The impact of viral mutations on antibodies used to treat COVID-19
Jonathan Li, @DrJLi
Jesse Bloom, @jbloom_lab
Tyler Starr, @tylernstarr
Allison Greaney, @AllieGreaney
Manish Choudhary, @Dr_MChoudhary
These slides at https://slides.com/jbloom/covid-19-clinical-antibodies
Classical way to identify escape mutations is to grow virus with antibody in lab
"... novel spike mutants rapidly appeared after in vitro passaging in the presence of individual antibodies..."

From the paper describing REGN-CoV2 cocktail
Limitation of this approach is you only find some of the possible escape mutants
We express SARS-CoV-2 receptor binding domain (RBD) on surface of yeast
RBD
fluorescent antibody
fluorescent tag on RBD
Library of yeast, each expressing different RBD mutant

We map escape mutations by sorting for RBD variants that don't bind antibody
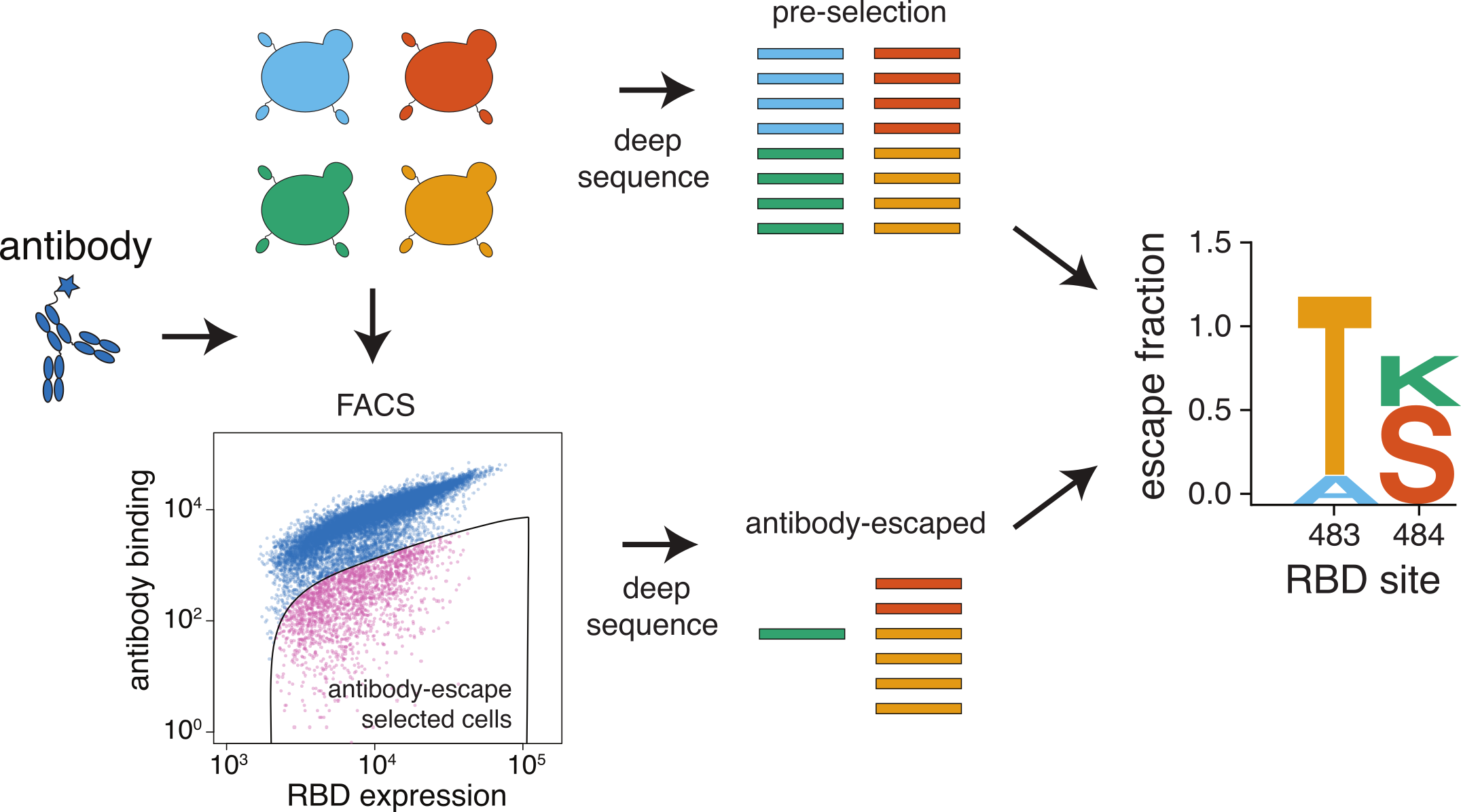
In maps, tall letters indicate strong escape mutations
Sort done at single antibody concentration, which determines sensitivity!
Escape map for REGN10987
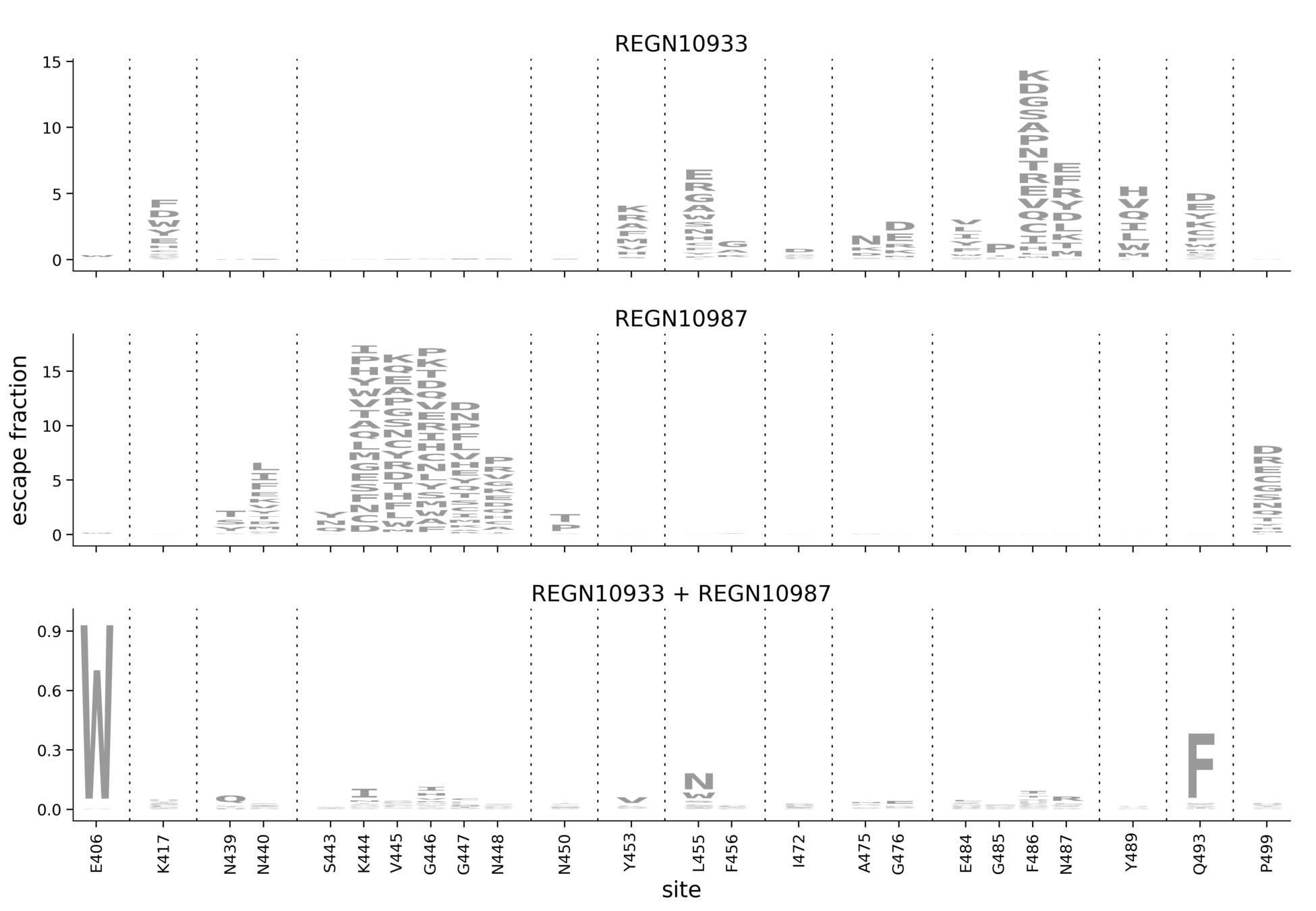

