Association between SARS-CoV-2 and metagenomic content of samples from the Huanan Seafood Market
Jesse Bloom
Fred Hutch Cancer Center / HHMI
These slides at https://slides.com/jbloom/huanan-market-samples
I will summarize this preprint, which addresses following question:
What is association between number of SARS-CoV-2 reads and number of reads from different animals in environmental samples from Huanan Seafood Market?


Downloaded full dataset posted by Chinese CDC on the NGDC
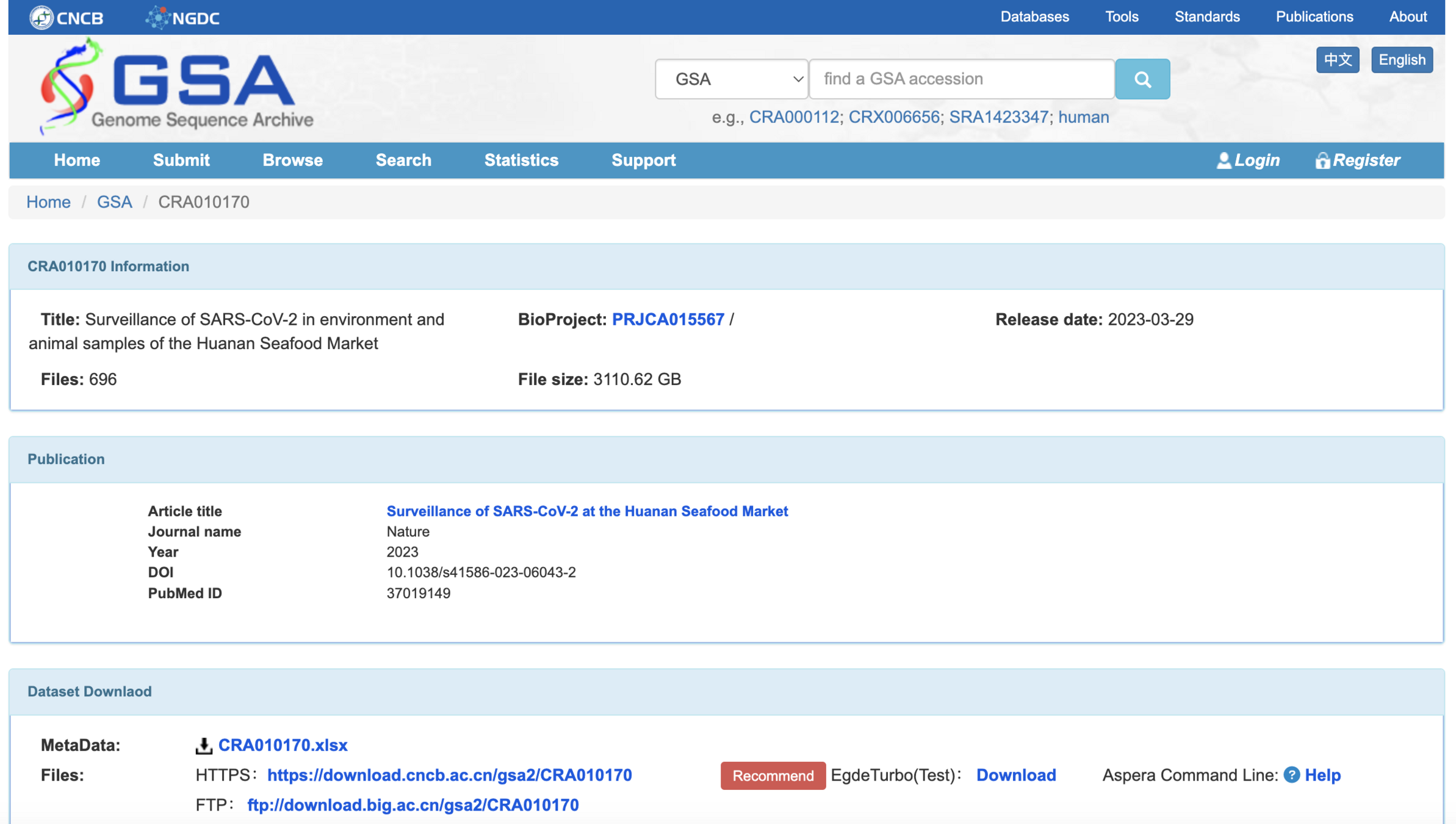
Confirmed NGDC data are a superset of the data analyzed by Crits-Christoph et al
This sheet shows that all FASTQ files analyzed by Crits-Christoph et al have identical matches in the files uploaded to the NGDC
Quantified mitochondrial composition among chordate species for samples
Among mammalian species, compositions I determined are highly correlated to those reported by Crits-Christoph et al
Quantified mitochondrial composition among chordate species for samples
The chordate and mammalian mitochondrial composition for each metagenomic sample can be examined using interactive plots above.
