugfix of afforestation limit. `c32_max_aff_area_glo` renamed to `s32_max_aff_area_glo` in default.cfg.
- **34_urban** added set urban_scen34 and the switch c34_urban_scenario
- **35_natveg** corrected naming of Frontier Forests (FF) to Intact Forest Landscapes (IFL) and changed input data for BH_IFL implementation.
- **35_natveg** implementation of land protection moved to new module `22_land_conservation`
- **38_factor_costs** Sticky free/dynamic switch was removed and the realization was split into two realizations: the new per_ton_fao_may22 (free) and sticky_feb18 (dynamic). vm_cost_prod(i,kall) is now vm_cost_prod_crop(i,req) for crops factor costs. The results are now given differentiating between capital and labor for all realizations (new set req).
- **42_water_demand** Added new input data on pumping costs for India, equation to calculate water costs and scalars for policy shocks
- **44_biodiversity** Improved documentation, simplification of equations and flexible options for price on biodiversity loss
- **56_ghg_policy** additional scenarios for c56_emis_policy
- **56_ghg_policy** Deactivated GHG emission policies were not accounted for in the MACCs module. This has been corrected by an extension of the interface `im_pollutant_prices`, which now has an additional dimension for emission sources `emis_source`. In this context some equations in `56_ghg_policy` have been simplified (sets: `emis_source_reg`, `emis_source_cell`). Also, GHG emissions from peatlands have been fully integrated into `56_ghg_policy`.
- **70_livestock** vm_cost_prod(i,kall) is now vm_cost_prod_livst(i,req) for livestock factor costs (req indicates differentiation between capital and labor) and vm_cost_prod_fish(i) for fish.
- **80_optimization** Updated solver settings
- **config** Update of regional and grid inputs from 4.68 to rev4.69, and additional files to 4.17. Removed free/dynamic sticky switch, and added scalars used in mixed_regional factor costs realization.
- **config** added s13_max_gdp_shr setting for tech cost upper bound as share of GDP PPP
- **config** included switch for non-agricultural water demand (s42_watdem_nonagr_scenario) in scenario_config.csv
- **config** included SHAPE SDP scenarios in scenario_config.csv
- **config** Update default tau realization from endo_jan18 to endo_jan22
- **config** Added new SSP scenario switch for pasture suitability cfg$gms$c31_past_suit_scen
- **config** Added new switch to limiting calibration to relative or absolute managed pastures yields: cfg$gms$s31_limit_calib.
- **inputs** updated non-agricultural water use scenarios (watdem_nonagr_grper.cs3)
- **modules** Moved interface `vm_carbon_stock` from 52_carbon to 56_ghg_policy
- **scripts** replaced redundant files config.log and config.Rdata with a config.yml
- **scripts** updated FSEC modeling start script and added FSEC calibration scripts
- **scripts** clean up of the recalibrate_realizations.R script, project_LAMACLIMA_WP4.R, and sticky.R to remove sticky dynamic/free switch.
- **scripts** bugfix in disaggregation.R, disaggregation_BII.R with respect to urban scenario
- **scripts** added FSEC modeling start script (global runs)
- **scripts** The disaggregation_LUH2.R was extended to include the changes used to generate ISIMIP3b maps for LUH harmonization. The largest changes are: 1) The convertLUH function now breaks the grid level magpie objects by groups of years, then creates the raster for the groups and aggregates them to create the final map at a quarter of a degree resolution (this speeds up the process). 2) The mapping between LUH and MAgPIE is now defined by country and magpie-LUH types (not 1 to 1 anymore). 3) The split of MAgPIE's pasture land type between pasture and rangeland changed. Rangeland is assumed to stay constant after 2015, and changes in MAgPIE's pasture are due to managed pasture. 4) IFs were added so if a certain map already exists in the output folder, it will not generate it once again. 5) Flooded land now corresponds to a share of rice cropland, based on historical values. 6) To speed calculations, yields are read at the cell level, the crops are aggregated based on the new MAgPIE-LUH mapping, and then disaggregated to grid level.
- **scripts** added new disaggregation script to provide grid cell level BII
- **scripts** removed test script "irrig_dep_test" from "start" folder to "extra" folder
- **scripts** Added script projects/paper_grassland.R
- **scripts** scripts/output/extra/emulator.R Remove dependency on deprecated R package "magpie"
### added
- **core** macros for linear and sigmoidal time interpolation
- **22_land_conservation** added new module and realisation for land conservation. The realisation also includes a new WDPA initialisation data set (from 1995 to 2020) for protected areas under legal protection, meeting IUCN and CBD protected area definitions. The module also adds the interface `pm_land_conservation`.
- **30_crop** new module realizations including crop rotation scenarios by strict constraints (`rotation_apr22`) and by penalties (`penalty_apr22`)
- **36_employment** added new module to calculate agricultural employment. Includes one realization (`exo_may22`) in which employment is calculated based on the total labor costs (for crop and livestock production)
- **38_factor_costs** mixed_reg_feb17 realization added. This realization includes differences in productions costs between irrigated and rainfed crops, with the option of regional differentiation as well. per_ton_fao_may22 realization added. This realization corresponds to the old sticky_feb18 free.
