Interpreting the evolution of
SARS-CoV-2 in context of Omicron
Jesse Bloom
Fred Hutch Cancer Research Center / HHMI
Slides at https://slides.com/jbloom/sars-cov-2-evolution-short
Disclosures
- I am on the scientific advisory boards of Flagship Labs 77 and Oncorus
- I consult for Moderna
- I am an inventor on Fred Hutch licensed patents related to deep mutational scanning of viral proteins
- My lab has unfunded research collaborations with Vir Biotechnology
Only some viruses rapidly evolve to escape antibody neutralization
- Measles virus: Does not evolve to escape immunity. People are infected at most once in their lives. A vaccine developed in the 1960s still works today.
- Influenza virus: Evolves to escape immunity. People are infected every ~5 years. The vaccine needs to be updated annually.

We decided to look at another human coronavirus: CoV-229E causes common colds and has been circulating in humans since at least 1960s.
Reconstructing evolution of CoV-229E spike

We experimentally generated CoV-229E spikes at ~8 year intervals so we could study them in the lab:
- 1984
- 1992
- 2001
- 2008
- 2016
Note "ladder-like" shape of tree
Evolution of CoV-229E spike erodes neutralization by human antibody immunity
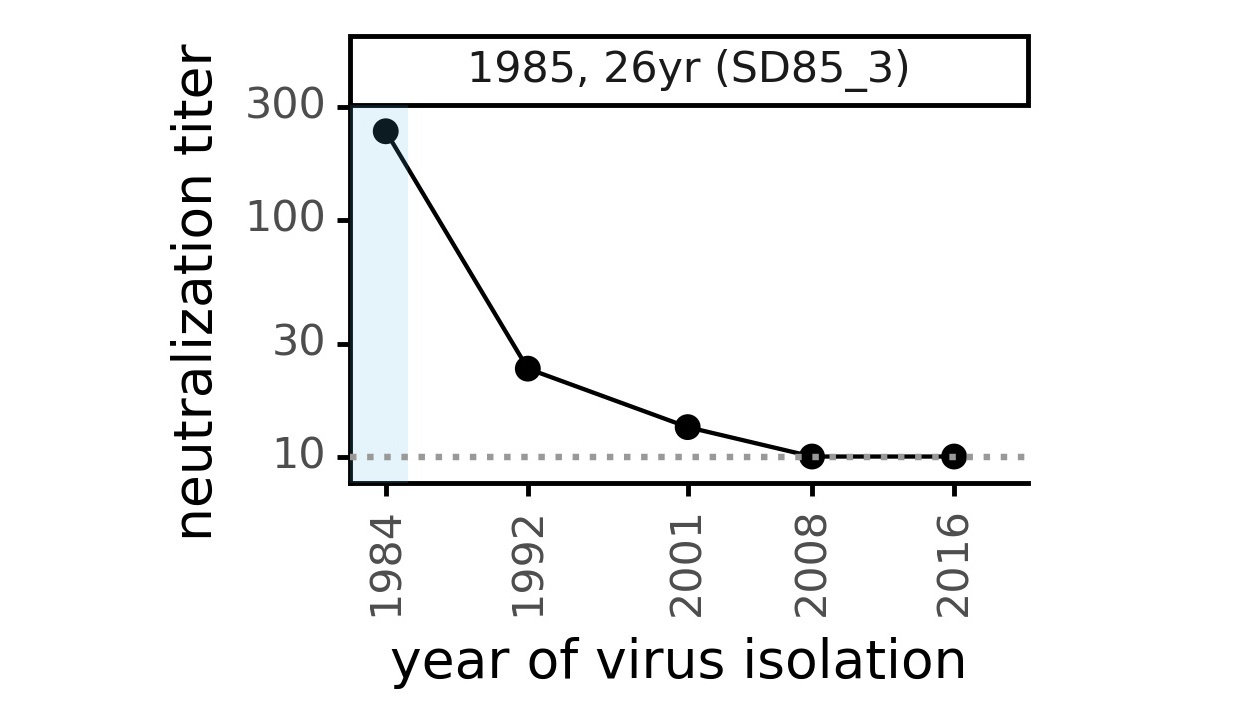
Serum collected in 1985 neutralizes virus with spike from 1984, but less effective against more recent viruses.
Viral evolution erodes antibody immunity of different people at different rates

