Viral evolution and COVID-19 vaccines
Jesse Bloom
Fred Hutch Cancer Center / HHMI
These slides at https://slides.com/jbloom/awaji-cov-vax
Broad evolutionary patterns in human coronavirus evolution relevant to vaccines
CoV-229E causes common colds and has been circulating in humans for a long time.
How do other human coronaviruses evolve?
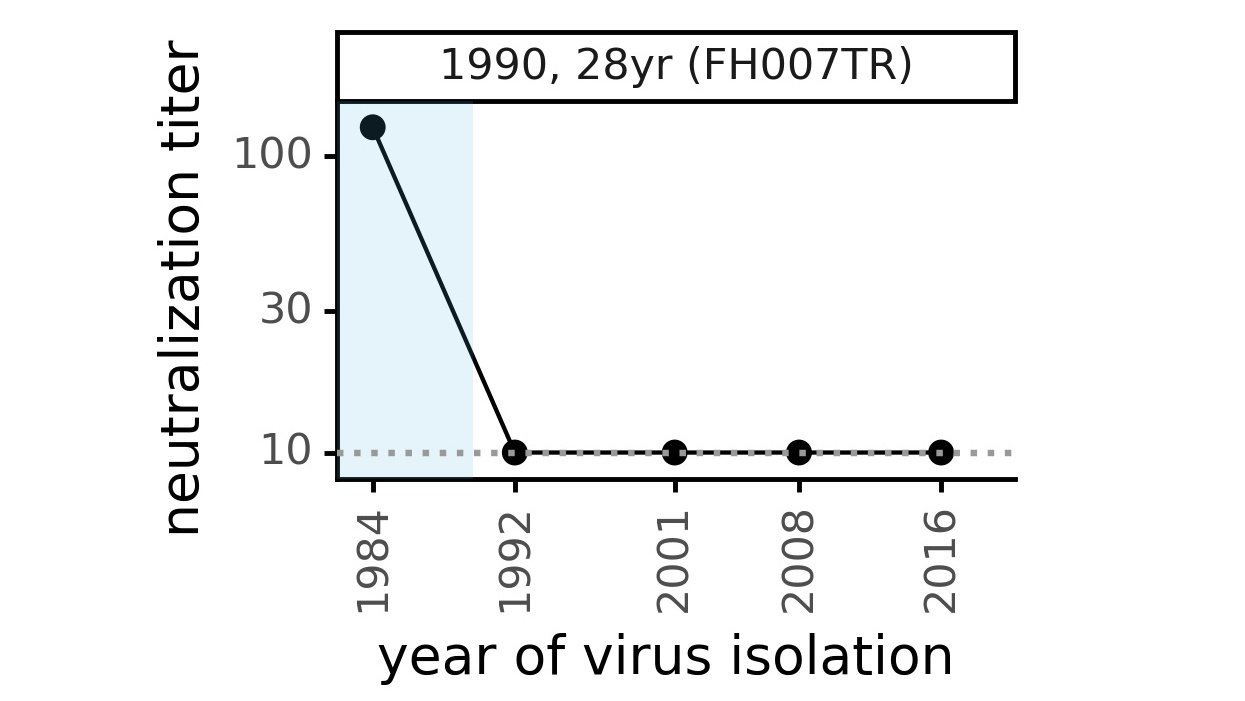
Evolution of CoV-229E spike

We experimentally generated CoV-229E spikes at ~8 year intervals to study in the lab:
- 1984
- 1992
- 2001
- 2008
- 2016
Evolution of CoV-229E spike erodes neutralization by human serum antibodies
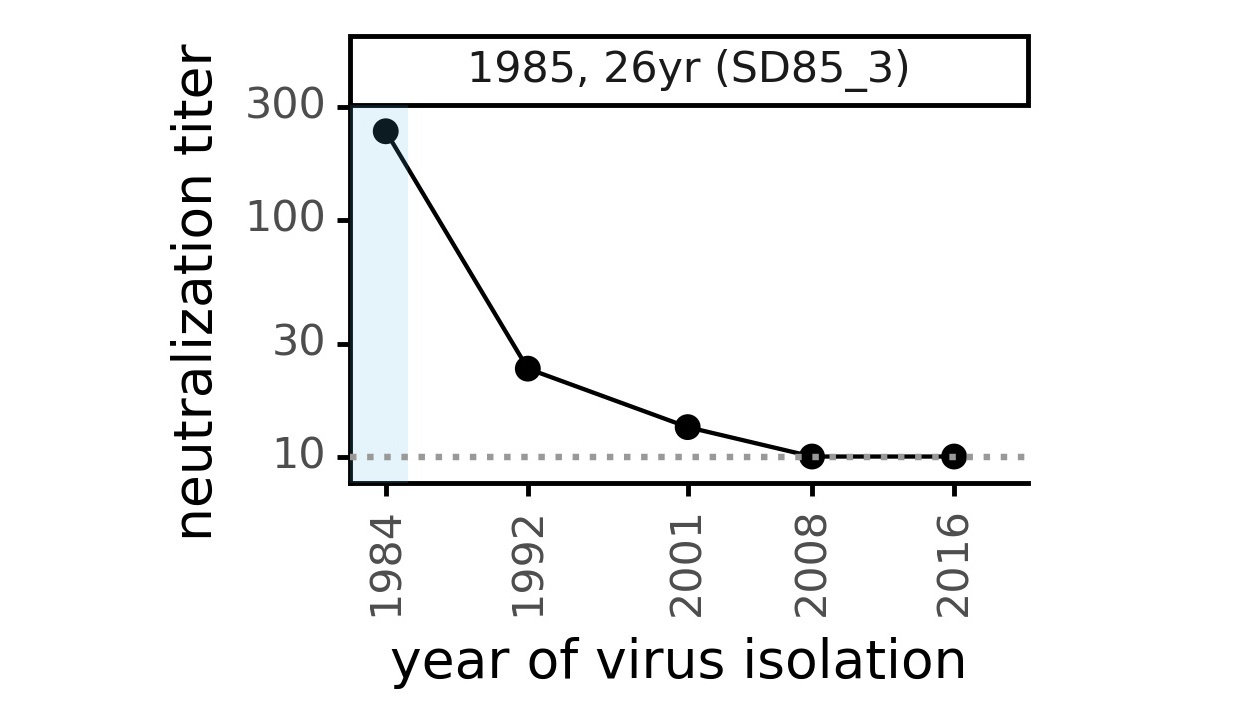