- **38_factor_costs** added switch `c38_fac_req` to choose between global and regional crop factor requirements. The default is "glo" (which corresponds to the previous implementation)
- **44_biodiversity** added new realization `bii_target`, which calculates the BII at the level of 71 biomes and allows to set targets for BII (e.g. no decrease in the future)
- **52_carbon** added land carbon sink adjustment factors, needed in R post-processing
- **70_livestock** added switch `c70_fac_req_regr` to choose between global and regionally calibrated regression to calculate livestock factor requirements. The default is "glo" (which corresponds to the previous implementation)
- **config** added option for CO2 emission pricing `cfg$gms$c56_carbon_stock_pricing`
- **config** added cfg$gms$s70_past_mngmnt_factor_fix with default 2005 (previous default was 2010). The previous setting caused a strong spike in CO2 emissions from pasture expansion in SSA. With 2005, this can be avoided.
- **inputs** New input files added:
f13_pastr_tau_hist.csv -> historical tau for managed pastures.
f31_pastr_suitability.cs3 -> Managed pasture suitability
f31_LUH2v2.cs3 -> LUH2v2 land classes separating rangelands from managed pastures
f31_grassl_yld.cs3 -> Rangelands and managed pastures grass yields
f31_grass_bio_hist.cs3 -> Historical grass biomass demand
- **modules** New dimension in `vm_carbon_stock` for different carbon stock types (actual, previousLandPattern, previousCarbonDensity)
- **scripts** output/projects/FSEC_StevenLord.R to create output for Steven Lord in the FSEC context
- **scripts** output/projects/FSEC_costs.R to create costs ouput for the FSEC project
- **scripts** output/projects/FSEC_dietaryIndicators.R to create output datasets for the FSEC project
- **scripts** output/projects/FSEC_nitrogenPollution.R to create output datasets of nitrogen pollutants for the FSEC project
- **scripts** Extended dissagregation.R script to replace single "past" land class by LHU range and pastr classes when grassland_apr22 realization is used.
- **scripts** `start/projects/test_rotations.R` testscript for different rotation scenario settings
### removed
- **38_factor_costs** mixed_feb17 and fixed_per_ton_mar18 realizations removed because they are not being used at the moment.
### fixed
- **09_drivers** bugfix concerning the use of the switch c09_gdp_scenario for defining population assumptions
- **09_drivers** introduced new sets for PAL and demography scenarios to account for only partial coverage of available socio-economic gdp and population scenarios
- **18_residues** off realization; missing variable declarations
- **34_urban** exo_nov21 realization; bugfix in calculation of biodiversity value
- **41_area_equipped_for_irrigation** static realization. bugfix in presolve.gms: f41_irrig_luh("y1995",j)
- **44_biodiversity** fixed accounting for time step length in realization `bv_btc_mar21`
- **50_nr_soil_budget** off realization; missing variable declarations
- **56_ghg_policy** Some equations in `56_ghg_policy` have been simplified. Also, GHG emissions from peatlands have been fully integrated into `56_ghg_policy`.
- **56_ghg_policy and config** removed switch `s56_reward_neg_emis`
- **59_som** static realization; avoid division by zero
- **62_material** exo_flexreg_apr16 realization; avoid division by zero
- **80_optimization** nlp_par realization; bugfix i2 in submission loop
- **inputs** included data for Sudan
- **scripts** calibration; set NA values to 1
- **scripts** fixed misleading warning in check_config
- **scripts** fixed configuration error in FSEC output scripts, FSEC_dietaryIndicators.R and FSEC_environmentalPollutants.R
- **scripts** scripts/start/extra/emulator.R Throw an error if no file can be found to take the GHG prices from
## [4.4.0] - 2021-12-13
### changed
- **additional_data** NDCs for Chinas afforestation now start earlier (1995) in line with observed afforestation.
- **config** comments added for correct use of nitrogen switches
- **inputs** new default LPJmL version with growing season adaptation (gsadapt) on
- **51_nitrogen** parameter change in rescaled_jan21, now including regionalized climate-dependent leaching factors
- **config** Update default configuration to new input data (especially cellular inputs) including all module realization updates (14_yield, 22_processing, 30_crop, 38_factor_costs, 39_landconversion). Moreover, climate impatcs (cc options for biophysical inputs) are activiated as default. New best_calib calibration routine is activated as default.
- **config** peatland module on by default (cfg$gms$peatland <- "on")
- **config** update default setting for 2nd generation bioenergy demand and GHG prices
- **config** update default setting for the 42_water_demand module (to all_sectors_aug13)
- **scripts** output/extra/disaggregation.R updated to account for country-specific set-aside shares in post-processing
- **scripts** output/extra/disaggregation.R updated to account for sub-categories of "forestry"
- **scripts** Default recalibration routine does not read in previous calibration factors anymore
- **09_drivers** Update sets in drivers to include new SDP and Ariadne GDP and Pop scenarios
- **21_trade** In the exo and off realization, equations corrected to be consistent with the mapping between supreg h and regions i. Bugfixes in trade exo and off realizations. Added scaling factor for exo realization.
- **inputs** Update of GDP and population scenarios based upon recent historic data from WDI (complemented with growth rates given by the James2019 dataset), short term projections until 2025 from IMF (for GDPpc) and WB (for pop) and reconverge to the original SSP GDPpc levels by 2100.
- **inputs** Update of all input data that are based on FAO, using the most up-to-date version of FAOSTAT datasets available at the date of input calculations via automated download.
- **inputs** Update of additional data to rev4.07
- **scripts** scripts/start/projects/project_LAMACLIMA.R -> scripts/start/projects/project_LAMACLIMA_WP4.R
- **58_peatland** "On" realization: Degraded peatland is estimated differently, based on an additional calibration factor.
