Interpreting evolution of SARS-CoV-2 and other viruses
Jesse Bloom
Fred Hutch Cancer Center / HHMI
These slides at https://slides.com/jbloom/chicago2024

The Faroe Islands


"Measles had not prevailed on the Faroes since 1781, then it broke out early in April 1846."
"Of the 7782 inhabitants, about 6000 were taken with measles."
"Of the many aged people still living in the Faroes who had measles in 1781, not one was attacked the second time."
Panum is describing immune memory, which provides lifelong protection from measles.
But we are repeatedly infected by some other viruses. Typical person infected by influenza ~5 years. Why?
History offers natural experiment with influenza like Panum's study of measles
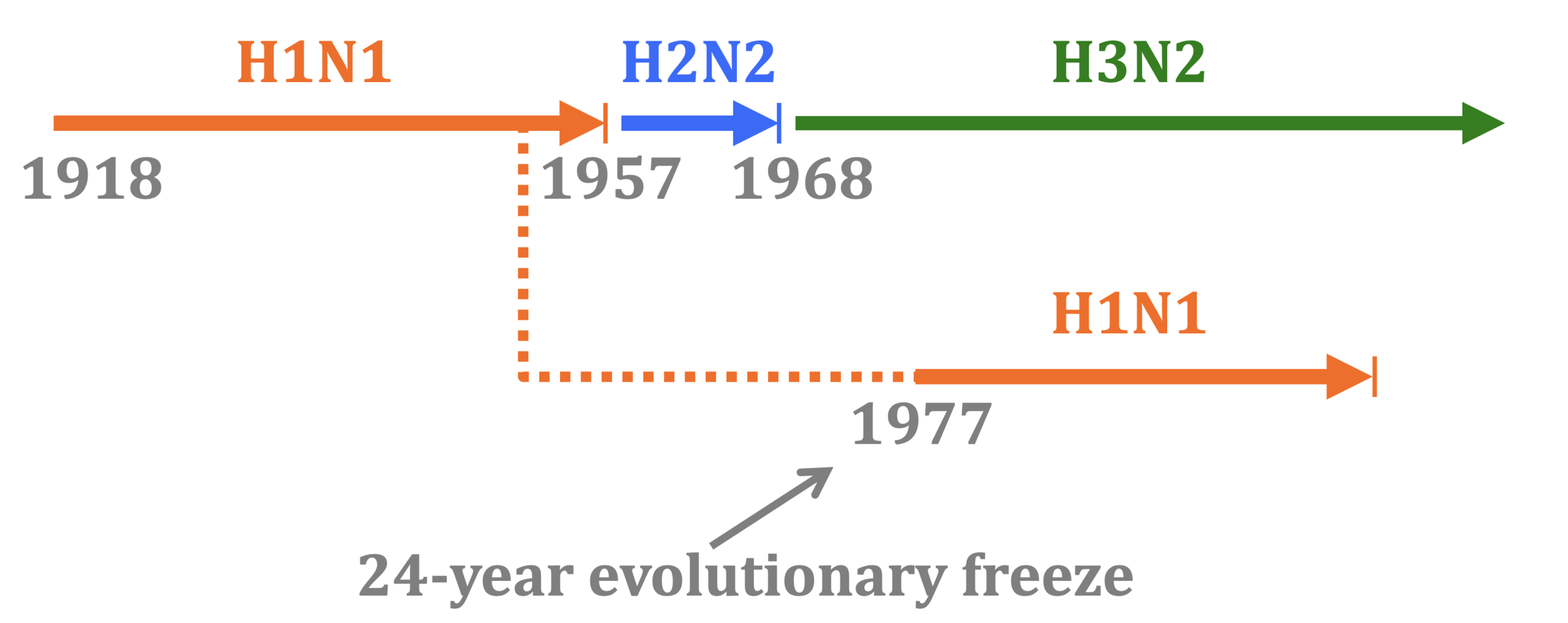
History offers natural experiment with influenza like Panum's study of measles
In 1977, old H1N1 strain from ~1954 was inadvertently re-released and caused pandemic. So re-introduction of identical virus after a few decades.