Ideally vaccines would elicit evolution-resistant antibodies (like those made by person at right) rather than evolution-sensitive antibodies (like those made by person at left)
Evolution erodes neutralization by sera from different people at different rates
We now know SARS-CoV-2 evolves to erode neutralizing antibodies, just like CoV-229E
neutralization from original COVID-19 vaccine
Original vaccine induced high neutralizing antibody titers against early viral strains
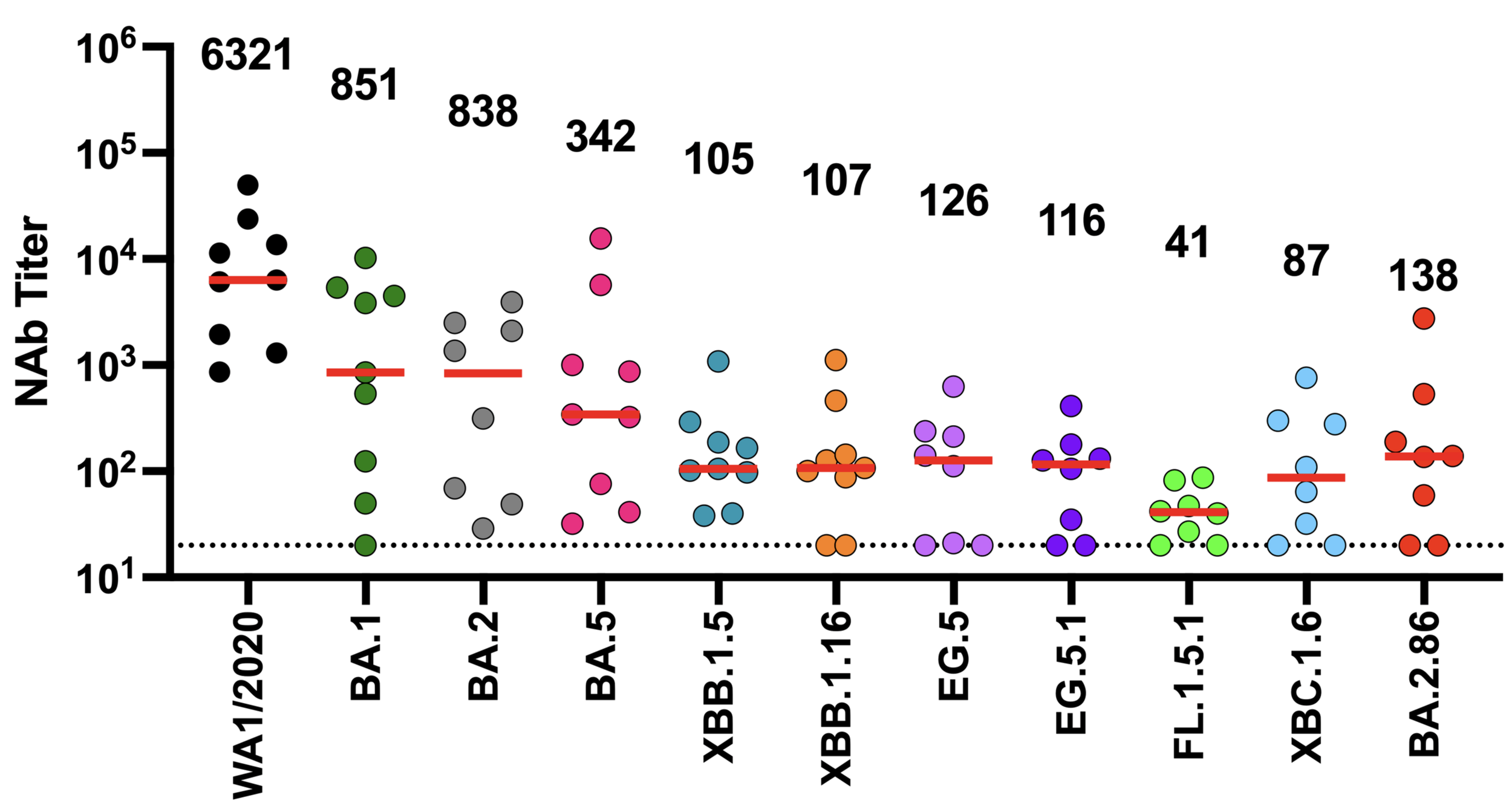
We now know SARS-CoV-2 evolves to erode neutralizing antibodies, just like CoV-229E
newer viral variants
neutralization from original COVID-19 vaccine

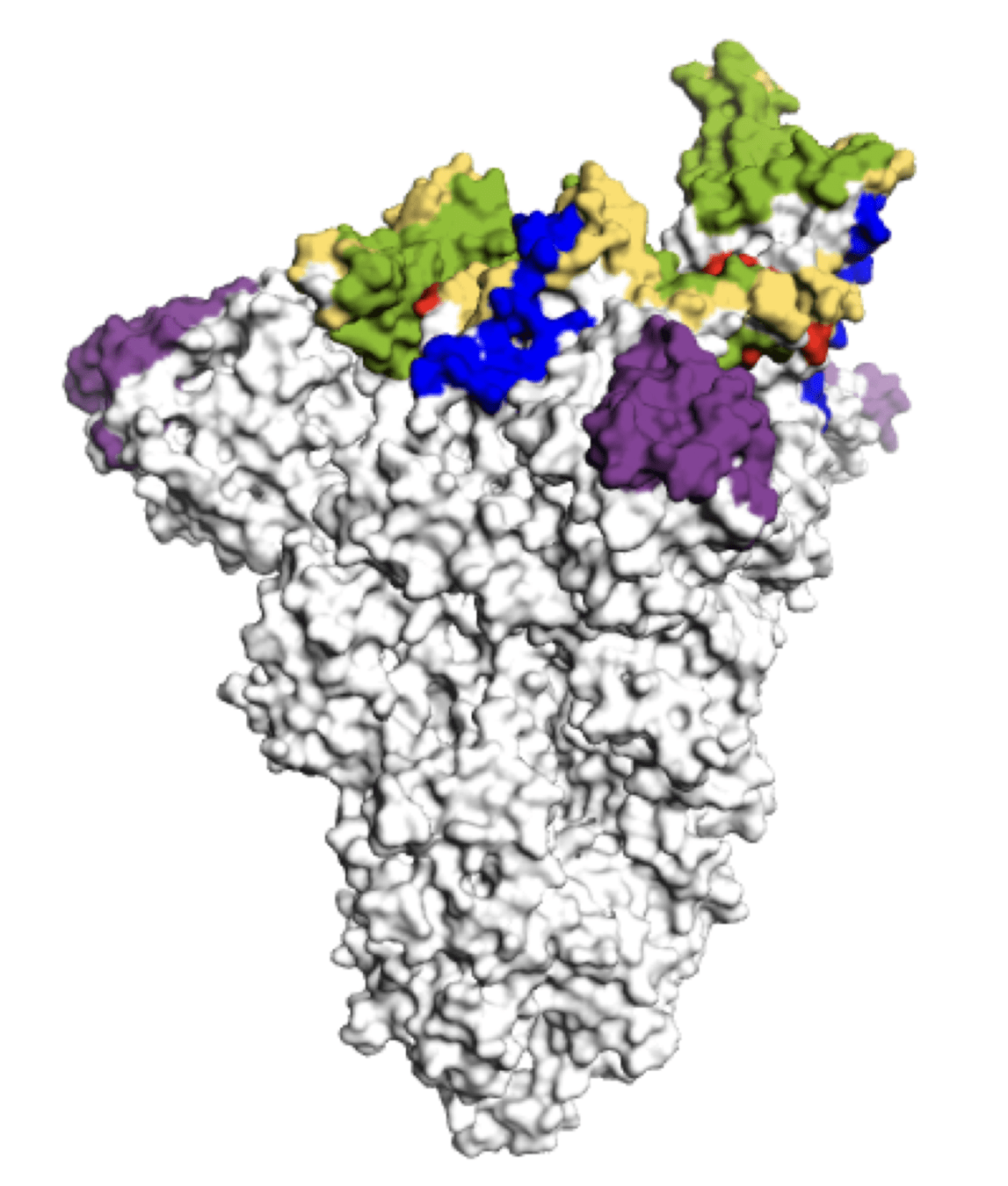
Main regions where neutralizing antibodies bind
Molecular basis of this antigenic evolution?

