Antigenic evolution of human coronaviruses
Jesse Bloom / Katie Kistler
Fred Hutch Cancer Center & HHMI
Disclosures
Jesse Bloom is:
- on the scientific advisory boards of Apriori Bio and Oncorus
- an inventor on Fred Hutch licensed patents related to deep mutational scanning of viral proteins
- has unfunded research collaborations with Vir Biotechnology
Outline
- The long-term antigenic evolution of CoV-229E (Jesse Bloom)
- Evolutionary patterns in human coronaviruses (Katie Kistler)
- Antigenic evolution of SARS-CoV-2 to date (Jesse Bloom, as time permits)
Outline
- The long-term antigenic evolution of CoV-229E (Jesse Bloom)
- Evolutionary patterns in human coronaviruses (Katie Kistler)
- Antigenic evolution of SARS-CoV-2 to date (Jesse Bloom, as time permits)
Only some viruses evolve to rapidly erode immunity
- Measles virus: Does not evolve to escape immunity. People infected at most once in their lives. Vaccine developed in 1960s still works today.
- Influenza virus: Evolves to escape immunity. People infected every ~5 years. Vaccine needs to be updated annually.
We decided to look at evolution of a human coronavirus: CoV-229E causes common colds and has been circulating in humans for a long time.
Reconstructing evolution of CoV-229E spike