- **43_water_availability** changed scaling factor
- **10_land** Converted "v10_landreduction" to interface "vm_landreduction", used in "modules/39_landconversion/calib"
- **52_carbon** Removed interface "vm_carbon_stock_change", no longer needed
- **scripts** recalibrate_realizations.R and recalibrate.R adjusted for land conversion cost calibration + default time steps for convenient validation of results
- **scripts** start_functions adjustments for land conversion cost calibration
- **scripts** start.R added SLURM medium as choice
- **scripts** yield calibration, "best" setting uses factors from iteration with lowest standard deviation
- **14_yield** read-in file f14_yld_calib.csv if exists. Set default calibration factors to 1 in case f14_yld_calib.csv does not exist
- **13_tc** different educated guess for vm_tau in 1995
- **scaling** Update of scaling factors. removed duplicates
- **32_foresty** Avoid division by zero (observed under higher regional resolutions)
- **35_natveg** Avoid division by zero (observed under higher regional resolutions)
- **70_livestock** Avoid division by zero (observed under higher regional resolutions)
- **60_bioenergy** Minimum dedicated 2nd generation bioenergy demand assumed in each region raised from 0.01 to 1 mio. GJ per yr, and added as option in the config file (s60_2ndgen_bioenergy_dem_min)
- **config** Remove elements from the parameter list of start_run(), instead include them as regular settings in the default.cfg.
- **scripts** Add option to take ghg prices from different file than the regular reporting file (used in the REMIND coupling)
- **60_bioenergy** Switch off fixing the bioenergy demand to SSP2 until 2020 if MAgPIE runs coupled (to REMIND) or for emulator runs (to derive biomass supply crurves).
- **56_ghg_policy** Switch off fixing the GHG prices to SSP2 until 2020 if MAgPIE runs coupled (to REMIND) or for emulator runs (to derive biomass supply crurves).
- **scripts** start/test_runs.R added SSP1, SSP2 and SSP5 as default test runs
### added
- **34_urban** New exo_nov21 exogenous realization of urban land expansion
- **21_trade** Missing interface parameter for failing exo realization runs
- **59_som** exogenous pathway for vm_nr_som via f59_som_exogenous
- **config** Addition of a new scenario column (Tland) in scenario_config.csv
- **config** Added option c32_max_aff_area, which allows to provide a file with regional limits for afforestation
- **14_yield** parameter created to save historical cellular yields and to be used in the sticky realization of 38_factor_costs and in the 17_production module
- **17_production** switch added to decide if initialization of cellular crop production is needed or not. Also, a parameter to calculate initial production based on input cellular crop patterns and semicalibrated yields (potential yields calibrated to FAO values).
- **scripts** Added calibration script to generate default calibration for different factor costs realization
- **scripts** scripts/output/extra/disaggregation_LUH2.R script for exporting spatial output in LUH2 format (NetCDF)
- **37_labor_prod** labor productivity module with two realizations: off and exo
- **38_factor_costs** new realization "sticky_labor", based on "sticky_feb18" but accounting for changes in labor productivity
- **15_food** Added additional solve with CONOPT3 in case of modelstat 7
- **scripts** Added script "landconversion_cost.R" for land conversion cost calibration in scripts/calibration, for matching historic cropland in 2015
- **39_landconversion_cost** added new realization "calib", which uses the calibration factors derived by "landconversion_cost.R"
- **scripts** Added start script for yield and land conversion cost calibration "recalibrate_all.R"
- **scripts** added script validation_short.R with aggregated crop types (cutting the PDF size in half) -> replaces validation.R in default.cfg
- **scripts** added start script "scripts/start/Rprofile.R" for adding a R snapshot to the ".Rprofile" file
- **config** file "land_carbon_sink_adjust_grassi.mz" added to cfg$files2export$start
- **config** Inclusion of LAMACLIMA scenarios in scenario_config.csv
- **output.R** added SLURM standby maxMem and SLURM priority maxMem; needed for some output scripts (e.g. disaggregation_LUH2.R)
### removed
- **32_foresty** Removed static realization
- **35_natveg** Removed static realization
- **scripts** lpjml_addon script is removed and all calls within dependend starting scripts
- **scripts** output/extra/disaggregation_transitions_.R moved to deprecated folder
- **scripts** output/extra/disaggregation_cropsplit.R moved to deprecated folder
- **14_yield** Removed `biocorrect` and `dynamic_aug18` realizations
- **20_processing** Removed `substitution_dec18` realization
- **30_crop** Removed `endo_jun13` realization
- **scripts** scripts/start/extra/highres.R
- **39_landconversion_cost** removed realizations "global_static_aug18" and "devstate"
### fixed
- **80_optimization** Improved solve logic in "nlp_apr17" and "nlp_par" realization, multiple bugfixes and switch to solvelink=3 in "nlp_par"
- **58_peatland** fixed rare infeasibility in "on" realization
- **10_land** fixed rare infeasibility in "landmatrix_dec18" realization
- **38_factor_costs** For the sticky_feb18 realization correction in initial capital stocks, use of production initial values, and 05USDppp units changed to 05USDMER for sticky so it matches the units of the other realizations
- **80_optimization** Bug fixes in the nlp_par (parallel optimization) and improved code to collect failing handles.