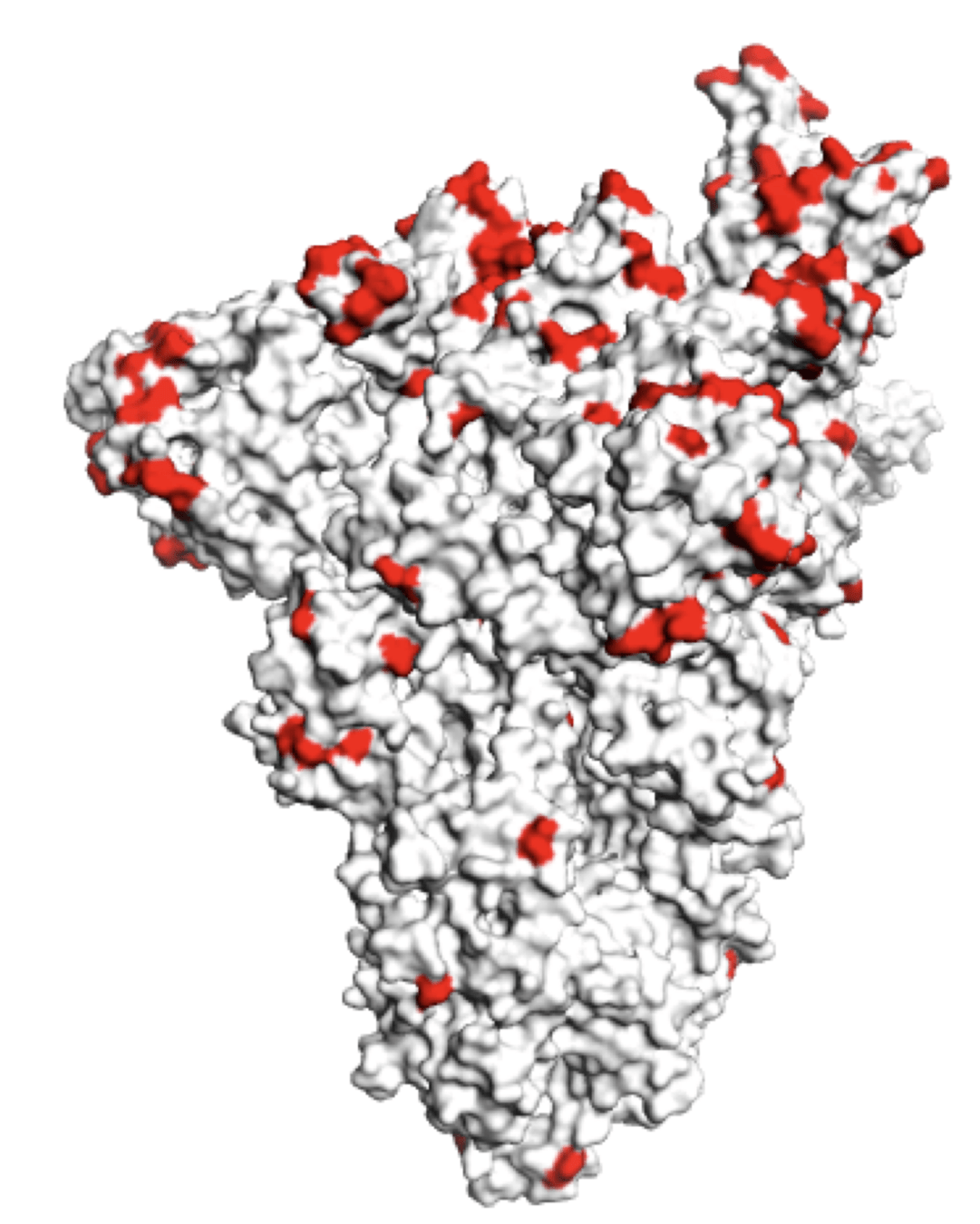

Molecular basis of this antigenic evolution?Mutations where antibodies bind
Main regions where neutralizing antibodies bind
Sites of mutations in recent (BA.2.86) SARS-CoV-2 strain relative to early 2020 strain
In SARS-CoV-2 evolution, new variants replace old ones (just like for CoV-229E)
Sites of evolutionary change in the spike of CoV-229E over the last four decades
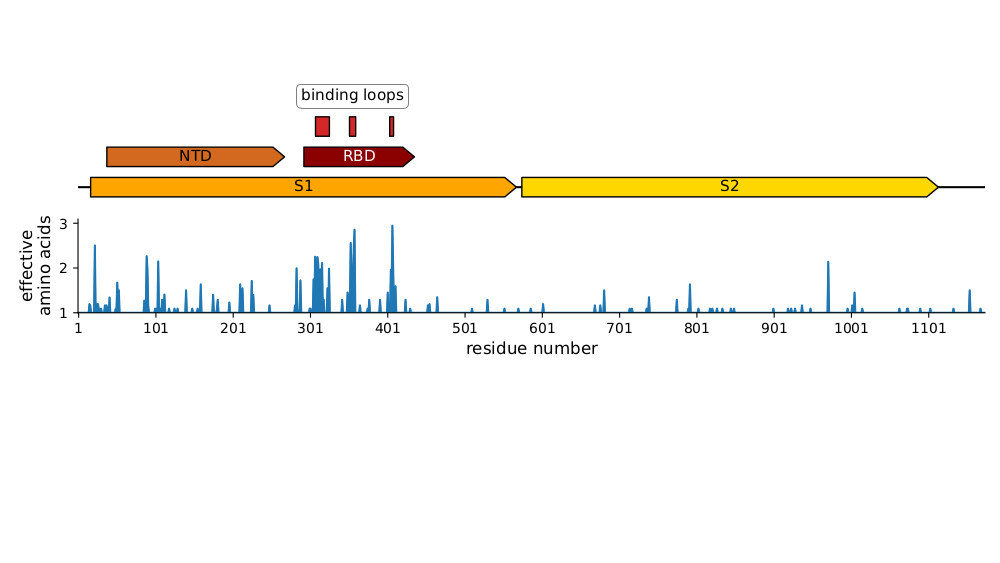