"One boy from Hong Kong had a transient febrile illness from 15 to 18 January. On Sunday 22 January, three boys were in the college infirmary… 512 boys (67%) spent between three and seven days away from class."

"Of about 130 adults who had some contact with the boys, only one, a house matron, developed similar symptoms."
Influenza infection also elicits multi-decade immunity, but only if virus is evolutionarily frozen
Some human RNA respiratory viruses evolve to escape immunity
Why some viruses evolve to escape immunity while others don't is a deep question outside scope of this talk. See here for some possible explanations.
Rate of viral antigenic evolution
Measles
Influenza

CoV-229E causes common colds and has been circulating in humans for a long time.
The typical person is infected every ~3 to 5 years.
How do other human coronaviruses evolve?
Evolution of CoV-229E spike

We experimentally generated CoV-229E spikes at ~8 year intervals so we could study them in the lab:
- 1984
- 1992
- 2001
- 2008
- 2016
Evolution of CoV-229E spike erodes neutralization by human serum antibodies
Evolution erodes CoV-229E neutralization by different sera at different rates

Ideally vaccines would elicit evolution-resistant neutralizing antibodies (like those naturally made by person at right) rather than evolution-sensitive antibodies (like those naturally made by person at left)
Phylogenetic tree shape & vaccine strategy
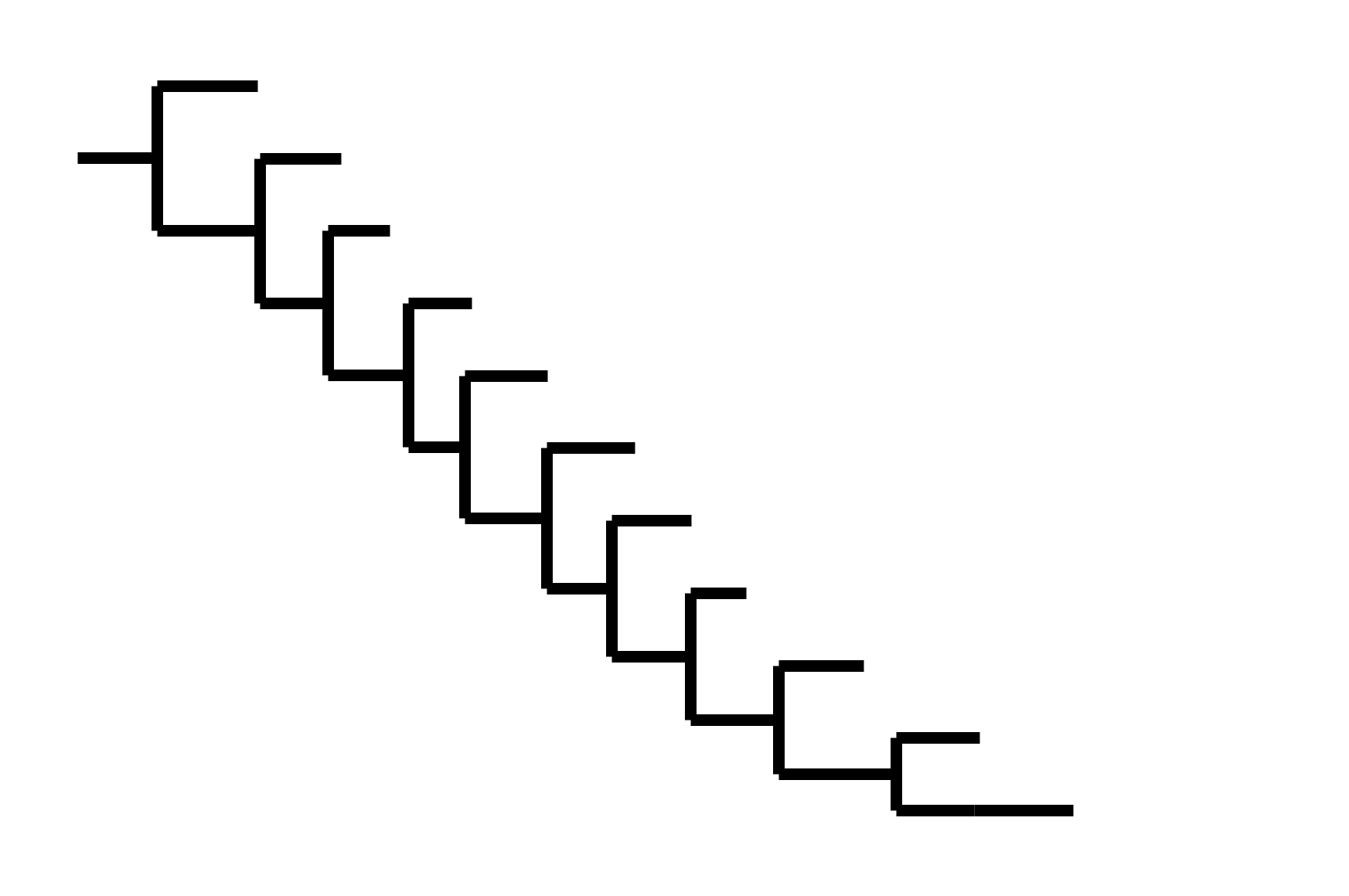
CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.
Phylogenetic tree shape & vaccine strategy

CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.
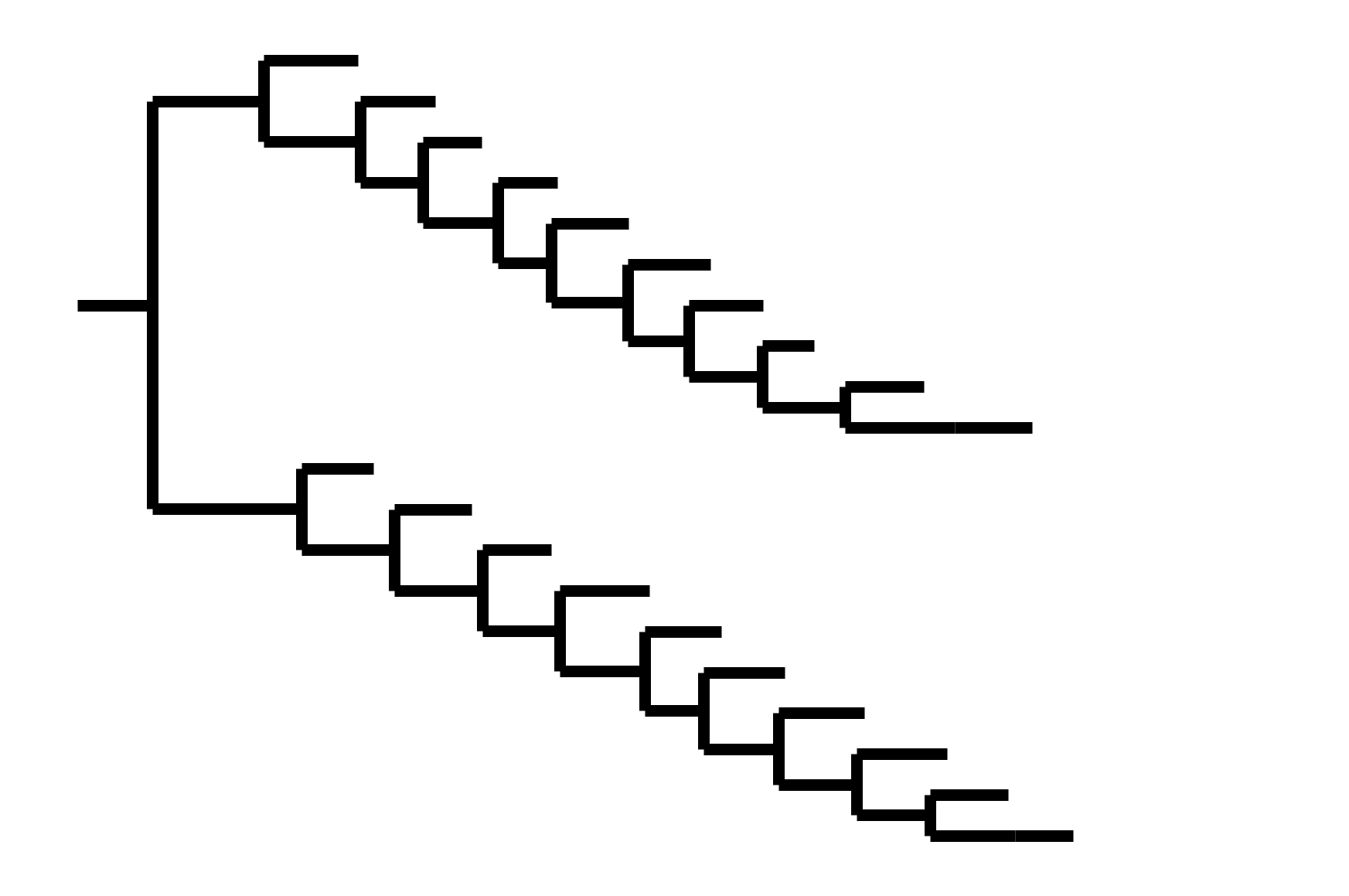
CoV-OC43 split into two ladder-like lineages. Influenza B evolves this way too. It's theoretically possible to pick well-matched bivalent vaccine.
Phylogenetic tree shape & vaccine strategy

CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.

CoV-OC43 split into two ladder-like lineages. Influenza B evolves this way too. It's theoretically possible to pick well-matched bivalent vaccine.
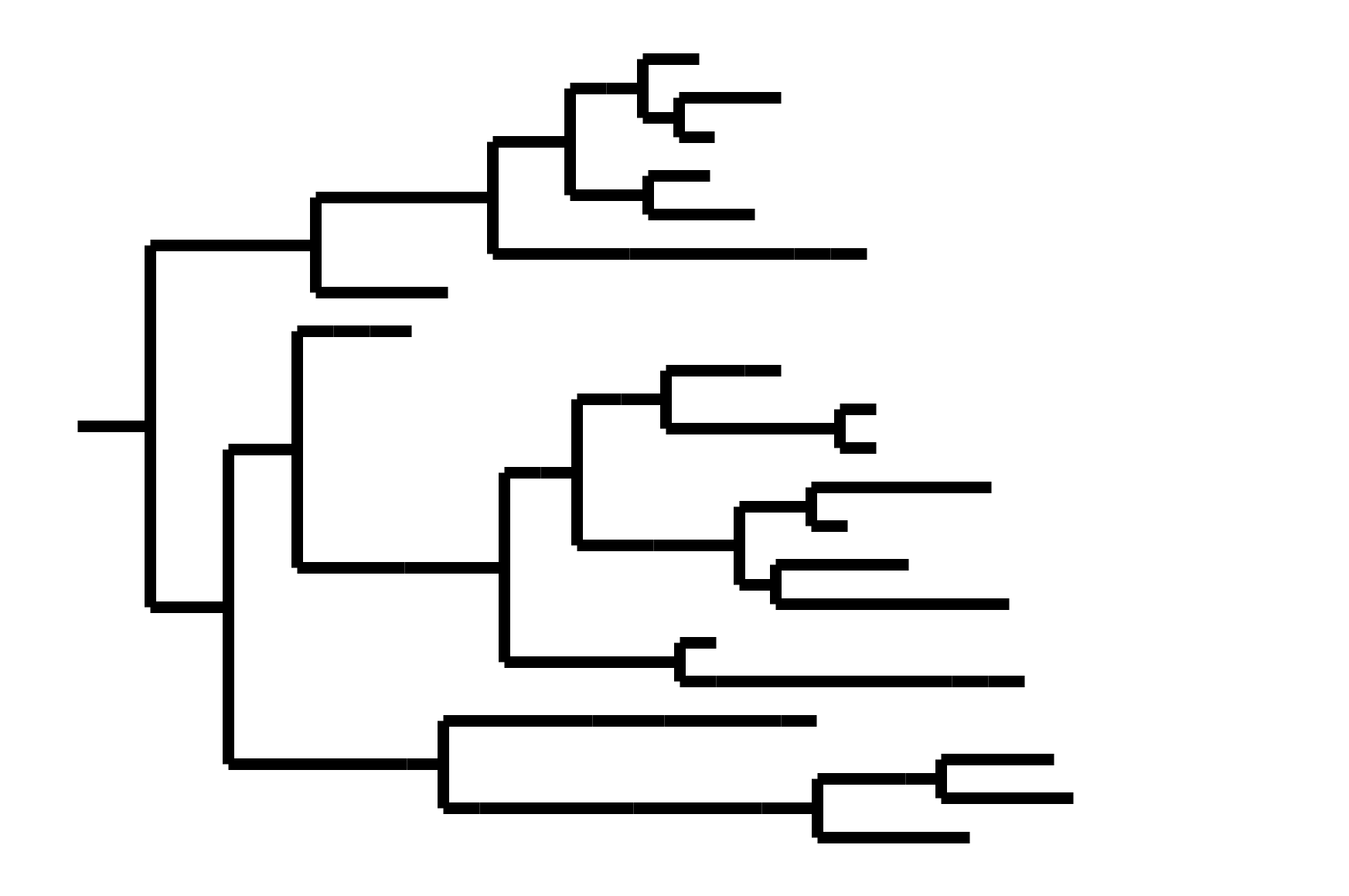
In non-ladder-like tree, there can be high standing genetic variation. Makes picking vaccine strains difficult.
Can we measure molecular phenotypes that shape viral fitness?
All enveloped viruses have one or more entry proteins that bind receptor and fuse the viral and cell membranes
- SARS-CoV-2 spike
- influenza hemagglutinin
- HIV envelope protein
- Lassa virus glycoprotein
- Nipah virus G and F proteins
- RSV G and F proteins
Several viral entry protein molecular phenotypes are under strong selection
- Receptor binding
- Cell entry
- Escape from antibodies