We experimentally generated CoV-229E spikes at ~8 year intervals so we could study them in the lab:
- 1984
- 1992
- 2001
- 2008
- 2016
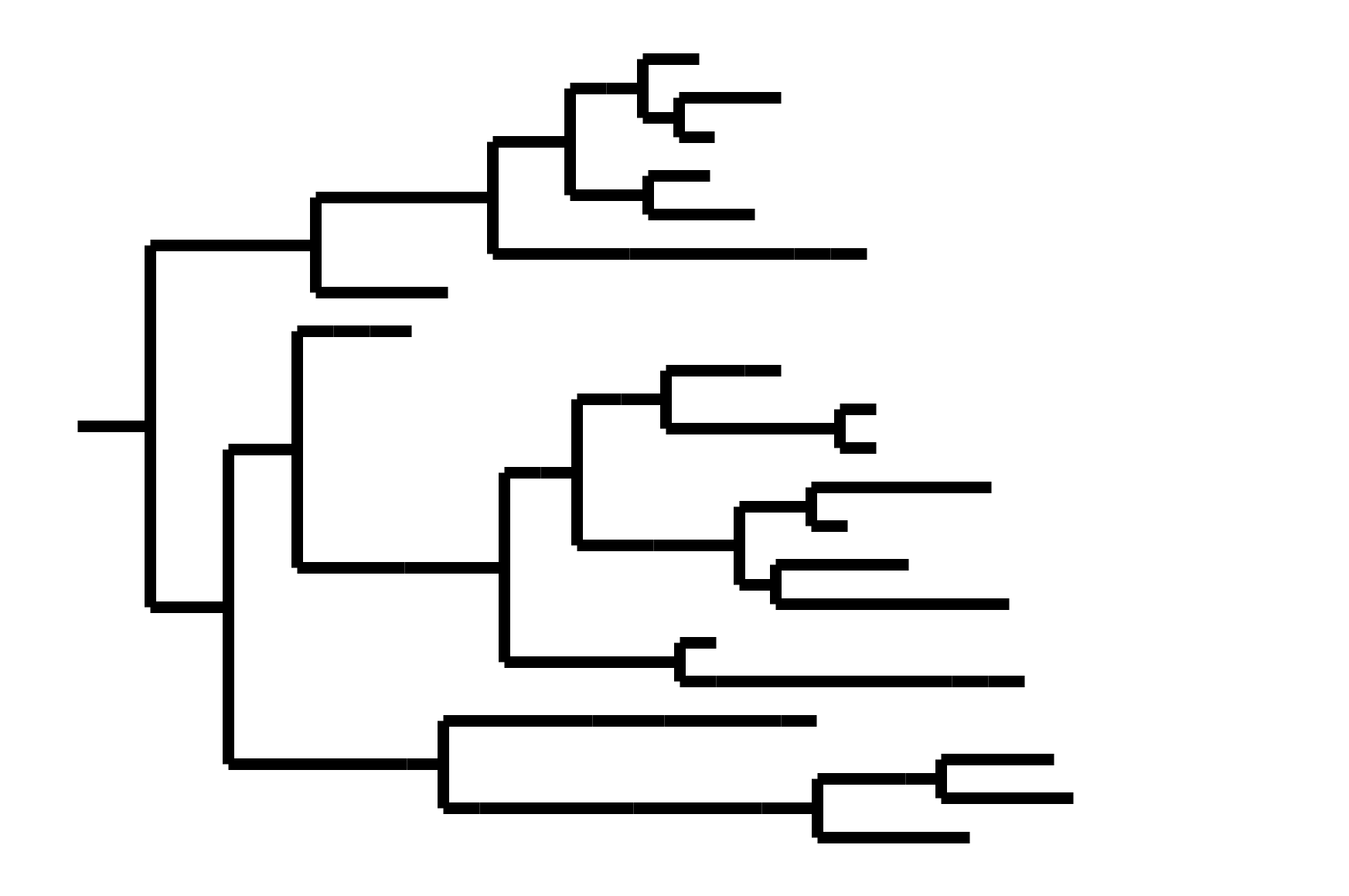
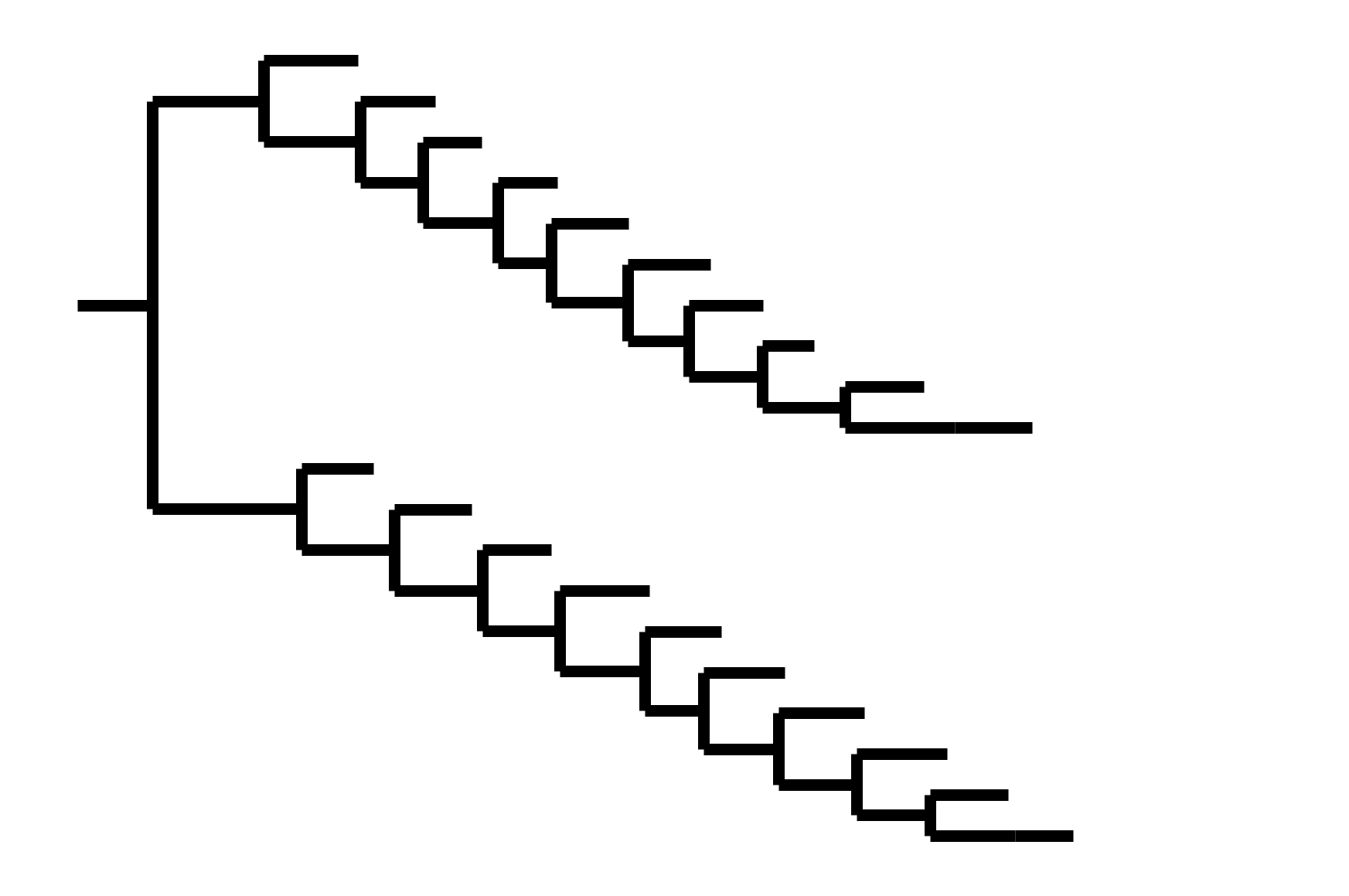
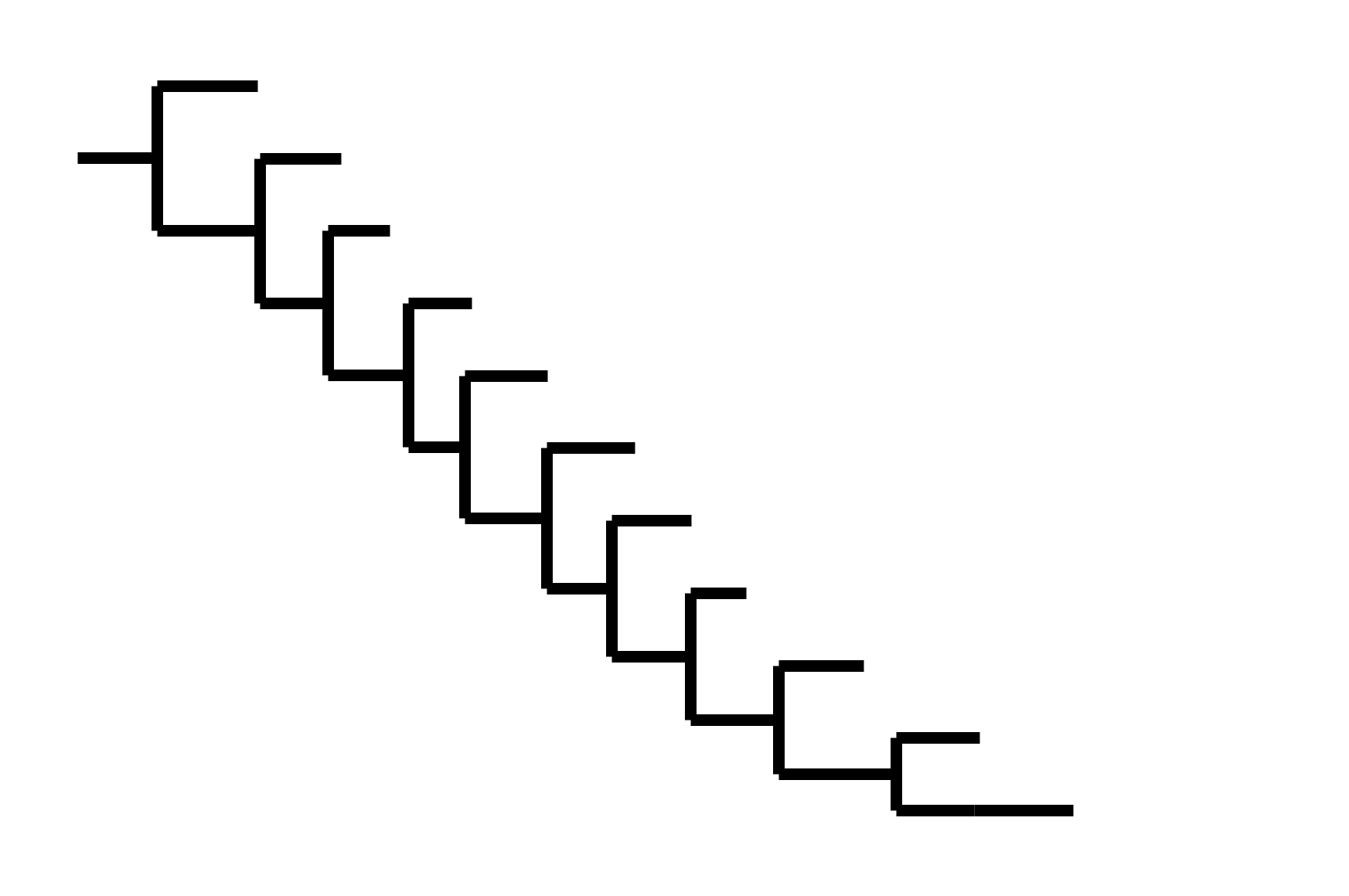
Note "ladder-like" shape of tree
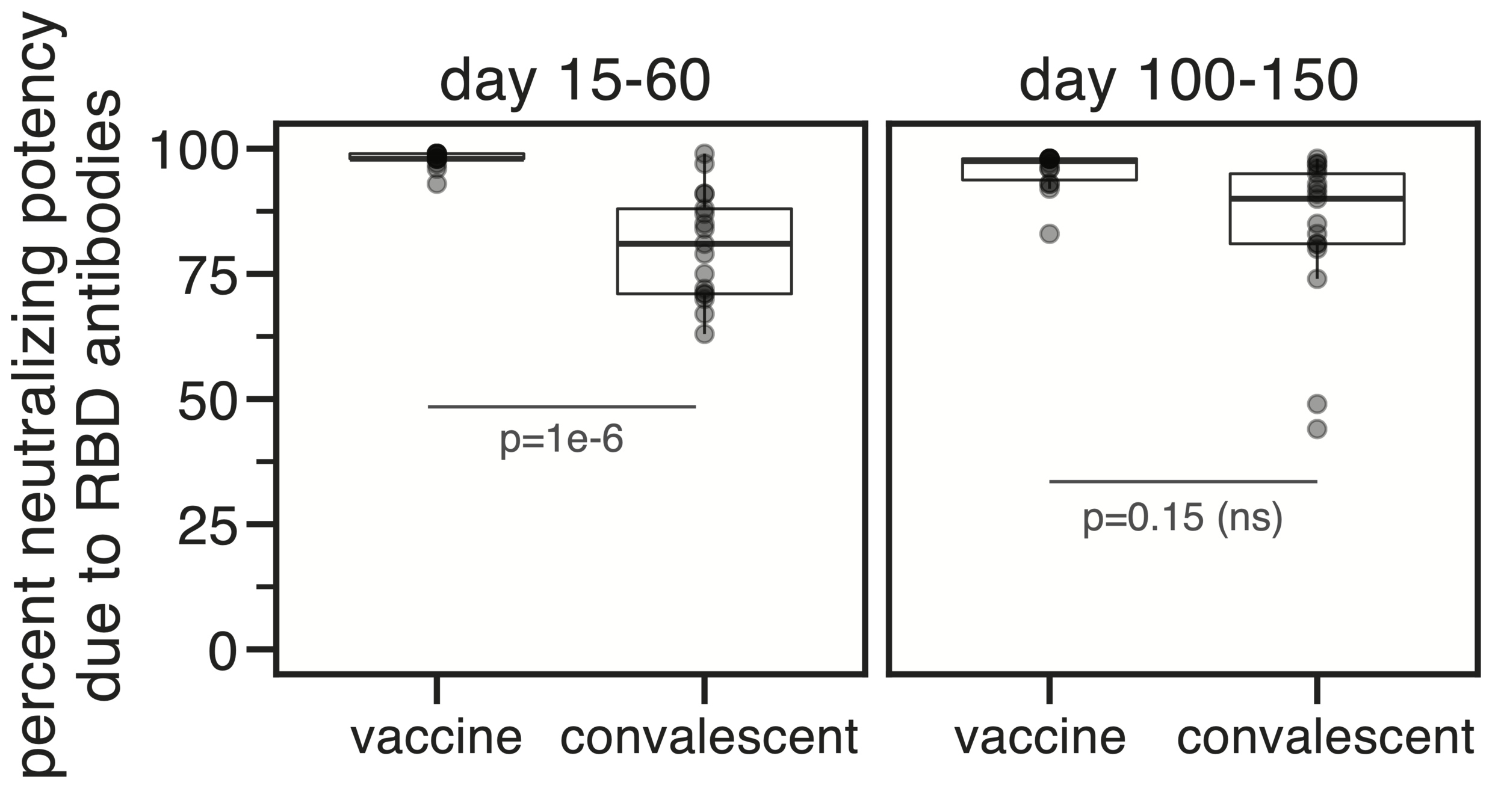
Evolution of CoV-229E spike erodes neutralization by human serum antibodies
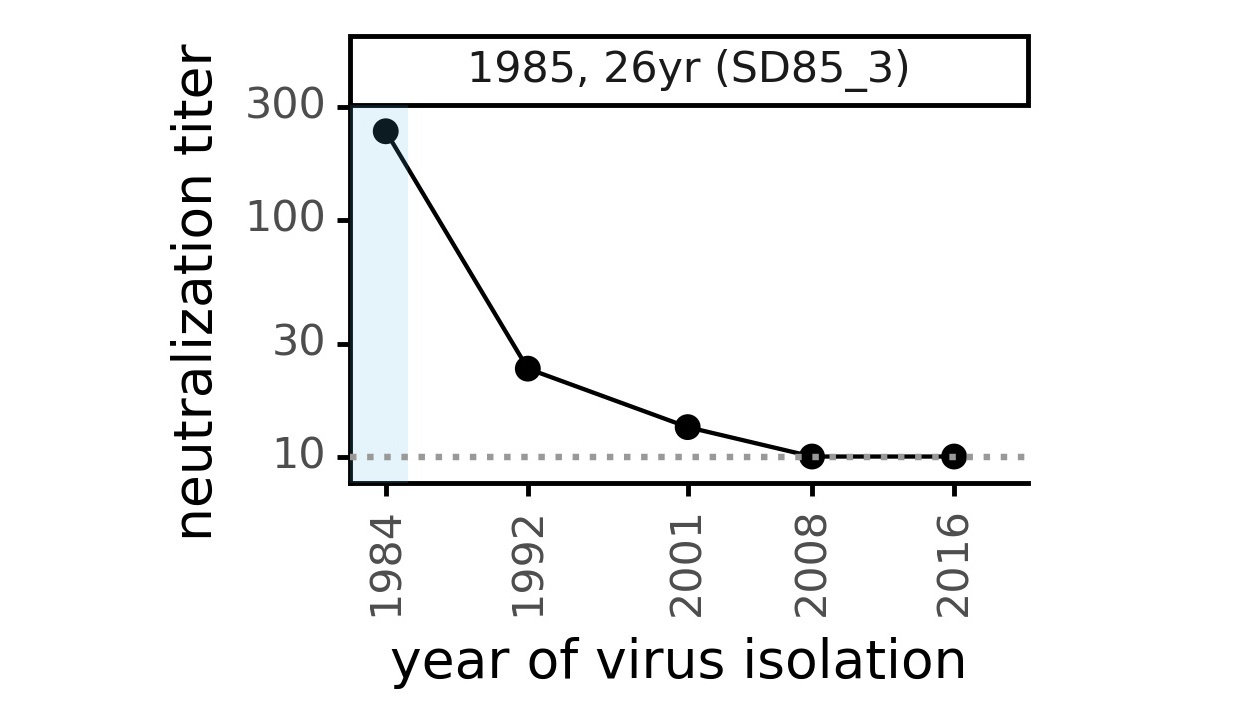
Serum collected in 1985 neutralizes virus with spike from 1984, but less effective against more recent viruses.
Viral evolution erodes antibody immunity of different people at different rates