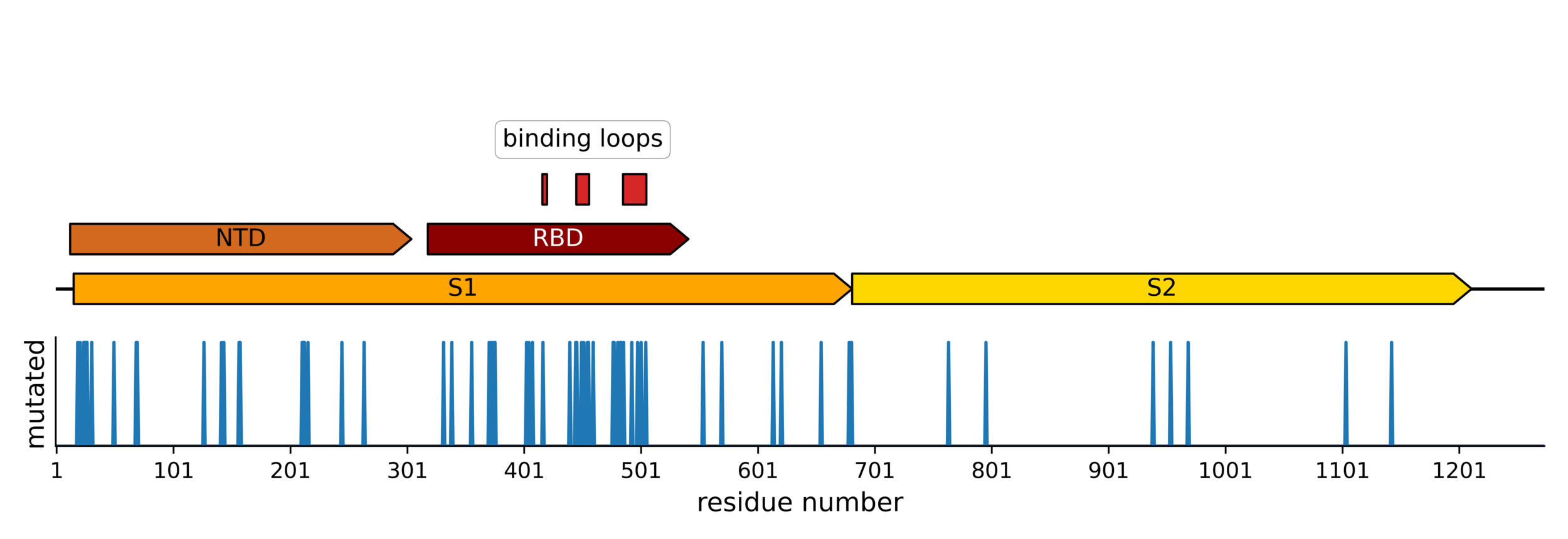
Sites of mutations in the current KP.3.1.1 SARS-CoV-2 variant relative to the early Wuhan-Hu-1 strain
CoV-229E and SARS-CoV-2 acquire mutations in analogous regions of spike
The most intense evolutionary selection is in the receptor-binding domain (RBD)
Phylogenetic tree shape & vaccine strategy
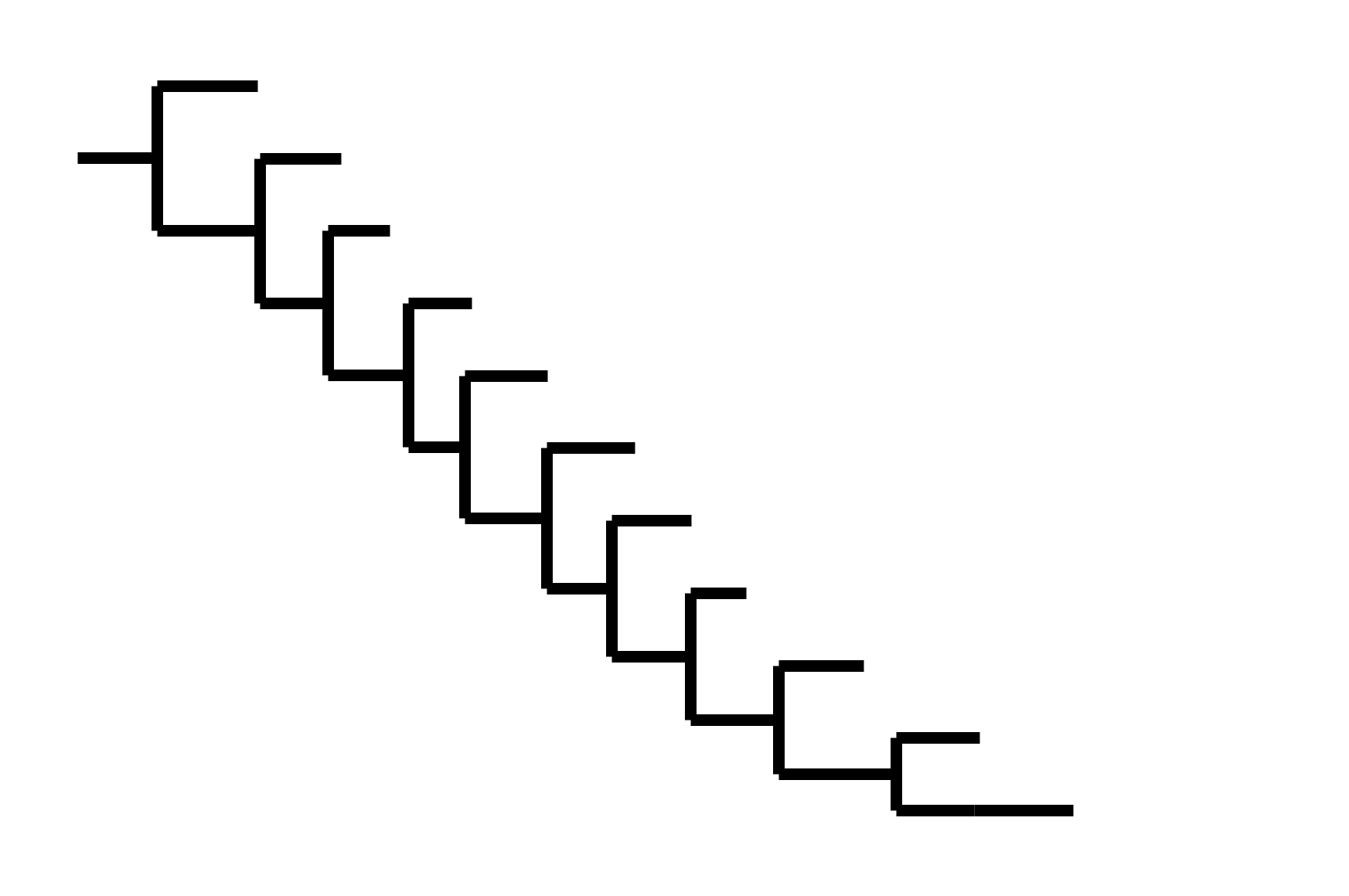
CoV-229E has ladder-like tree: new variants displace old ones.