- **32_foresty** Avoid division by zero in q32_establishment_dynamic_yield
- **35_natveg** fixed land protection to SSP2 default (WDPA) for historic period
- **15_food** New iteration needs to be started before setting food prices for curr_iter15
- **scripts** scripts/output/extra/highres.R bugfixes
- **38_factor_costs** units in sticky_feb18
- **32_foresty** Global afforestation limit s32_max_aff_area was not effective in case of parallel optimization -> added option c32_max_aff_area, which allows to provide a file with regional limits for afforestation;
- **73_timber** plausible cost for balance variable in case of s73_timber_demand_switch = 0 to avoid cost distortion
- **56_ghg_policy** choose the correct scenario for fixing the GHG prices until sm_fix_SSP2
## [4.3.5] - 2021-09-02
### changed
- **13_tc** added switch to ignore historic tau patterns in historic time steps (new default)
- **16_demand** Moved most of cropping related set definitions (k, kve, kcr) from **16_demand** to **14_yield**
- **32_foresty** Added option to choose a rotation length calculation criteria
- **35_natveg** Calculation of land protection policies revised and moved from presolve.gms to preloop.gms
- **38_factor_costs** Realization `sticky_feb18` extended to differentiate capital requirements between regions and their specific development status (GDP) in each time step of the magpie run. The changes in the `sticky` realization also include an additional switch so it can be operated as `dynamic` (change of each region capital share at each time step) or `free` (capital shares equal to zero and equivalent to the `fixed_per_ton_mar18` realization). Bugfix in the yearly update of the variable input requirements. Addition of the time dimension and clean up of names of parameters used in the realization. Removal of the management factor (this factor was not being used, it was being cancelled out in previous calculations). Correction of the costs, they are given in 05USDppp.
- **39_landconversion** lower costs for expansion of forestry land
- **58_peatland** Peatland area is initialized in 1995 based on levels for the year 2015, and hold fixed depending on `s58_fix_peatland`. This provides a better proxy for peatland area and associated GHG emissions for the historic period, which where assumed zero in previous versions.
- **80_optimization** **nlp_par** parallelizes now on superregional level `h` instead of regional level `i` as before.
- **script** Added forestry run script which used LPJmL addon
- **script** New standard for cluster to region mapping (rds-files) is used in all scripts. If old spam files are provided by input data, rds-mapping file is created.
- **script** updated test run script. Update of the sticky run script.
- **start scripts** improved function for GAMS set creation from R and outsourced it to package `gms`
- **inputs** Changed file format from cs2 to cs2b for cellular input files with a single data column
- **scenario_config** added RCPs as columns for use with setSceanrio function. This required the addition of "gms$" in the 1st column.
### added
- **73_timber** Added construction wood demand scenarios based on Churkina et al. 2020
- **script(s)** Added scripts to replicate runs for Mishra et al. 2021 (in review : https://doi.org/10.5194/gmd-2021-76)
- **13_tc** Added new interfaces for tau factor of the previous time step (`pcm_tau`)
- **14_yield** Added new realization `managementcalib_aug19` that is able to calibrate yield data coming from uncalibrated crop models (e.g. LPJmL yields for unlimited N supply). The yield calibration is either a purely multipicative factor or is limited to additive change in case of a underestimated FAO yield by the initial crop model yields (based on the switch `s14_limit_calib`). For pastures spillover of crop yield increases due to technological change from the previous time step are allowed and can be scaled using `s14_yld_past_switch`.
- **20_processing** Added new almost identical realization that excludes a calibration of the oil crop demand for oils (Note: old realization can be removed, when old yield realizations are deleted).
- **30_crop** Added new realization `endo_apr21`. The realisation includes new input data for available cropland and a new switch `c30_marginal_land`, which provides different options for including marginal land as cropland. Furthermore, a given share of the available cropland can be set aside for the provisioning of natures contribution to people and to promote biodiversity. The new switches `s30_set_aside_shr` and `c30_set_aside_target` are included to specify the share that should be set aside and the target year.
- **30_crop** Added new interface parameter historic croparea (`fm_croparea`)
- **30_crop** Added new option `policy_countries30` for country specific set aside share
- **35_natveg** Added new option `"FF+BH"` for protected areas.
- **35_natveg** Added new option `policy_countries35` for country specific land protection
- **38_factor_costs** Added scaling factors for improving model run time
- **39_landconversion** new realization `devstate` in which global land conversion costs are scaled with regional development state (0-1)
- **41_area_equipped_for_irrigation** Added switch for using different input data including new LUH2v2 consistent initialisation pattern.
- **41_area_equipped_for_irrigation** Added scalar for depreciation rate that depreciates certain area in every timestep, defined by switch in config.
- **58_peatland** Added option for one-time and recurring costs of peatland degradation (USD05MER per ha)
- **calibration run** has two new features: 1. Upper bound to cropland factor can be added (`crop_calib_max`). 2. Best calibration factor (factor with the lowest divergence) can be picked individually for each regions based on all calibration factors calculated during the calibration run iteration (`best_calib`).
- **disaggregation** Added new disaggregation script that is in line with new crop realisation and can account for cropland availabilty on grid level during disaggregation (see `interpolateAvlCroplandWeighted()` in package `luscale` for further details).
- **sets** added superregional layer `h` as additional spatial layer and moved constraints in **13_tc** and **21_trade** partly to the superregional level.