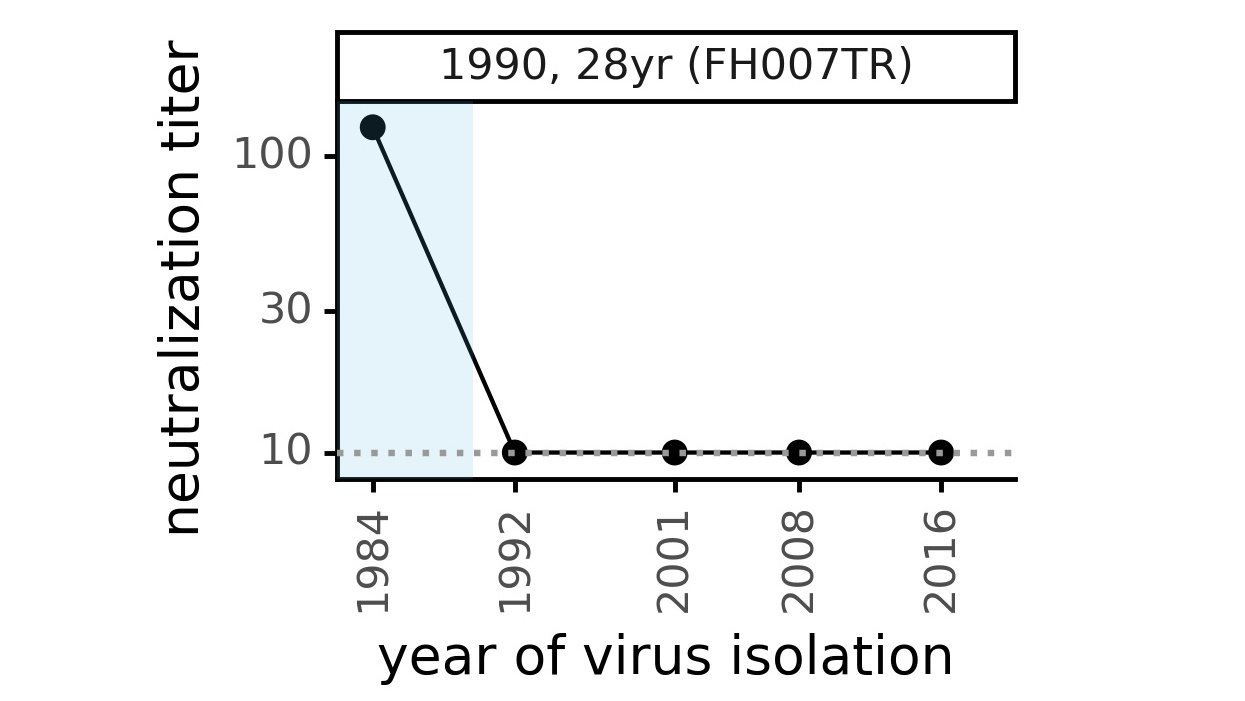
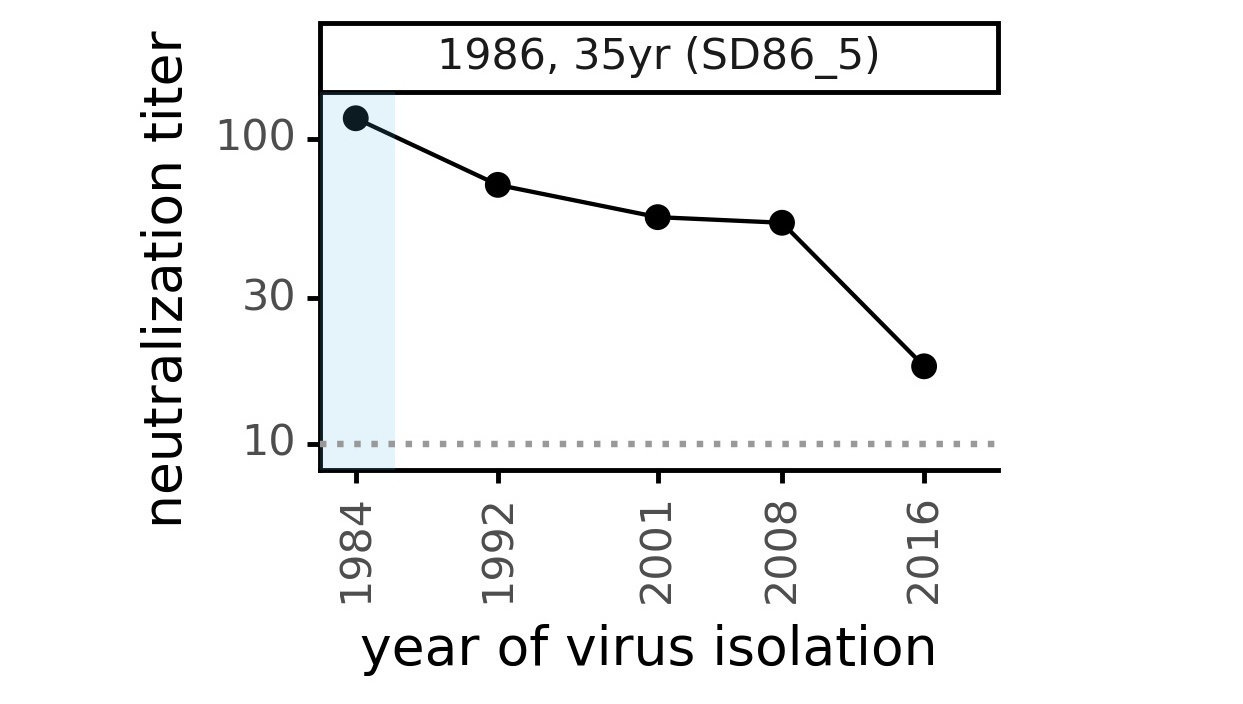
We are studying basis of these differences, as ideally vaccines would elicit more evolution-resistant sera as on the right.
Strongest evolutionary selection in RBD
Sites of evolutionary change in the spike of CoV-229E over the last four decades
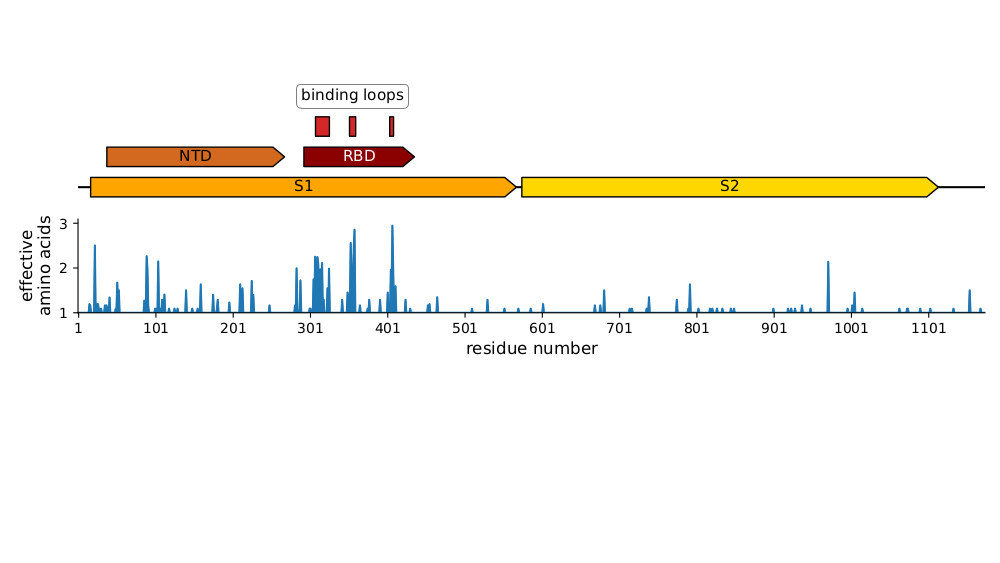
Strongest evolutionary selection in RBD
Sites of evolutionary change in the spike of CoV-229E over the last four decades