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.
Phylogenetic tree shape & vaccine strategy

CoV-229E has ladder-like tree: new variants displace old ones.
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.
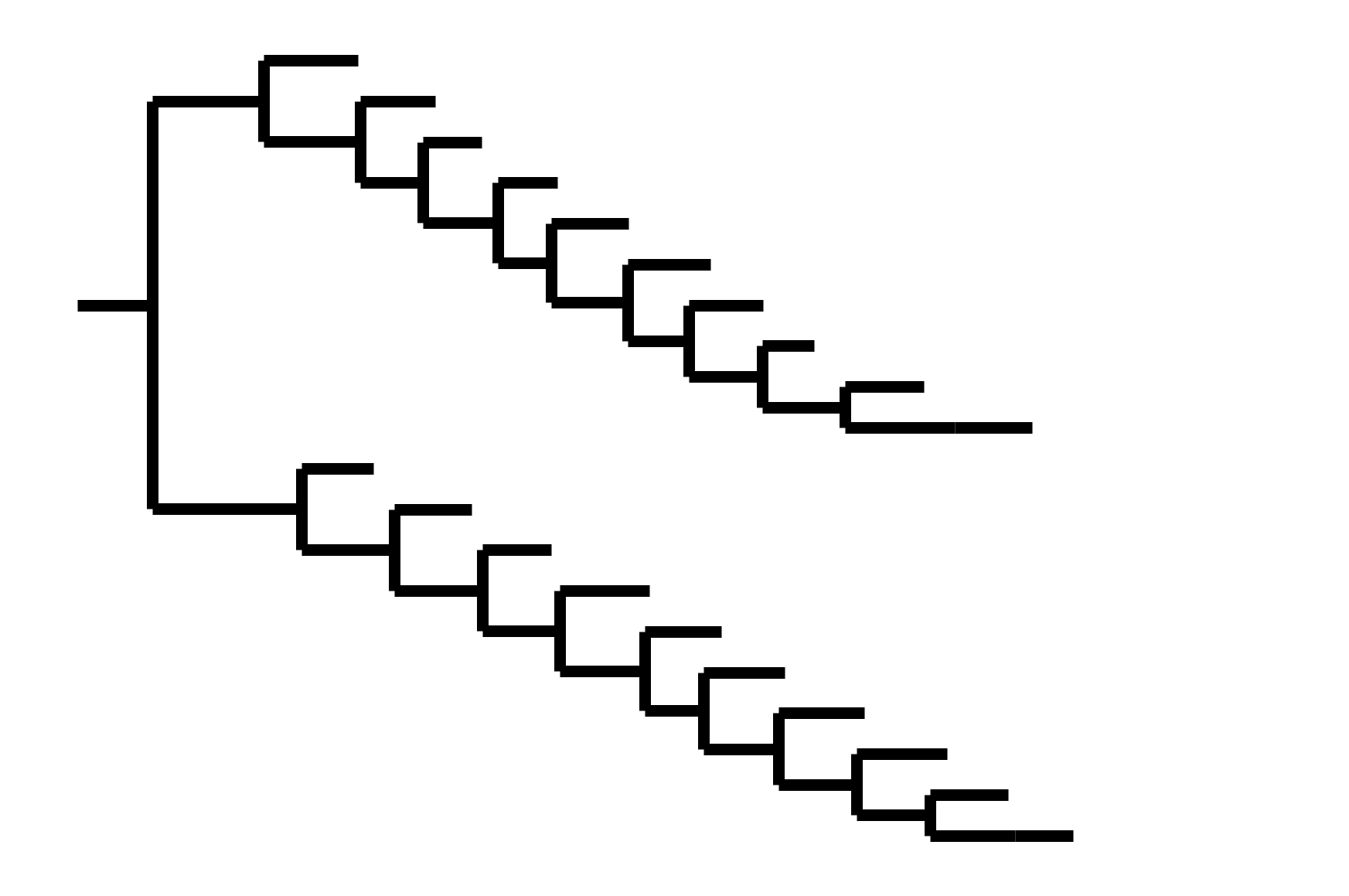
CoV-OC43 split into two ladder-like lineages. Influenza B evolved this way too (until very recently). It's theoretically possible to pick well-matched bivalent vaccine.
Phylogenetic tree shape & vaccine strategy

CoV-229E has ladder-like tree: new variants displace old ones.
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.

CoV-OC43 split into two ladder-like lineages. Influenza B evolves this way too. It's theoretically possible to pick well-matched bivalent vaccine.