Quantified SARS-CoV-2 content of samples
Most samples have little or no SARS-CoV-2 reads. Plot above allows selection of samples by date, source, etc.
There is little or no SARS-CoV-2 in samples with abundant material from potentially susceptible animals
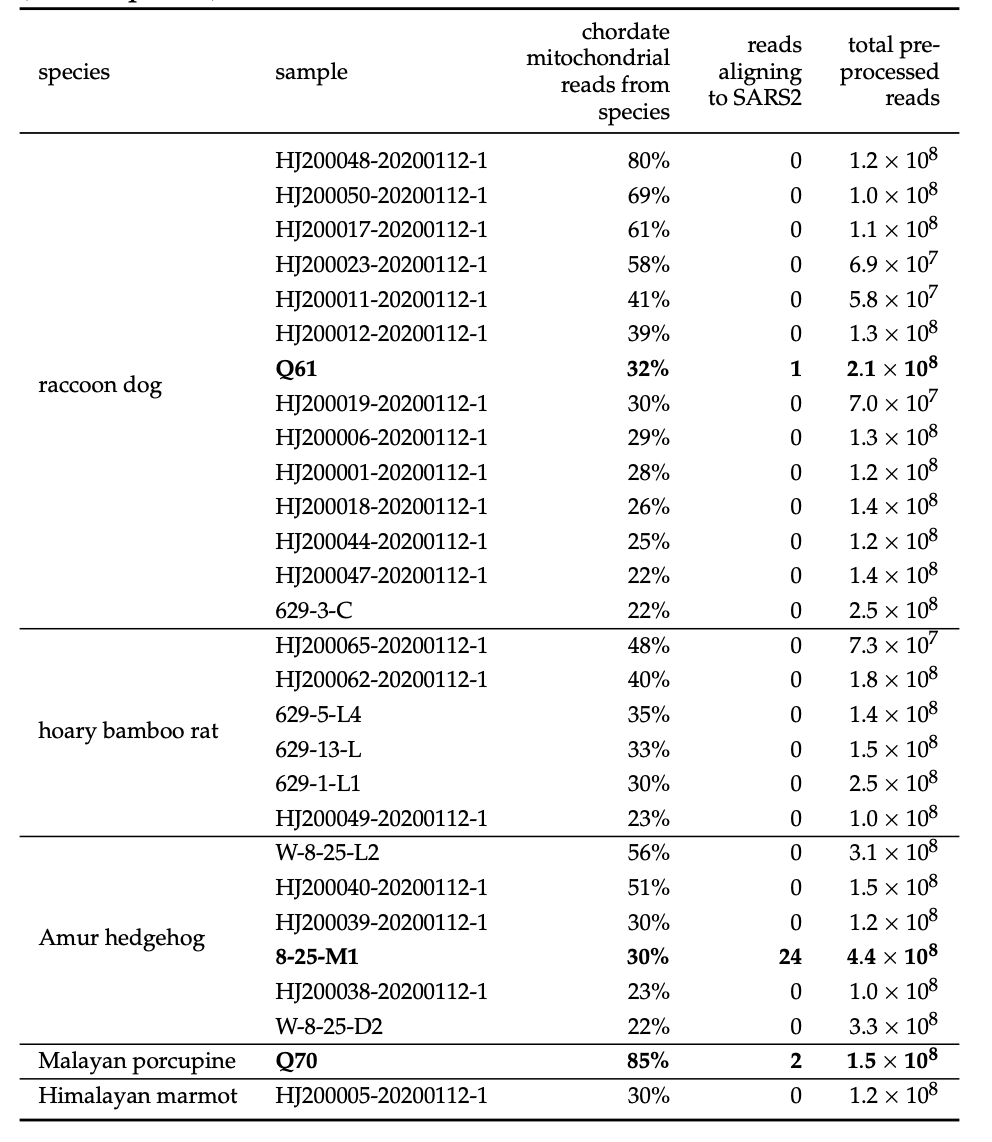


Association of SARS-CoV-2 vs animal reads
Interactive plot above enables you to look up relationship of SARS-CoV-2 and animal genetic material content for any species.