### removed
- **13_tc** Removed disfuctional setting c13_tccost = "mixed"
- **core** removed sets ac_young and ac_mature (no longer needed due to changes in 44_biodiversity)
### fixed
- **32_foresty** BII coefficients for CO2 price driven afforestation
- **32_foresty** growth curve CO2 price driven afforestation
- **32_foresty** NPI/NDC afforestation infeasibility
- **32_foresty** Correct distribution from LUH data to plantations and ndcs
- **35_natveg** option to fade out damage from shifting agriculture by 2030
- **44_biodiversity** ac0 included in pricing of biodiversity loss
## [4.3.4] - 2021-04-30
### changed
- **51_nitrogen** New calculations for emissions from agricultural residues (vm_res_ag_burn)
- **53_methane** New calculations for emissions from agricultural residues (vm_res_ag_burn)
- **citation file** added new contributors
### added
- **config** The set "kfo_rd" (livst_rum, livst_milk), which is used in the food substitution scenarios c15_rumdairy_scp_scen and c15_rumdairyscen, has been added to the default.cfg file. This allows for sensitivity scenarios (e.g. only livst_milk or only livst_rum).
- A new scenario (nocc_hist) was added to the cc/nocc switch. In this scenario, parameters affected by the cc/nocc switch in **14_yields**,**42_water_demand**,**43_water_availability**,**52_carbon**,**59_som** keep their historical/variable values up to the year defined by sm_fix_cc. Afterwards, sm_fix_cc values are kept constant for the overall run horizon.
### fixed
- **09_drivers** migration of sm_fix_SSP2 and sm_fix_cc declaration from the core declarations to the drivers module. This will allow to set the scalars properly .
- - **15_food** single-cell protein substitution scenarios included in intersolve.gms.
- **20_processing** The "mixed" scenario for single-cell protein production (c20_scp_type) was not working as expected. The corresponding code in 20_processing has been updated.
## [4.3.3] - 2021-03-30
### added
- **15_food*** added 3 sigmoid food substitution scenarios
- **44_biodiversity** New biodiversity module. The realization bv_btc_mar21 now allows to calculate an area-based biodiversity value across all land types. Switch `c44_price_bv_loss` to implement cost for biodiversity loss.
- **56_ghg_policy** Automatic sets for scenarios
- **60_bioenergy** Automatic sets for scenarios
- **70_livestock*** added 3 sigmoid feed substitution scenarios
- **scripts** added output script for disaggregation to GAINS regions
- **scripts** Automatic sets for 56_ghg_policy and 60_bioenergy
- **scripts** Added pre-commit hook
### fixed
- **60_bioenergy** Minimal bioenergy demand
## [4.3.2] - 2021-03-17
### changed
- **12_interest_rate** Interest fader changed to csv
- **15_food** better documentation of parameters over model iterations
- **15_food** added scenario switch for ruminant and dairy replacement by Single-Cell Protein
- **20_processing** added different options for Single-Cell Protein production
- **35_natveg** Fader for HalfEarth protection policy
- **50_nr_soil_budget** added necessary interfaces to 50_nitrogen module
- **70_livestock** added scenario switch for feed replacement (crop and forage) by Single-Cell Protein
- **scripts** Updated AgMIP output scripts.
- **runscripts** adapted to new input data and model version
- **tests** Replaced TravisCI with GithubActions
### added
- **15_food** Added the option to fade out livestock demand towards a target level in kcal/cap/day.
- **21_trade** Added scalar `s21_trade_bal_damper` and new set `k_trade_excl_timber`
- **29_ageclass** New age-class module
- **32_forestry** added new default realization
- **32_forestry** Simplified routine for plantation establishments. Added plantation area initialization based on MODIS data. Calibration to FAO growing stocks via carbon densities. New switches: `s32_distribution_type `, `s32_hvarea`, `s32_establishment_dynamic`, `s32_establishment_static`, `s32_max_self_suff`. New settings `c32_dev_scen`, `c32_incr_rate`, `c32_incr_rate`
- **35_natveg** Added new default realization
- **35_natveg** Added distribution in secondary forest based on Poulter et al. 2019. Added forest damages due to wildfire and shifting agriculture. Bugfix in forest protection calculations. New switches: `s35_secdf_distribution`, `s35_forest_damage`, `s35_hvarea`
- **35_natveg** Added HalfEarth scenario to protection scenarios
- **51_nitrogen** new module realization rescaled_jan21, which rescales n-related emissions with nitrogen surplus to account for lower emissions with higher NUE
- **52_carbon** Simplified routine for carbon stock calculations in timber plantations and cleanup of unused code.
- **56_ghg_policy** Added new scenario to emission policy
- **73_timber** Additive calibration with FAO data for roundwood demand. New switches: `c73_wood_scen`
- **73_timber** Added new realization `default` (modified version of previous realization)
- **default.cfg** New `forestry` scenario which simulates timber production in MAgPIE
- **scenario.csv** Added three plantation scenarios
- **scaling** Updated scaling across the modules
- **scripts** Updated to `forestry` script with general cleanup for publication. Added `forestry_magpie` script for generic forestry runs.
- **scripts** added output script for disaggregation of land transitions
### removed
- **32_forestry** Removed previous default realization
- **35_natveg** Removed previous default realization
- **73_timber** Removed previous default realization
### fixed
- **32_forestry** Bugfixes for "ac_est" and carbon treshold afforestation; removed plantations from "vm_cdr_aff".
- **core** bugfix m_fillmissingyears macro; was running over t before; now running over t_all_
## [4.3.1] - 2020-11-03
### added
- **main** Added Dockerfile for running MAgPIE in a container
### fixed
- **35_natveg** Bugfix "v35_secdforest_expansion"
- **52_carbon** Bugfix "p52_scaling_factor" for climate change runs
- **73_timber** New scenario switch `c73_wood_scen`.