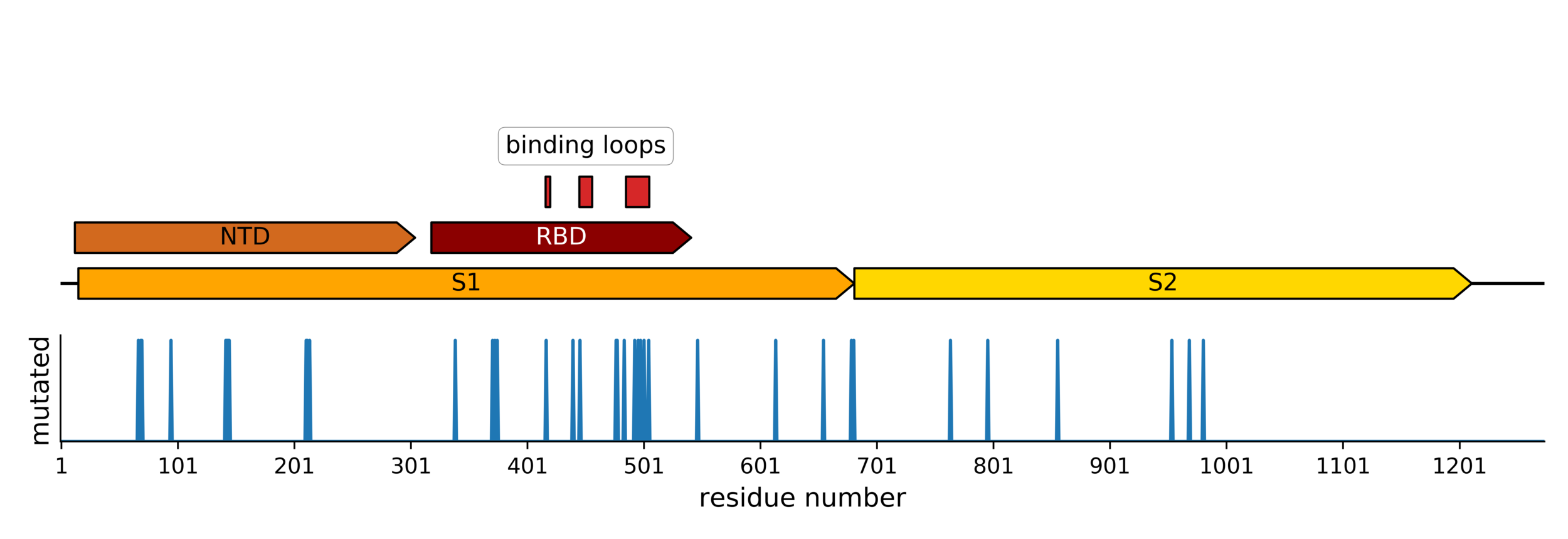
Sites of mutations in SARS-CoV-2 Omicron (BA.1) spike relative to Wuhan-Hu-1
Main difference is SARS-CoV-2 also fixing transmissibility-enhancing spike mutations that affect proteolytic processing and stabilize defects cause by furin-cleavage site
RBD is under strongest selection because it is main target of neutralizing antibodies
Outline
- The long-term antigenic evolution of CoV-229E (Jesse Bloom)
- Evolutionary patterns in human coronaviruses (Katie Kistler)
- Antigenic evolution of SARS-CoV-2 to date (Jesse Bloom, as time permits)
Four seasonal CoVs are endemic in humans
Seasonal CoV tree topologies suggest different modes of evolution
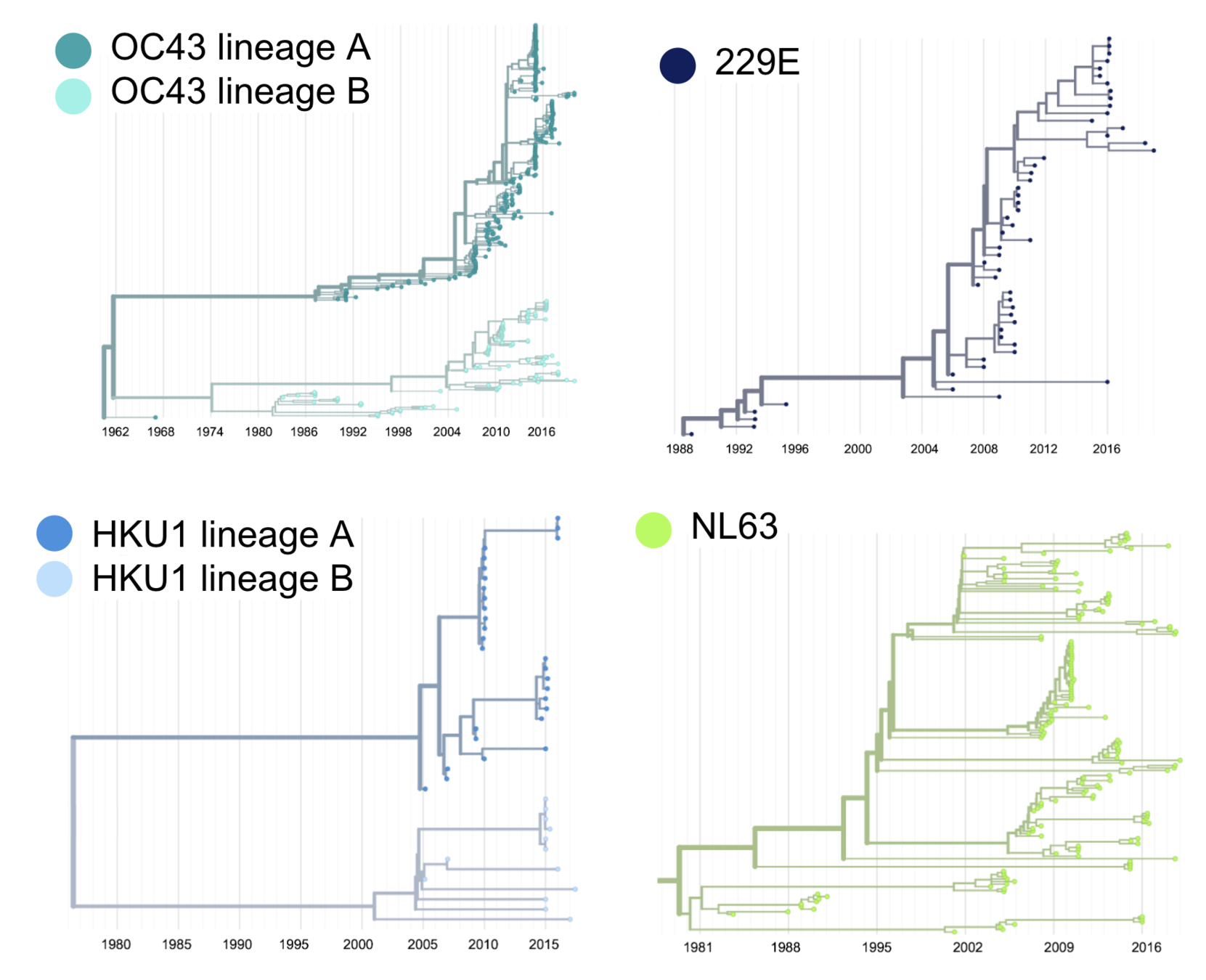
OC43 and 229E have ladder-like topologies, similar to influenza A