Associations for all animals
Plot summarizing correlations for all animals
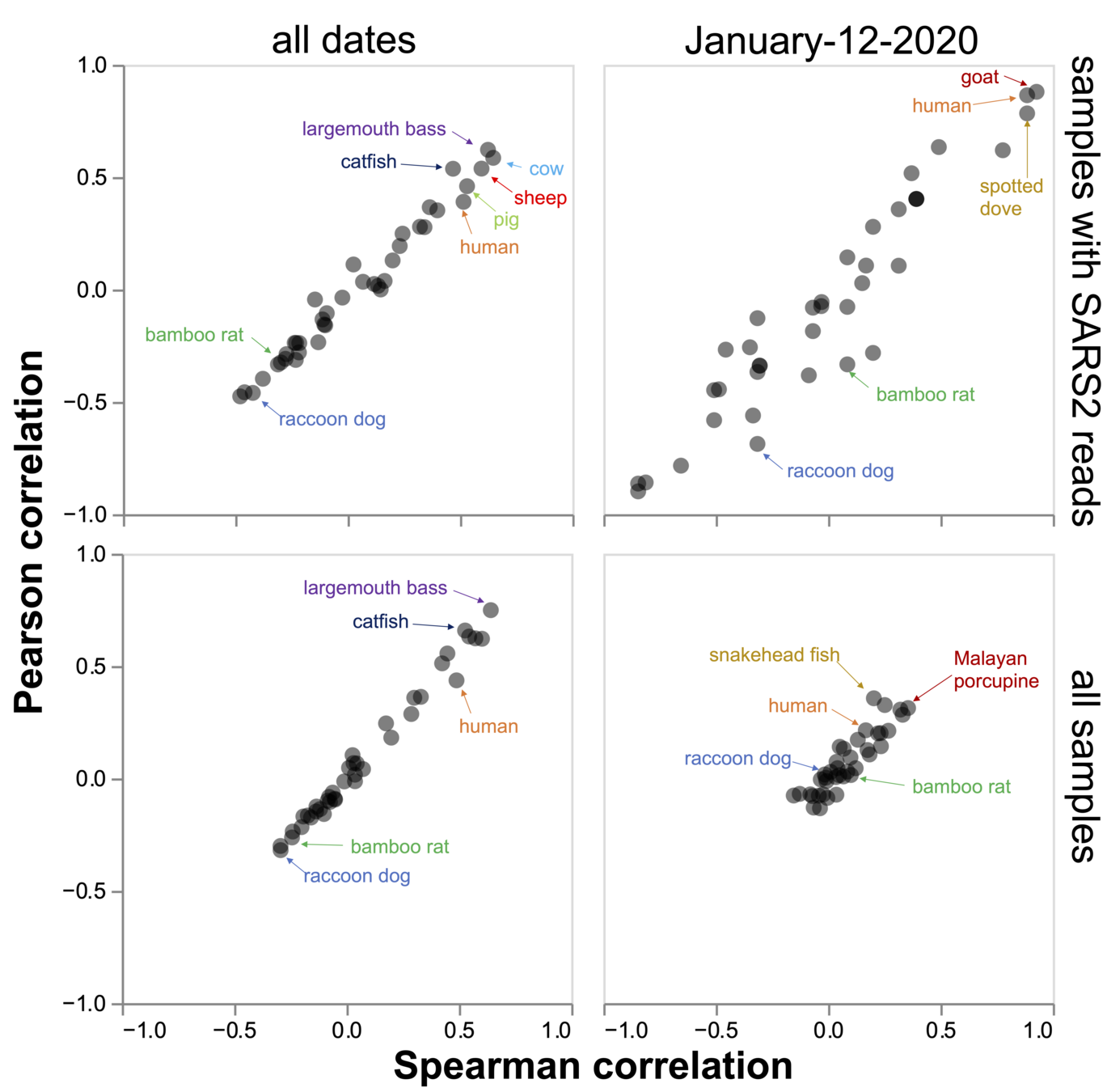
There are many confounders, so there could conceivably be infected animals but no correlation (confounders include sampling time differences among samples, sampling bias, sampling too late, etc).
However, if there is no consistent correlation between SARS-CoV-2 and material from potentially infected animals, then the samples cannot be taken as providing evidence animals were infected. These results indicate content of samples is uninformative for answering this question.
Interactive version of plot on previous slides. Choose options at bottom and mouse over points in scatter plot for identities of species
Plot summarizing correlations for all animals
My overall interpretation
Many samples with the most virus derive their animal genetic material from species that were clearly not infected.
No consistent correlation between the SARS-CoV-2 content and genetic material from susceptible species.