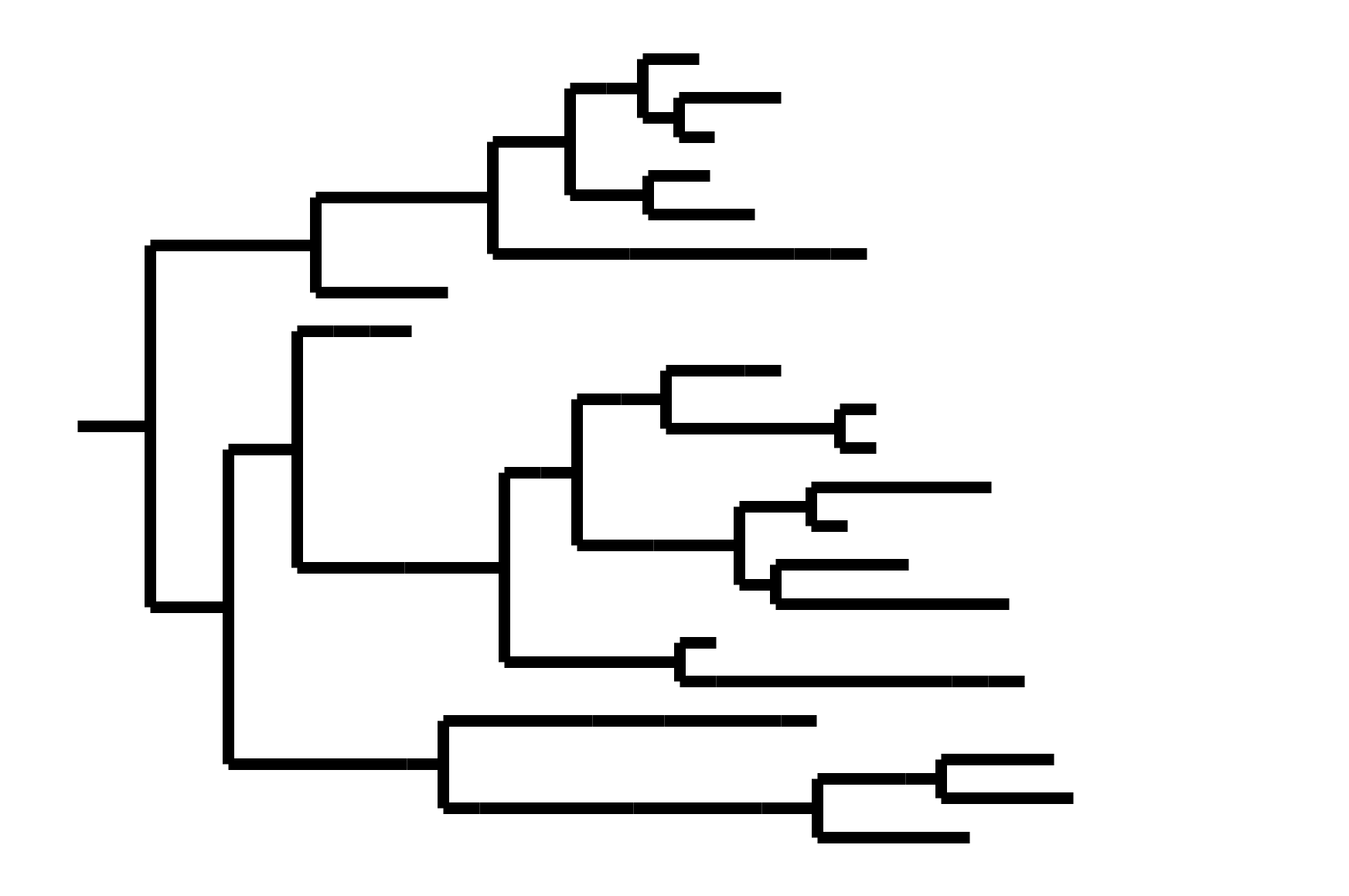
In non-ladder-like tree, there can be high standing genetic variation. Makes picking vaccine strains difficult.
Mutational drivers of SARS-CoV-2 antigenic evolution
Types of mutations relevant to SARS-CoV-2 antigenic evolution:
- Direct antibody-escape mutations in the RBD
- Direct antibody-escape mutations in the NTD
- Mutations that modulate RBD "up" versus "down" conformation
Types of mutations relevant to SARS-CoV-2 antigenic evolution:
- Direct antibody-escape mutations in the RBD
- Direct antibody-escape mutations in the NTD
- Mutations that modulate RBD "up" versus "down" conformation
The RBD is antigenically very important
As shown on an earlier slide, the RBD is the most intense region of evolutionary selection in both SARS-CoV-2 and CoV-229E.
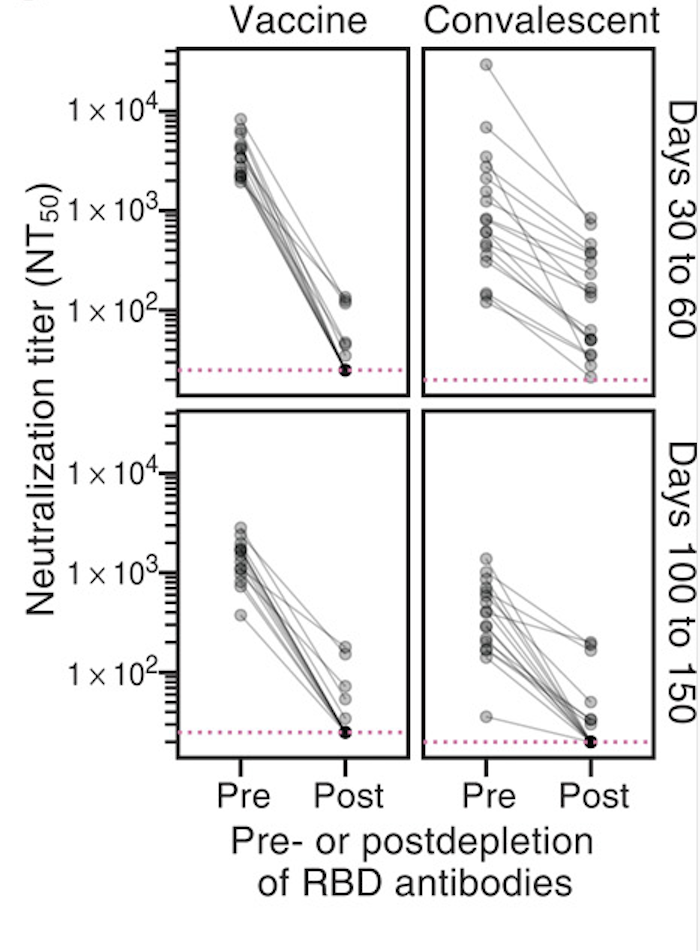
RBD-binding antibodies are responsible for most neutralizing activity in sera from humans vaccinated or infected with the original SARS-CoV-2 viral strain (see plot at right).