## [4.3.0] - 2020-09-15
### added
- **38_factor_costs** Added the new "sticky" realization to the factor costs module. The realization "sticky_feb18" favors expansion in cells with preexisting farmland and capital based on capital investment decisions.
- **modules** added endogenous implementation of local biophysical (bph) impacts of afforestation to existing realizations in modules 32_forestry (dynamic_oct19) and 56_ghg_policy (price_jan20). default = off
- **73_timber** Added timber module which brings the ability of producing woody biomass for timber plantations and natural vegetation. Default = off. New switch: `s73_foresight`. New scalars : `s73_timber_prod_cost`, `s73_timber_harvest_cost`,`s73_cost_multiplier`,`s73_free_prod_cost`
- **32_forestry** New realization `dynamic_may20` for forestry land use dynamics. This builds up on previous forestry realization for afforestation. New switches: `s32_fix_plant`, `c32_interest_rate`. New scalars : `s32_plant_share`, `s32_forestry_int_rate`, `s32_investment_cost`, `s52_plantation_threshold`.
- **35_natveg** New realization `dynamic_may20` for natural vegetation land use dynamics. New forest protection scenario.
- **52_carbon** Added interface which is used for calculating additional investment needed in plantations when carbon stocks are lower than a specified threshold. New scalar: `s52_plantation_threshold`.
- **57_maccs** Added MACCs from Harmsen PBL 2019
- **15_food** Added the option to include calories from alcohol consumption in healthy and sustainable EAT-Lancet diets.
- **scripts** added start script for making timber production runs (forestry.R).
### changed
- **scripts** updated selection routine for start and output scripts
- **scripts** replaced lucode dependency with newer packages lucode2 and gms
- **32_forestry** include new datasets of the bph effect of afforestation / replaced the bph ageclass switch with a fade-in between ac10 and ac30 in (dynamic_may20)
- **13_tc**, **39_landconversion**, **41_area_equipped_for_irrigation** and **58_peatland**. For the current time step, the optimization costs only include now the annuity of the present investment. magpie4's reportCosts() function was modified to consider these changes.
### removed
- **scripts** deleted outdated start and output scripts
### fixed
- **32_forestry** Rotation length calculation based on correct marginals of growth function in timber plantations. Clearer calculations for harvested area for timber production.
- **35_natveg** Clearer calculations for harvested area for timber production.
- **52_carbon** Fix to the Carbon densities received from LPJmL for timber plantations.
## [4.2.1] - 2020-05-15
### added
- **modules** added option of regional scenario switches in modules 12_interest_rate, 15_food, 42_water_demand, 50_nr_soil_budget, 55_awms, 56_ghg_policy, 60_bioenergy
- **58_peatland** added peatland module. Two realizations: off (=default) and on.
- **80_optimization** added realization for parallel optimization of regions in combination with fixed trade patterns.
- **metadata** added .zenodo.json metadata file for proper metadata information in ZENODO releases
### changed
- **12_interest_rate** merged the two realizations (glo_jan16 and reg_feb18) into one (select_apr20) with same functionality and add on of option to choose different interest rate scenarios for different regions selected via country switch select_countries12
- **scripts** streamlined and improved performance of NPI/NDC preprocessing
### fixed
- **56_ghg_policy and 60_bioenergy** update of GHG prices and 2nd generation bioenergy demand from SSPDB to most recent snapshot
- **NPI/NDC policy calculations** revision of calculation method
## [4.2.0] - 2020-04-15
This release version is focussed on consistency between the MAgPIE setup and the [REMIND model] and result of a validation exercise of the coupled (REMIND 2.1)-(MAgPIE 4.2) system.
### added
- **config** Added new socioeconomic scenario (SDP) to scenario_config.csv (which include all switches to define among others the SSP scenarios). For the parametrization of the new SDP (Sustainable Development Pathway) scenario, the list of scenario switches was extended to account for a broad range of sustainability dimensions.
- **10_land** added new land realization landmatrix_dec18 to directly track land transition between land use types
- **15_food** stronger ruminant fade out in India
- **15_food** Added exogenous food substitution scenarios that can be selected via settings in the config-file, defining speed of convergence, scenario targets and transition periods (applied after the food demand model is executed). Among these scenarios are the substitution of livestock products with plant-based food commodities and the substitution of beef or fish with poultry. The food substitution scenarios are based on the model-native, regression-based calculation of food intake and demand.
- **15_food** Added exogenous EAT Lancet diet scenarios: It is now possible to define in the config-file exogenous diet scenarios that replace the regression-based calculation of food intake and demand. Possible settings are the target for total calorie intake (e.g. according to a healthy BMI) and variants of the EAT Lancet diet (e.g. in addition to the flexitarian a vegetarian or vegan variant).
- **15_food** Added exogenous food waste scenarios which can be defined via settings in the config-file, including scenario targets for the ratio between food demand and intake and the year in which full transition to the target should be achieved.
- **30_crop** added crop specific land use initialization pattern (used as interface for other modules)
- **50_nr_soil_budget and 55_awms** Additional inputs for the GoodPractice Scenario.
- **52_carbon** Added new forest growth curve parameters based on Braakhekke et al. 2019. Growth curves are now differentiated between natural vegetation (default) and plantations.