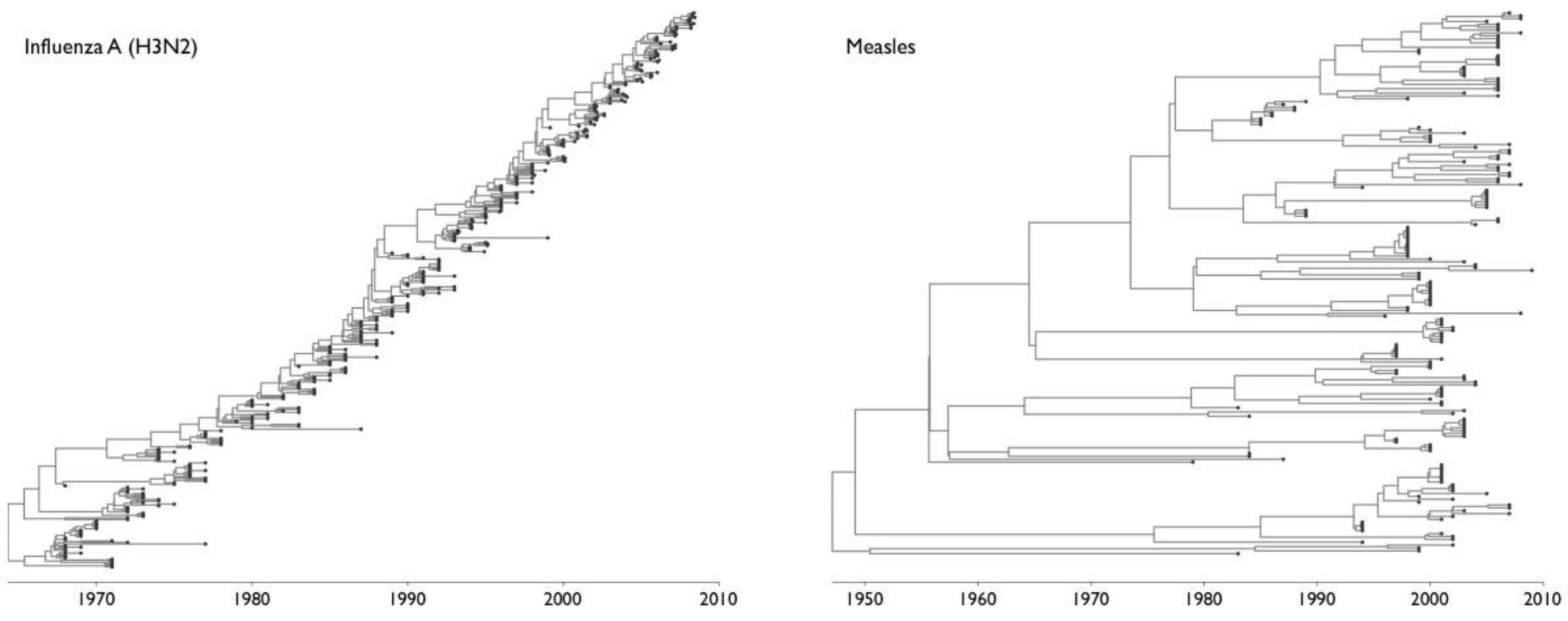
Ladder-like topologies indicate continual replacement of variants by more fit ones, an evolutionary process driven by antigenic drift in influenza
The NL63 tree is bushier, like measles