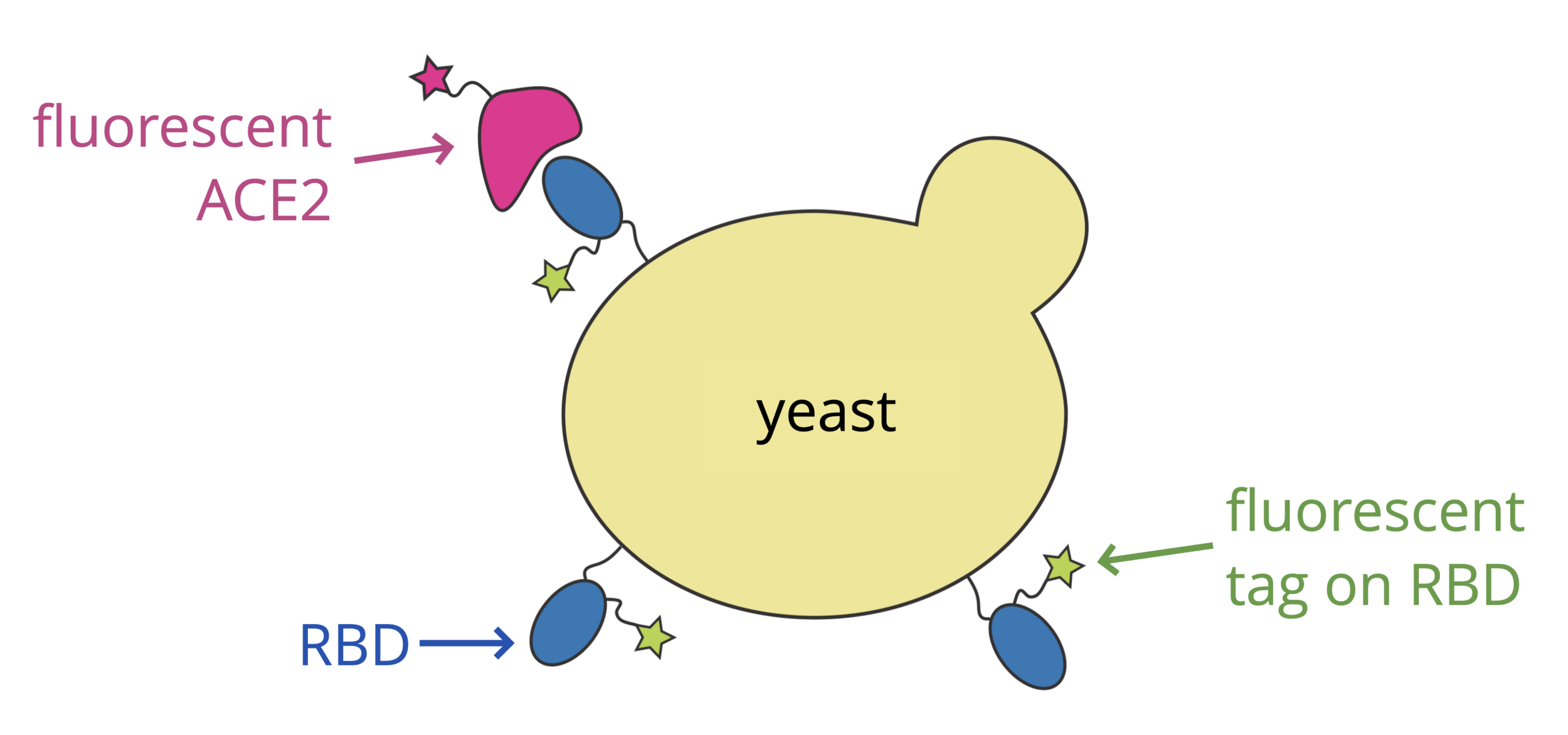
Identifying RBD mutations that escape binding by neutralizing antibodies
Libraries of RBD mutants displayed on yeast can be used to measure how all mutations affect binding by neutralizing antibodies (Greaney et al, 2021)
Yunlong Cao et al have used this approach to identify escape mutations from thousands of human antibodies (Cao et al, 2022; Yisimayi et al, 2024; Jian et al, 2024)
We have aggregated these mutations into an "escape-calculator" which helps identify which sites in RBD are the major targets of antibodies: https://jbloomlab.github.io/SARS2-RBD-escape-calc/

BA.2 had mutations at key sites of RBD escape in original RBD (eg, sites 484, 417)
XBB had mutations at key sites of RBD escape in BA.2 (eg, sites 346, 445, 446, 486)

RBD sites that currently most escape binding by neutralizing antibodies
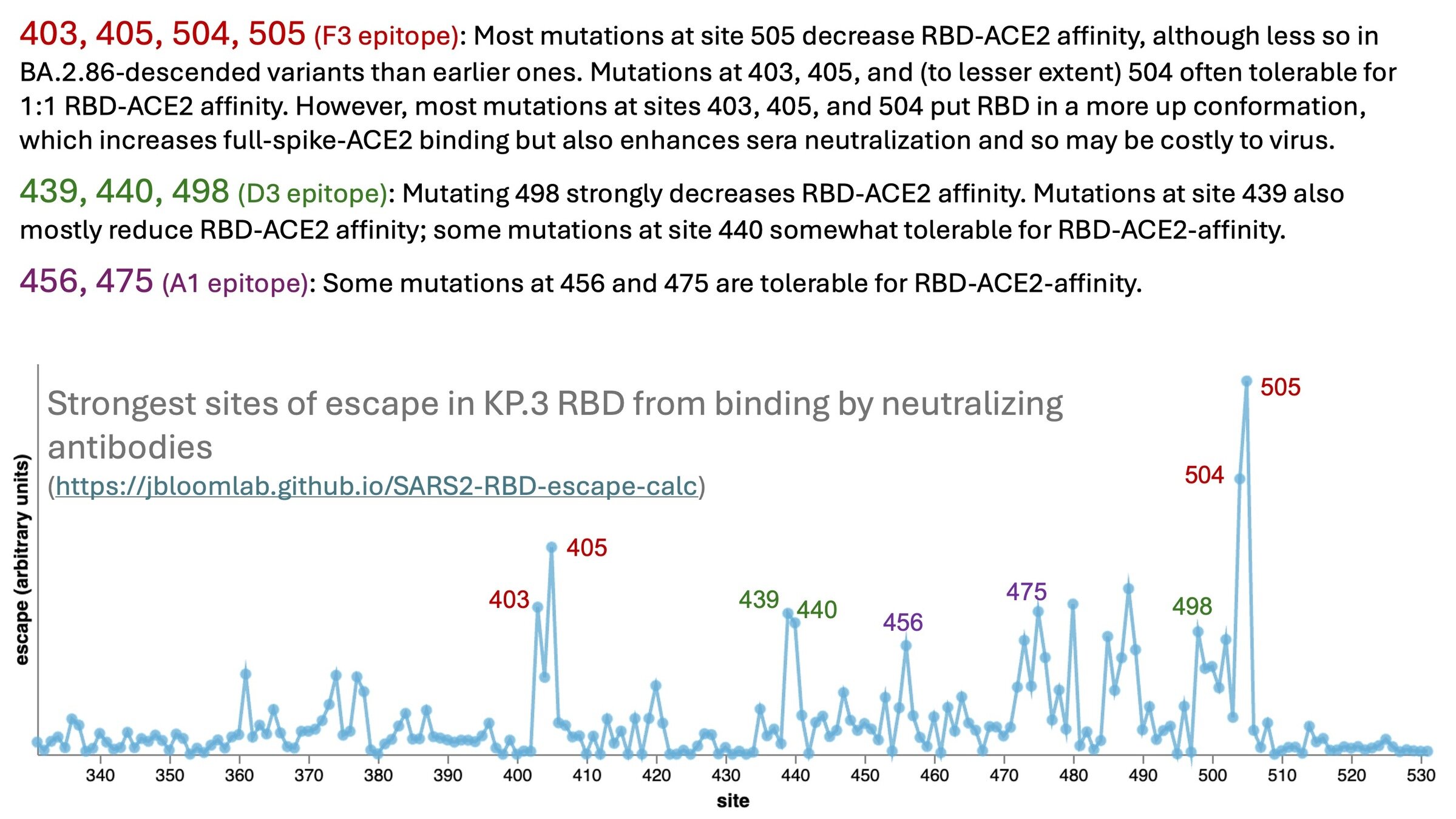
Mutations at epitope with sites 403, 405, 504, 505 currently escape binding by RBD antibodies, but mutations at these sites have pleiotropic effects on RBD up-down (see here)
Types of mutations relevant to SARS-CoV-2 antigenic evolution:
- Direct antibody-escape mutations in the RBD
- Direct antibody-escape mutations in the NTD
- Mutations that modulate RBD "up" versus "down" conformation
Although most neutralizing activity targets RBD, NTD antibodies also important
As described earlier in this talk, NTD is also a hotspot of mutations in both SARS-CoV-2 and CoV-229E. Many SARS-CoV-2 NTD mutations are in antigenic supersite.