- **59_som** added new realization static_jan19 (new default) including all soil carbon related calculations. Before all carbon pools were updated in the specific land use type modules. This still holds true for the above ground pools (vegetation and litter carbon)
- **.gitattributes** file added to set line ending handling to auto for all text files
- **scaling** added scaling.gms files for several modules to improve optimization (based on gdx::calc_scaling)
- **scripts** added output scripts for global soil carbon maps (SoilMaps.R).
### changed
- **config** new default ghg emission pricing policy "redd+_nosoil" in c56_emis_policy. Includes all pools included in the previous default "SSP_nosoil", and in addition "forestry".
- **13_tau** lower bound for vm_tau for historical time steps
- **50_nr_soil_budget** atmospheric deposition is now estimated on the cluster-level instead of the region level to improve spatial patterns.
- **56_ghg_policy** updated scenarios in f56_emis_policy: none, all natural (called 'ssp') and all land use change emissions (pure co2) being included in greenhouse gas pricing. ssp and all also featuring additional scenarios excluding soil carbon pricing (marked with '_nosoil' postscript).
- **56_ghg_policy** Several changes regarding afforestation: use of detailed formula for incentive calculation instead of simplified Hotelling formula, 50 year planning horizon (instead of 80 years), phase-in of GHG prices deactivated by default (now done in REMIND), CO2 price reduction factor deactivated by default, introduced buffer reduction factor of 20% for afforestation.
- **59_som** updated cellpool_aug16 realization to use new interfaces from land module on land use type specific land expansion and reduction as well as crop type specific land initialization pattern. Additionally added irrigation as stock change factor sub-type. N fertilizer from soil organic matter decomposition is truncated after threshold to avoid unrealistically high fertilization rates.
- **80_optimization** write extended run information in list file in the case that the final solution is infeasible
- **modules** modular structure updated from version 1 to version 2
- **line endings** changed to unix-style for all text files
### fixed
- **modules** Fixing of all parameters to SSP2 values until 2020 (switch sm_fix_SSP2) for having identical outcomes in all scenarios (SDP, SSP1-5) until 2020.
- **21_trade** Bugfix kall instead of k in exo realization; Bufix begr/betr trade in default realization; Bugfix sets in free realization
- **32_forestry** NPI/NDC afforestation targets are now counted towards the global afforestation limit, which can be set for specific scenarios via the switch *s32maxaff_area* and constrains the potential for carbon-price induced endogenous afforestation.
- **56_ghg_policy** bugfix full soil carbon loss in default setting, renamed it from ssp to ssp_nosoil, indicating, that soil carbon losses are not priced.
- **56_ghg_policy** bugfix afforestation: vmbtm_cell was a free variable for some sources and pollutants, which could result in GHG cost neutral shifting of age classes to ac0 (e.g. from ac55 to ac0).
- **80_optimization** added fallback routine for CONOPT4 failure (fatal system error)
## [4.1.1] - 2020-03-09
This version provides the model version used for the publication starved, stuffed and wasteful. It provides a few technical updates compared to the 4.1 release, which include
### added
- **scripts** a startscript that allows the exchange of model parameters as a sensitivity analysis
### changed
- **core** allow for flexible calibration period of the model, which allows for uncalibrated runs of the past for validation purposes
- **15_food** Parameters for bodyheight regressions were included explicitly as input parameters
- **config** updated input data of the drivers and food demand regressions
### fixed
- **15_food** Precision of iteration convergence criterium for magpie-demandmodel-iteration is calculated more precisely, avoiding unnecessary iterations.
## [4.1.0] - 2019-05-02
This release version is focussed on consistency between the MAgPIE setup and the [REMIND model] and result of a validation exercise of the coupled REMIND-MAgPIE system.
### added
- **80_optimization** added support for GAMS version 26.x.x
- **scripts** added new start and output scripts
- **license** added exception to the applied AGPL license to clarify handling of required GAMS environment, solver libraries and R libraries
### changed
- **56_ghg_policy** apply reduction factor on CO2 price to account for potential negative side effects; lowers the economic incentive for CO2 emission reduction (avoided deforestation) and afforestation
- **56_ghg_policy** non-linar phase-in of GHG prices over 20 year period
- **56_ghg_policy** multiply GHG prices with development state to account for institutional requirements needed for implementing a GHG pricing scheme
- **40_transport** introduced transport costs for monogastric livestock products
- **NPI/NDC scripts** added forest protection policy for Brazilian Atlantic Forest in default NDC and NPI scenarios
- **NPI/NDC scripts** harmonized the starting year of the NDC policies 2020.
- **interpolation scripts** changed output files to seven magpie land use types, added additional cropsplit script for more detailed cropland output
- **15_food** clean-up and cosmetic changes (correction of comments, parameter names, structure of code); update BMI share calculations with the values of the last consistent MAgPIE/food-demand-model iteration
### fixed
- **42_water_demand** bugfix environmental flow policy harmonization for historic period
- **57_maccs** correction of cost calculation; Conversion from USD per ton C to USD per ton N and USD per ton CH4 was missing.
- **71_diagg_lvst** adjusted monogastric disaggregation for more flexiblity to avoid infeasibilities with EFPs (see 42_water_demand)
- **15_food** correction regarding the convergence measure of the iterative execution of the food demand model and MAgPIE; correction accounting for unusual time step length in body height calculations; body height regression parameters updated
## [4.0.1] - 2018-10-05
### fixed
- **FABLE** adapted FABLE-specific configuration so that it works with MAgPIE 4.0
## [4.0.0] - 2018-10-04
First open source release of the framework. See [MAgPIE 4.0 paper](https://doi.org/10.5194/gmd-12-1299-2019) for more information.