On this timescale, a bushier tree suggests lack of adaptive evolution. In measles, this fits with a known lack of antigenic evolution
Quantify seasonal CoV adaptive evolution
McDonald-Krietman assumption under neutrality (McDonald and Kreitman, 1991)
Adaptive fixations are excess of replacement fixations compared to neutral expectation
In RNA viruses (with high mutation rates), adaptive substitutions may be fixed or present at high-frequency (Bhatt et al, 2011)
Calculate rate of adaptive evolution in seasonal CoV spike S1
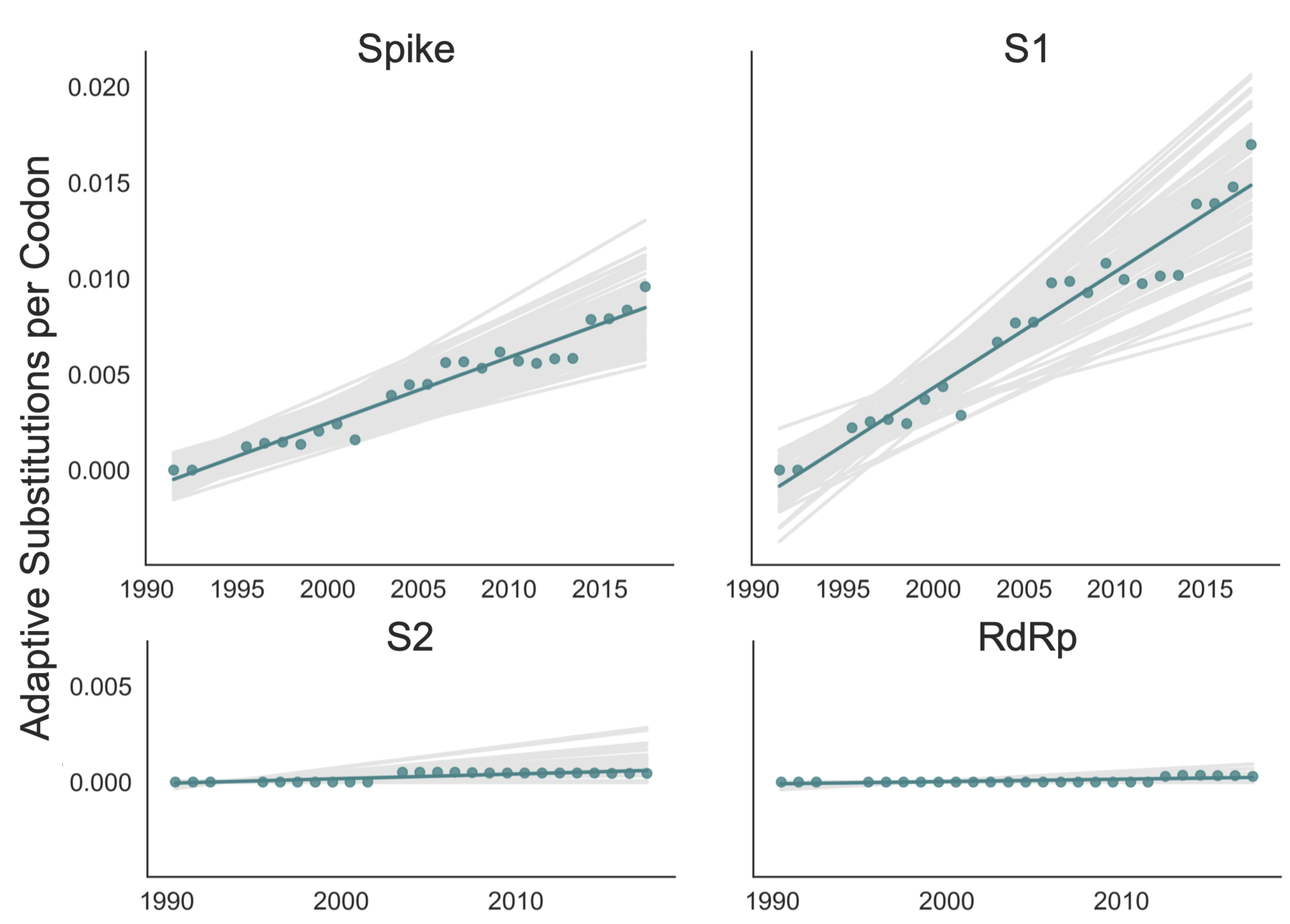
OC43 lineage A
adaptive subs per codon per year
adaptive subs in S1 each year
Compare rate of adaptation in seasonal CoVs to other endemic viruses
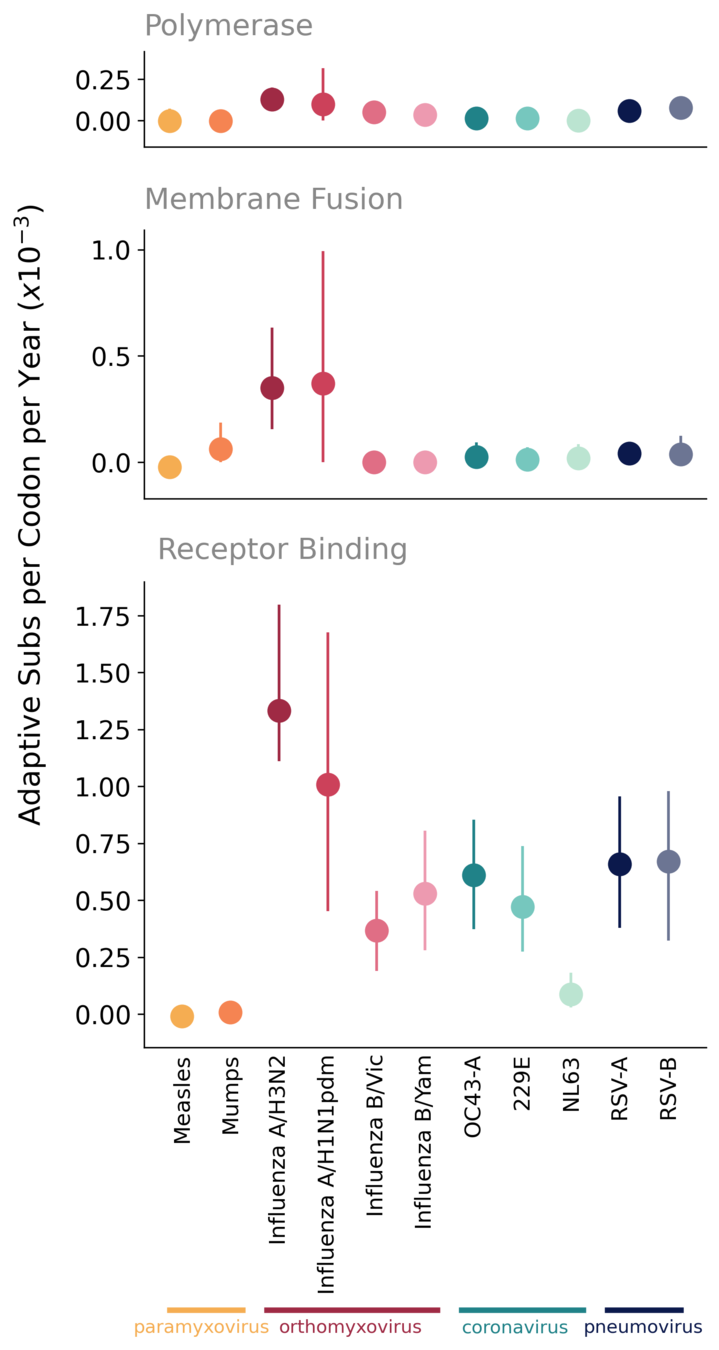