Deep mutational scanning of viral entry proteins for key molecular phenotypes
Considerations for experimental approach
- Comprehensive: measure as many relevant phenotypes as possible
- High-throughput: can measure effects of all amino-acid mutations
- Safe: we want to avoid making potentially dangerous viral mutants
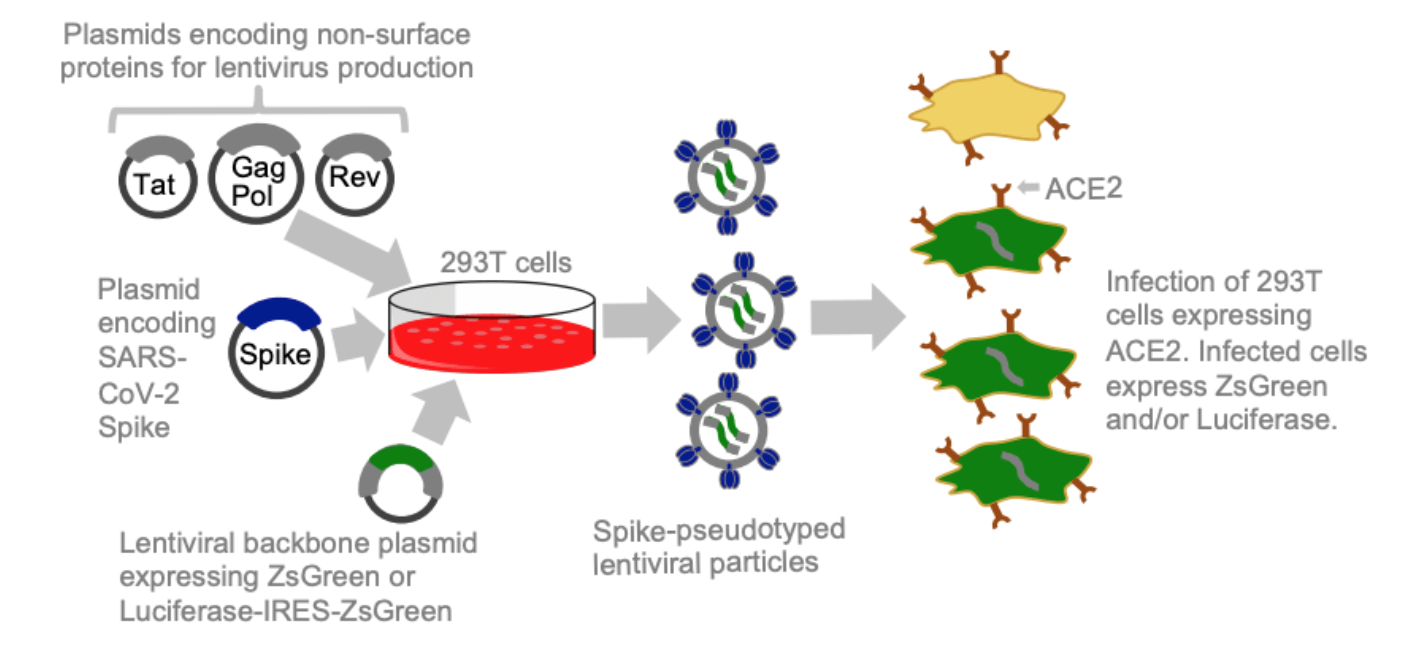
Lentiviral pseudotyping