[Unreleased]: https://github.com/magpiemodel/magpie/compare/v4.7.2...develop
[4.7.2]: https://github.com/magpiemodel/magpie/compare/v4.7.1...v4.7.2
[4.7.1]: https://github.com/magpiemodel/magpie/compare/v4.7.0...v4.7.1
[4.7.0]: https://github.com/magpiemodel/magpie/compare/v4.6.11...v4.7.0
[4.6.11]: https://github.com/magpiemodel/magpie/compare/v4.6.10...v4.6.11
[4.6.10]: https://github.com/magpiemodel/magpie/compare/v4.6.9...v4.6.10
[4.6.9]: https://github.com/magpiemodel/magpie/compare/v4.6.8...v4.6.9
[4.6.8]: https://github.com/magpiemodel/magpie/compare/v4.6.7...v4.6.8
[4.6.7]: https://github.com/magpiemodel/magpie/compare/v4.6.6...v4.6.7
[4.6.6]: https://github.com/magpiemodel/magpie/compare/v4.6.5...v4.6.6
[4.6.5]: https://github.com/magpiemodel/magpie/compare/v4.6.4...v4.6.5
[4.6.4]: https://github.com/magpiemodel/magpie/compare/v4.6.3...v4.6.4
[4.6.3]: https://github.com/magpiemodel/magpie/compare/v4.6.2...v4.6.3
[4.6.2]: https://github.com/magpiemodel/magpie/compare/v4.6.1...v4.6.2
[4.6.1]: https://github.com/magpiemodel/magpie/compare/v4.6.0...v4.6.1
[4.6.0]: https://github.com/magpiemodel/magpie/compare/v4.5.0...v4.6.0
[4.5.0]: https://github.com/magpiemodel/magpie/compare/v4.4.0...v4.5.0
[4.4.0]: https://github.com/magpiemodel/magpie/compare/v4.3.5...v4.4.0
[4.3.5]: https://github.com/magpiemodel/magpie/compare/v4.3.4...v4.3.5
[4.3.4]: https://github.com/magpiemodel/magpie/compare/v4.3.3...v4.3.4
[4.3.3]: https://github.com/magpiemodel/magpie/compare/v4.3.2...v4.3.3
[4.3.2]: https://github.com/magpiemodel/magpie/compare/v4.3.1...v4.3.2
[4.3.1]: https://github.com/magpiemodel/magpie/compare/v4.3.0...v4.3.1
[4.3.0]: https://github.com/magpiemodel/magpie/compare/v4.2.1...v4.3.0
[4.2.1]: https://github.com/magpiemodel/magpie/compare/v4.2.0...v4.2.1
[4.2.0]: https://github.com/magpiemodel/magpie/compare/v4.1.1...v4.2.0
[4.1.1]: https://github.com/magpiemodel/magpie/compare/v4.1.0...v4.1.1
[4.1.0]: https://github.com/magpiemodel/magpie/compare/v4.0.1...v4.1.0
[4.0.1]: https://github.com/magpiemodel/magpie/compare/v4.0...v4.0.1
[4.0.0]: https://github.com/magpiemodel/magpie/releases/tag/v4.0
[REMIND model]: https://www.pik-potsdam.de/research/transformation-pathways/models/remindcontentwise ....
What's new? - data updates (selection)
updated GDP & population scenarios (4.4.0)
updated FAO data (4.4.0)
new diet scenarios (4.2.0|4.3.0|4.6.0|4.7.1)
new Sustainable Development Pathway (SDP) scenarios (4.2.0 | 4.4.0)
new factor cost estimates (4.3.0|4.3.5|4.4.0)
updated Marginal Abatement Cost Curves - MACCS (4.6.0)
new data on areas equipped for irrigation (4.6.8)
updated transport costs (4.6.10)
What's new? - selected features
peatland simulation (4.2.1|4.3.5|4.4.0)

What's new? - selected features
dynamic forestry / timber production (4.3.0|4.3.2|4.3.5)

What's new? - selected features
sticky costs (4.3.0|4.3.5|4.4.0)
urban land expansion (4.4.0)
exogenous growth scenarios
simulation of set aside cropland (4.3.5)
(e.g. for its biodiversity implications)
updated yield calibration & updated LPJmL yield data (4.3.5)
new nitrogen emission realization (4.3.2|4.3.4|4.4.0)
added emissions from residue burning (4.3.4)
switched from 59k to 67k cells for spatial data (4.7.0)
What's new? - selected features
factor costs split into labor and capital (4.5.0 | 4.6.2)
new pasture realization with pasture / rangeland split (4.5.0|4.6.3)
land conservation module added (4.5.0)
new bilateral trade realization (4.6.4)
new land conversion cost calibration (4.4.0)
employment module added (4.5.0|4.6.0|4.6.2|4.7.0)
Further Reading
This presentation - slides.com/jandietrich/magpie-4-7
MAgPIE 4 framework paper - https://doi.org/10.5194/gmd-12-1299-2019
MAgPIE Repository | github.com/magpiemodel
R packages Repository | github.com/pik-piam
model requests | mxyzagpie@pik-poxyztsdam.de
contact me | dietrixyzch@pikxyz-potsdam.de