As expected, polymerases of endemic viruses are not evolving adaptively
Receptor-binding proteins exhibit ongoing adaptation, likely antigenic drift


In viruses that have been endemic in humans for many decades, ongoing adaptive evolution of the receptor-binding protein/ subunit is typically driven by immune evasion
OC43 and 229E S1 evolves adaptively, at a rate similar to influenza B


OC43 and 229E accumulate adaptive substitutions in S1 at a rate similar to influenza B viruses
No evidence of antigenic drift in NL63
No evidence of antigenic drift in NL63


NL63 preferentially infects children more than other seasonal CoVs
Vaccine considerations based on evolution of seasonal CoVs
OC43 and 229E will likely require regularly-updated vaccine components to combat antigenic evolution
Unclear whether HKU1 evolves in this way
OC43 and HKU1 have two lineages, likely requiring bivalent vaccine component
Sequence data suggest NL63 may be more antigenically stable
Outline
- The long-term antigenic evolution of CoV-229E (Jesse Bloom)
- Evolutionary patterns in human coronaviruses (Katie Kistler)
- Antigenic evolution of SARS-CoV-2 to date (Jesse Bloom, as time permits)
So far, SARS-CoV-2 tree not ladder-like
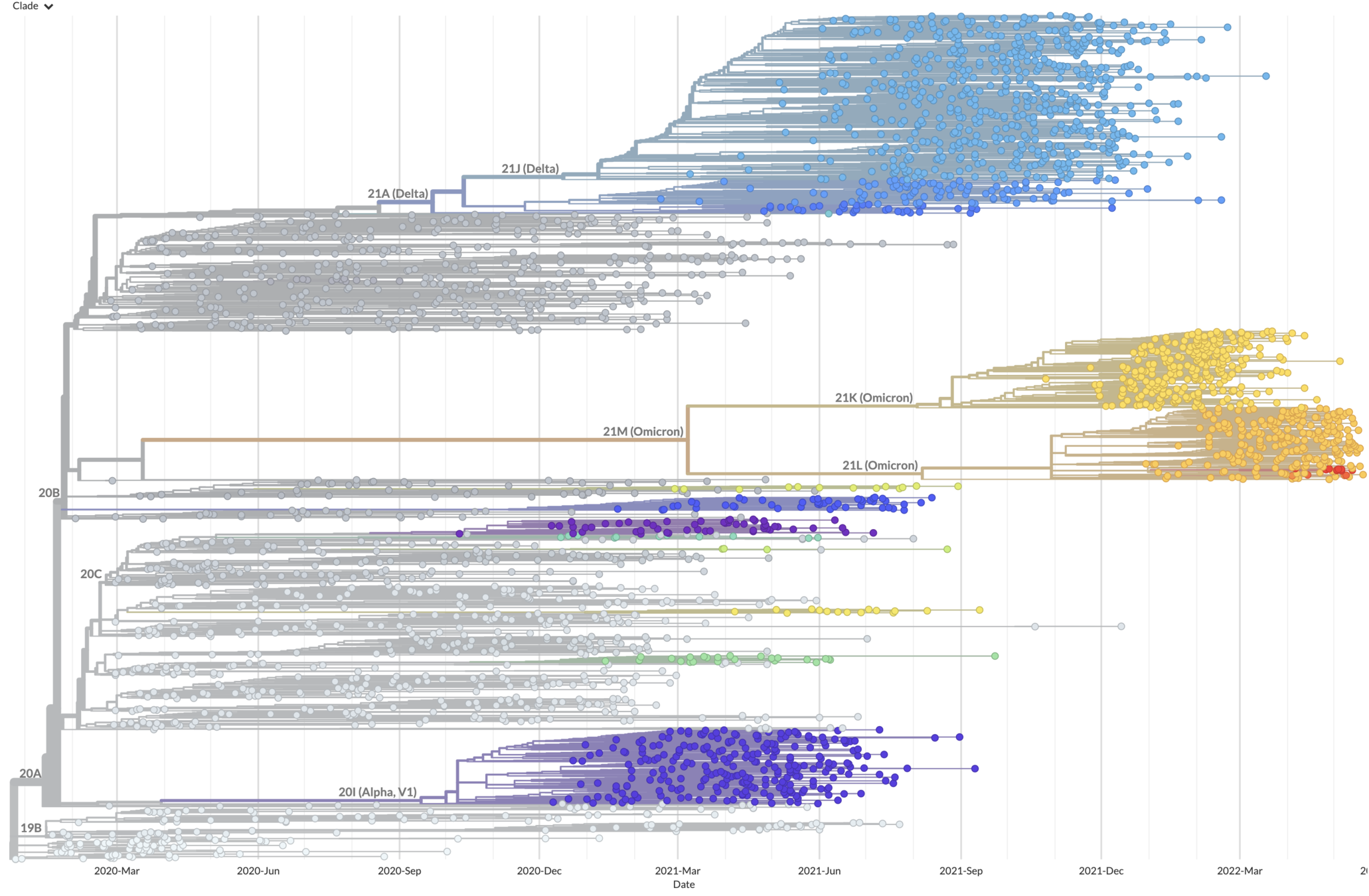
But early evolution may be non-typical as adaptation in antibody escape (will continue) & transmissibility (will plateau)