- Many enveloped viruses have entry proteins amenable to lentiviral pseudotyping.
- Safe and well-established way to study cell entry function of these proteins
- However, traditional pseudotyping does not create genotype-phenotype link.
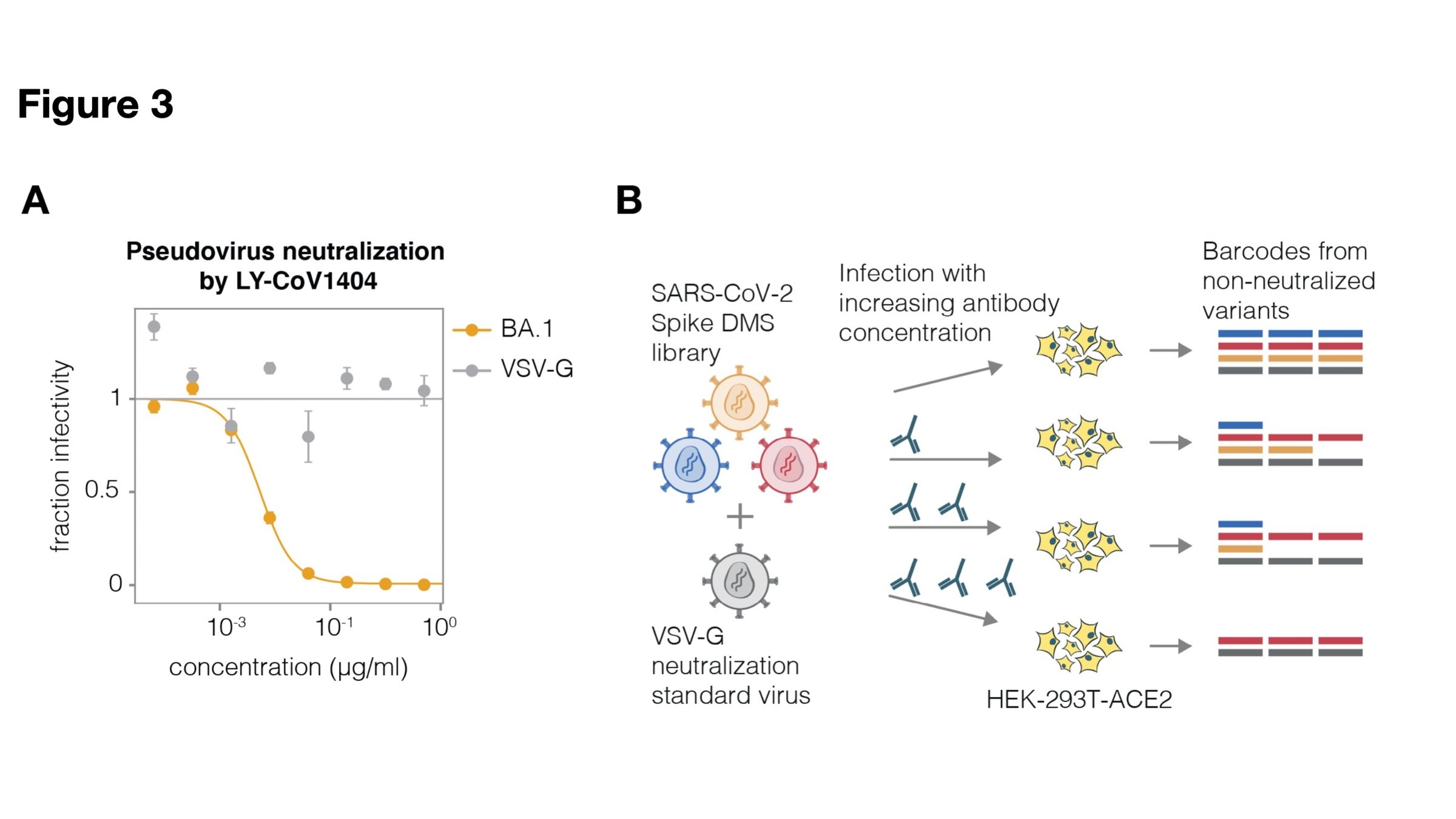
Two-step method to create genotype-phenotype linked pseudotype libraries
Two-step method to create genotype-phenotype linked spike-pseudotypes