Tall letters = strongly escape binding. For instance, mutations at 446 escape, but those at 484 don't. However, quantitative relationship of letter height to IC50 depends on FACS gate: we confidently capture mutations with >100 fold effects, but can miss smaller ones.
Image shows key sites of interest, click here for interactive map of all mutations.
We now have escape maps for many lead clinical antibodies
- REGN-COV2 cocktail (REGN10933 = casirivimab; REGN10987 = imdevimab)
- Paper available here
- Interactive maps available here
- LY-CoV555 (bamlanivimab) and occasional cocktail partner LY-CoV016 (etesivimab)
- Pre-print available here
- Interactive maps available here
- COV2-2130 and COV2-2196 (the two antibodies in AZD7442)
- Pre-print available here, collaboration with Crowe lab
- Interactive maps available here
Note: these are high-throughput maps generated using yeast display. We have found excellent correlation between the maps and pseudotyped viral neutralization assays (see papers above), but still recommend validating key conclusions.
In persistently infected patient, five RBD mutations after REGN-COV2 treatment
Patient described in Choi, Choudhary, ...,. Cernadas, Li. New England J Medicine (2020)
Note extensive genetic hitchhiking and competition among viral lineages; also described in within-patient evolution of influenza in persistent infections: Xue et al, eLife (2017)
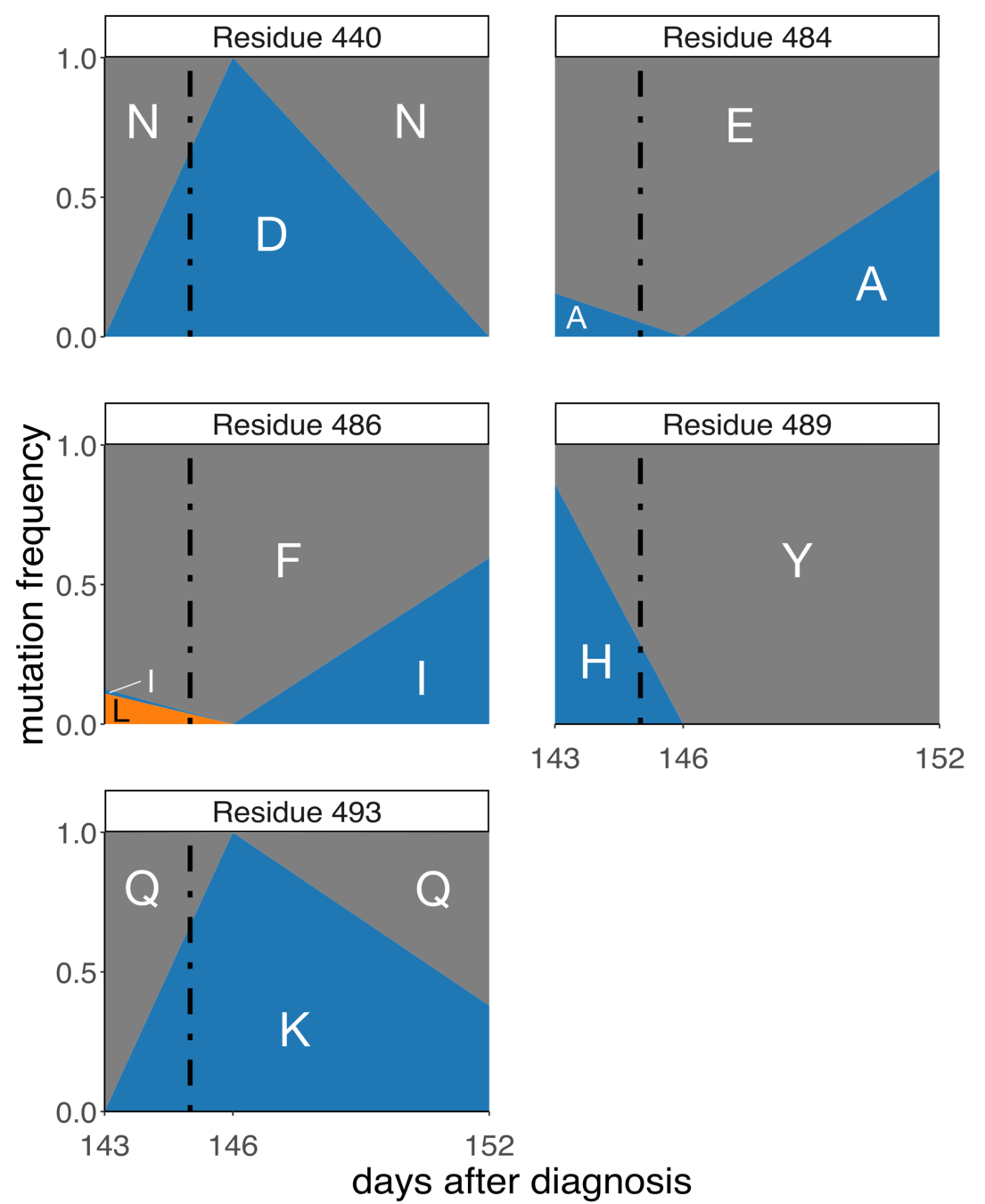
Importantly, our complete maps show that four of these mutations reduce binding by one or the other of the REGN-COV2 antibodies, only one of which was identified by Regeneron's escape selections. Illustrates value of complete maps.
Mutations in emerging variant lineages:

Studies validating these conclusions from the mapping:
- Wang et al (2021): B.1.351 fully escapes LY-CoV555, partially escapes REGN10933
- Hoffmann et al (2021): B.1.351 and P.1 fully escape LY-CoV555 and partially escape REGN10933
- Tada et al (2021): B.1.351 partially escapes RE; Y453F (mink) fully escapes REGN10933
- Widera et al (2021): B.1.351 and P.2 (another E484K variant) fully escape LY-CoV555
- Wang et al (2021): P.1 fully escapes LY-CoV555 and LY-CoV016, partially escapes REGN10933
Crowe lab (Vanderbilt):
- James Crowe
- Seth Zost
- Pavlo Gilchuk




Bloom lab (Fred Hutch)
-
Tyler Starr
-
Allie Greaney
-
Adam Dingens
-
Amin Addetia
-
Will Hannon
These slides at https://slides.com/jbloom/covid-19-clinical-antibodies
Li lab (Brigham & Women's):
- Jonathan Li
- Manish Choudhary