As described in my Sunday talk at this meeting, NTD antibodies can be major contributors to neutralization in young individuals infected by recent variants
Types of mutations relevant to SARS-CoV-2 antigenic evolution:
- Direct antibody-escape mutations in the RBD
- Direct antibody-escape mutations in the NTD
- Mutations that modulate RBD "up" versus "down" conformation
Image from Liu and Wilson (2022)
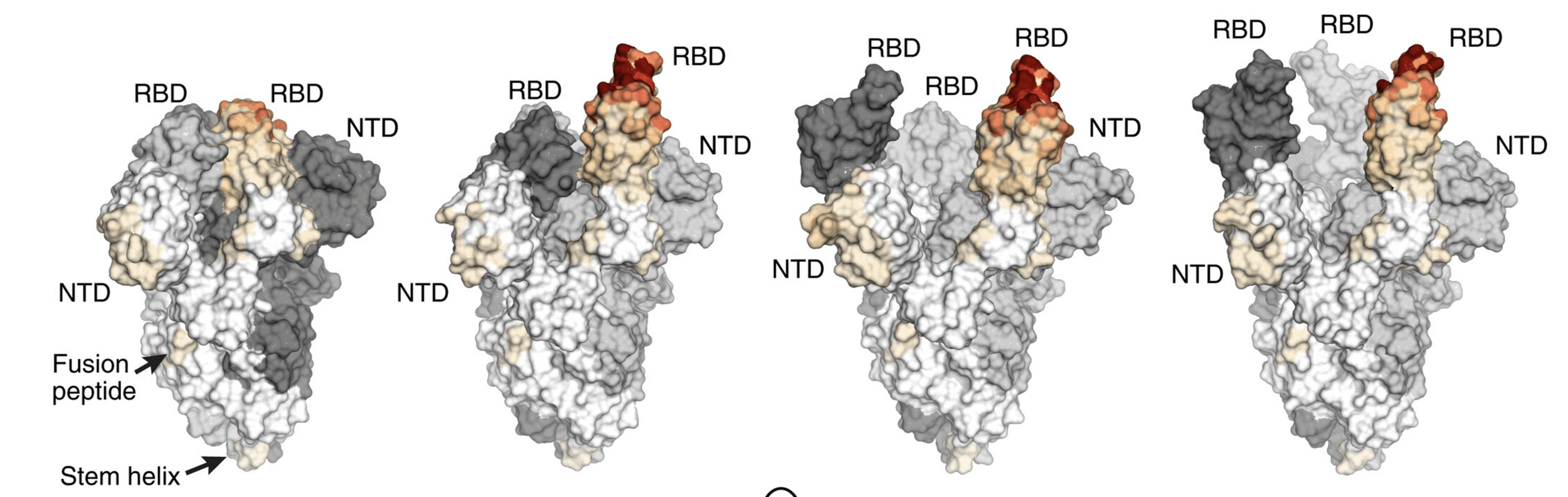
all-down RBDs
1-up RBD
2-up RBDs
3-up RBDs
RBD must go into up conformation to bind ACE2, but that increases antibody exposure
Most potent neutralizing epitopes are shown in red, and are more exposed in up RBD
Mutations that put RBD in down position reduce neutralization by polyclonal sera antibodies
Later SARS-CoV-2 variants have more down RBD than early variants (Zhang et al, 2024)
Mutations that put RBD more down have contributed to evolution of variants (Dadonaite et al, 2024), including NTD mutations that promote down RBD in recent KP.3.1.1 and XEC variants (Wang et al, 2024)
"Common-cold" coronaviruses that are highly adapted to humans have RBD mostly in down conformation (Pronker et al, 2023)
Summary
Human coronaviruses, including SARS-CoV-2, evolve to erode antibody neutralization.
Standing genetic diversity is low even though viral spike changes rapidly over time. This fact forms the basis for the strategy of regular vaccine strain updates.
Antigenic evolution is mostly due to mutations in the RBD and to a lesser extent NTD.
Initially SARS-CoV-2 antigenic evolution primarily involved mutations in RBD that escaped antibody binding. This remains important, but recently there has been increased role of mutations in NTD, including ones that shift RBD to more down position.
As exposure histories and imprinting become more diverse, the antibody pressure on SARS-CoV-2 imposed by the human population will become more heterogeneous.
Bloom lab




These slides: https://slides.com/jbloom/awaji-cov-vax
Thanks


Fred Hutch Cancer Center
Trevor Bedford
University of Washington
Helen Chu and HAARVI cohort
Neil King
David Veesler
Cincinnati Children's Hospital
Mary Staat
David Haslam
Allie Burrell


Tyler Starr (now at Utah)
Allie Greaney (now at UCSF)
Kate Crawford
William Hannon
Caelan Radford
Brendan Larsen


Bernadeta Dadonaite
Rachel Eguia