Libraries can measure how mutations affect phenotypes (eg, antibody neutralization)

We mapped how all tolerated mutations to XBB.1.5 spike affect three key phenotypes
Cell entry: how well pseudovirus enters 293T-ACE2 cells
Sera escape: how pseudovirus is neutralized by human polyclonal serum
ACE2 binding: how pseudovirus is neutralized by soluble ACE2
How well do these spike phenotypes predict human SARS-CoV-2 clade growth?
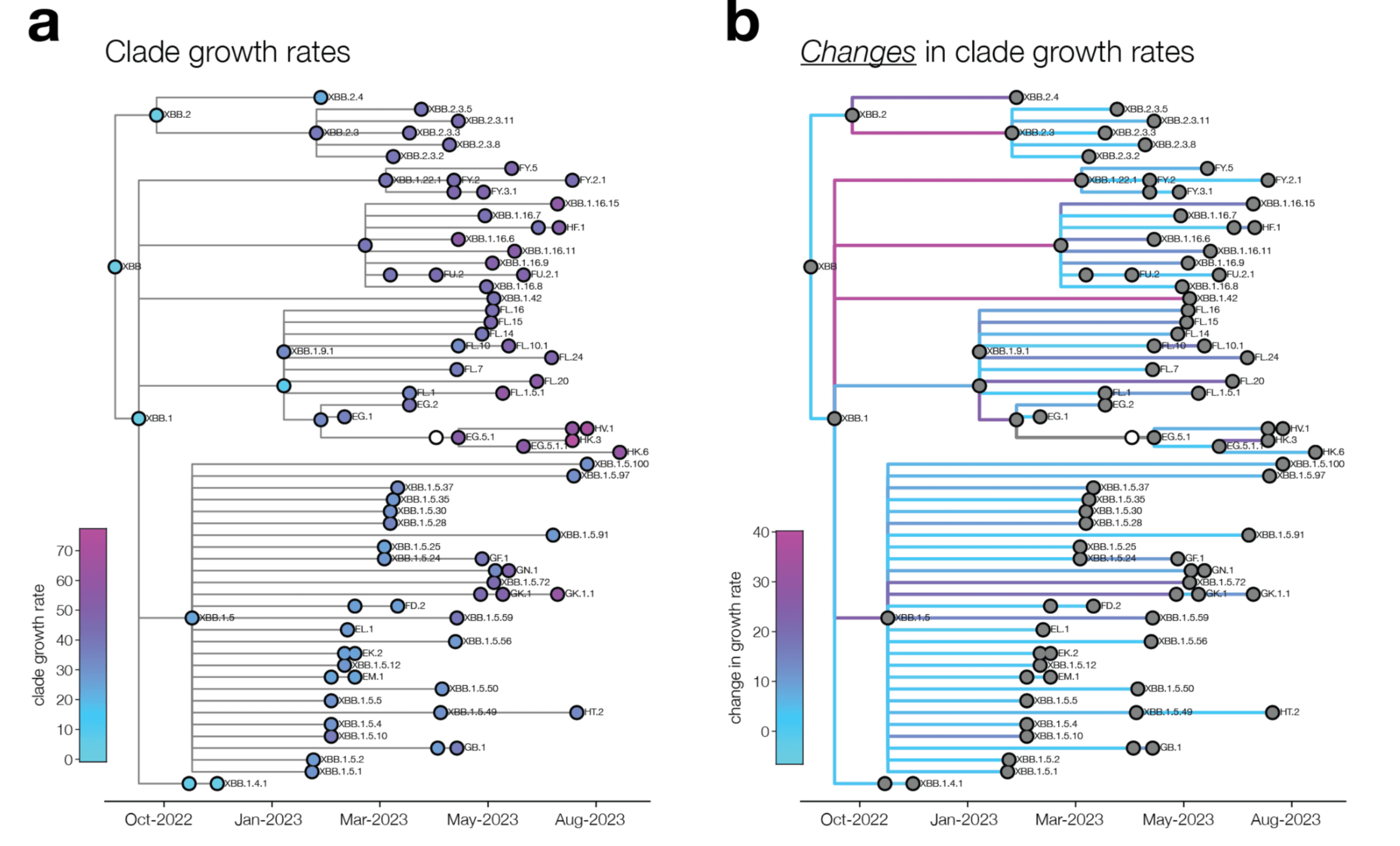
Measured spike phenotypes correlate with changes in SARS-CoV-2 clade growth
Multiple linear regression combining spike phenotypes predicts clade growth well
Extending this approach to H5N1 influenza hemagglutinin (HA)
HA molecular phenotypes relevant to pandemic risk
Pseudovirus library with all HA mutations
Conclusions
For human endemic (SARS-CoV-2) and potential pandemic (H5N1) viruses, we can safely measure how mutations to entry proteins affect key molecular phenotypes.
For SARS-CoV-2, these measurements can help predict success of variants in humans.
For H5N1, these measurements can inform surveillance of viral evolution.
Bloom lab
Bernadeta Dadonaite
Kate Crawford
Caelan Radford
Tyler Starr
Allie Greaney
Rachel Eguia
William Hannon
Jenny Ah
Fred Hutch Cancer Center
Trevor Bedford
John Huddleston
University of Washington
Helen Chu and HAARVI cohort
Neil King
David Veesler




Thanks


Pirbright Institute
Thomas Peacock
University of Pennsylvania
Scott Hensley
Louise Moncla
Jordan Ort
St Jude Children's Hospital
Richard Webby