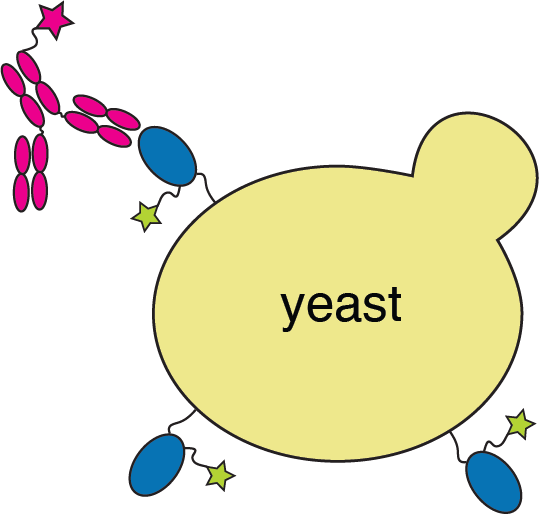
We can use deep mutational scanning to map all receptor-binding domain (RBD) mutations that escape antibody binding
RBD
fluorescently labeled antibody
yeast
fluorescent tag on RBD
For an interactive version of this escape map, see:
Escape map from one antibody (LY-CoV555, ie bamlanivimab). Peaks indicate sites where mutations escape binding.
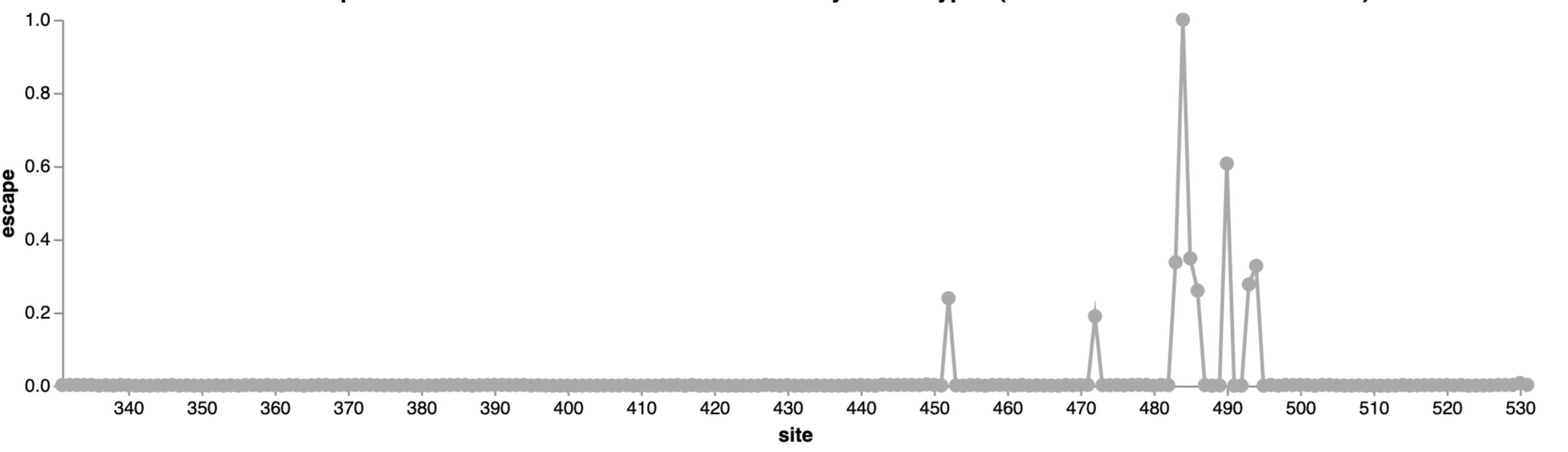
484
452
490
Infection / vaccination elicit polyclonal antibodies
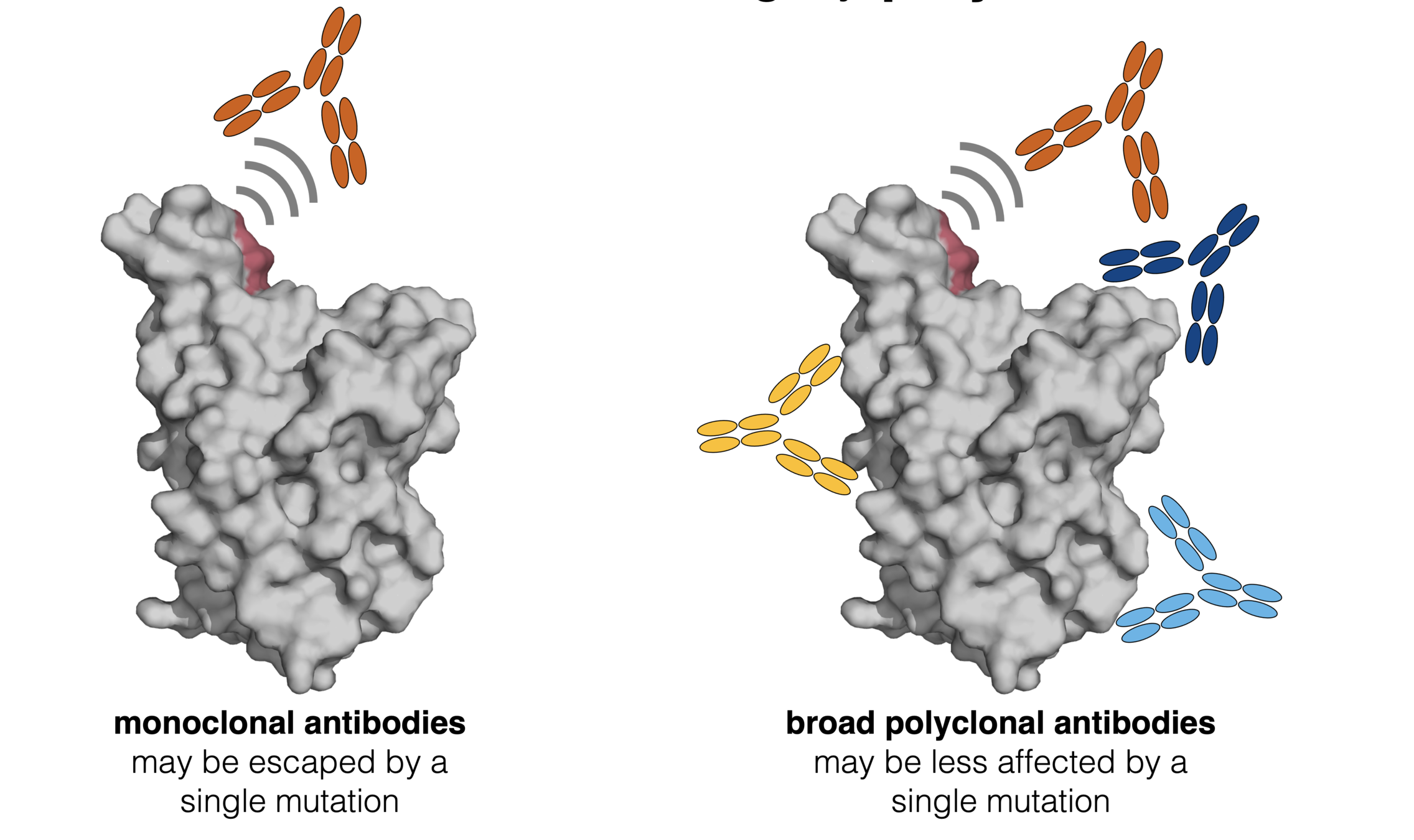
We add up escape maps for ~1,500 different human antibodies to understand which mutations impact polyclonal binding
Antibodies elicited by early SARS-CoV-2 that neutralize Wuhan-Hu-1 heavily focus on sites like 484, 417, 446

417
446
484
417
446
484

Omicron BA.1 is mutated at many of these sites, which is why it is neutralized substantially less well by current vaccines
Omicron BA.2 has some different RBD mutations than BA.1, but similar overall escape from antibodies from early SARS-CoV-2

Can identify mutations that mediate further escape. In Dec 2021 we predicted 486 as likely site of future evolution--in April 2022, mutation F486V was identified in Omicron BA.4 and BA.5.

486 is largest site of escape for antibodies not already escaped by mutations in BA.2


Explore yourself with online escape calculator: select antibodies by eliciting virus or strains they neutralize, click sites to mutate
https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/escape-calc/
Conclusions
- Human coronaviruses evolve to erode antibody immunity
- We can understand and perhaps even forecast this evolution
- Can these insights be combined with more rapid vaccine platforms?