There are potential confounders that cannot be adequately corrected for, but clearly by time of sampling, the virus was spread widely enough to be co-mingled with material from species that were definitely not infected.
We can conclude that co-mingling of viral and animal genetic material in these samples is unlikely to reliably indicate if any animals were infected.
Aside: we should revisit how samples were called positive vs negative
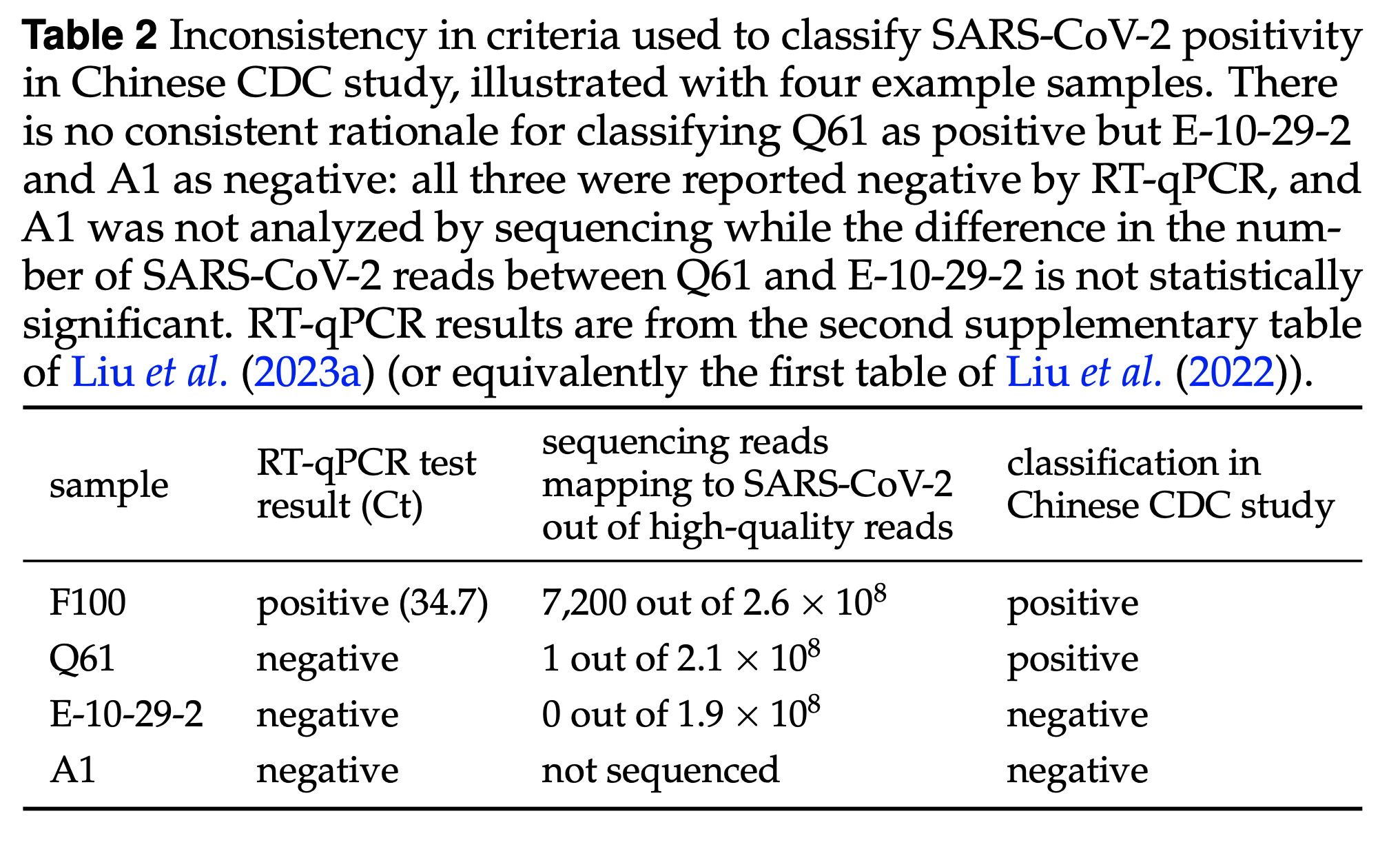
See here for plot of correlation of RT-qPCR and deep sequencing counts of SARS-CoV-2
Additional information
Preprint: https://www.biorxiv.org/content/10.1101/2023.04.25.538336v2
Twitter summary: https://twitter.com/jbloom_lab/status/1651428639676960769
Computer code: https://github.com/jbloom/Huanan_market_samples
Interactive plots: https://jbloom.github.io/Huanan_market_samples/