We are studying basis of these differences, as ideally vaccines would elicit more evolution-resistant sera as on the right.
Phylogenetic tree shape and vaccine strategy
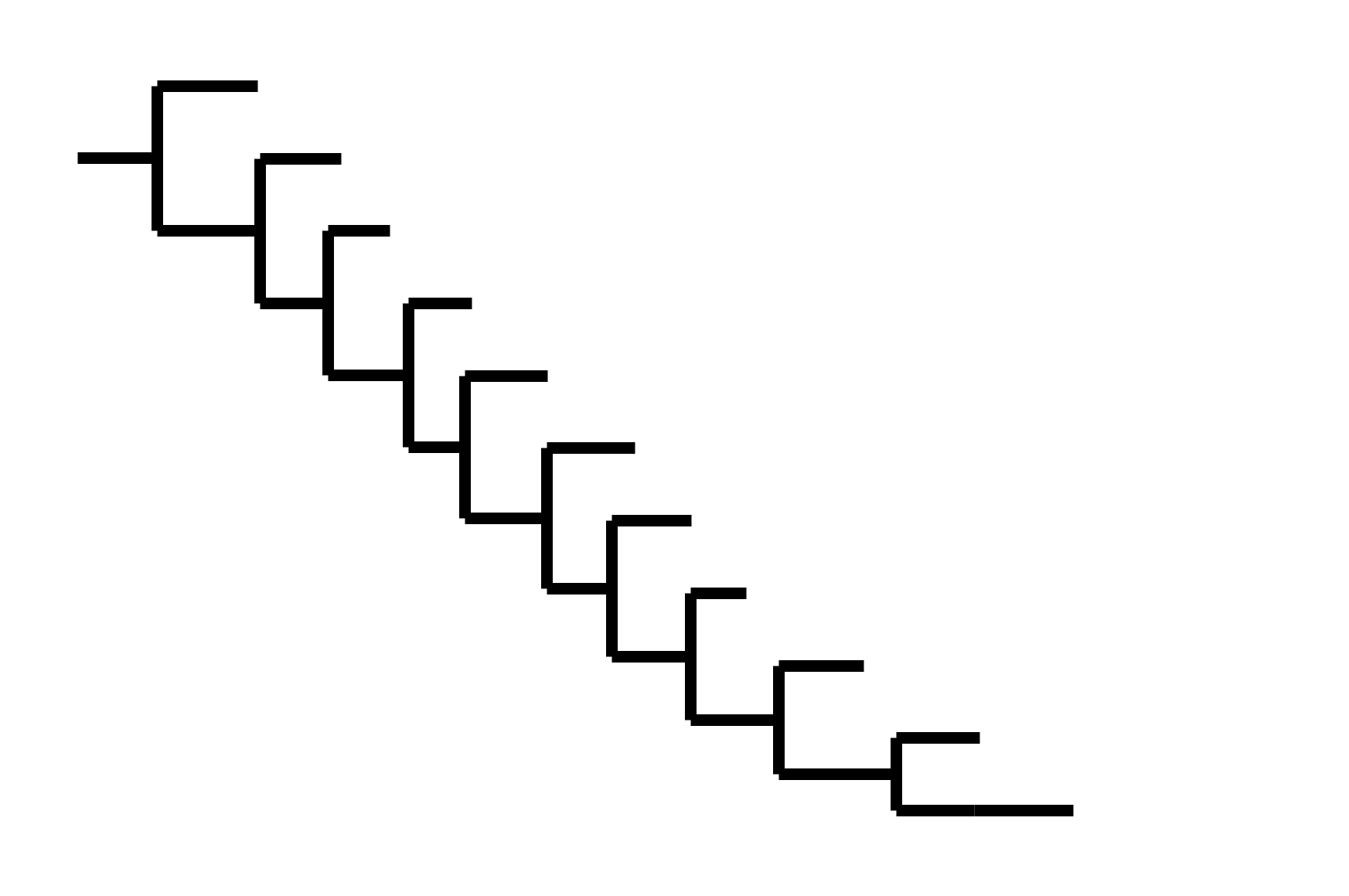
CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.
Phylogenetic tree shape and vaccine strategy
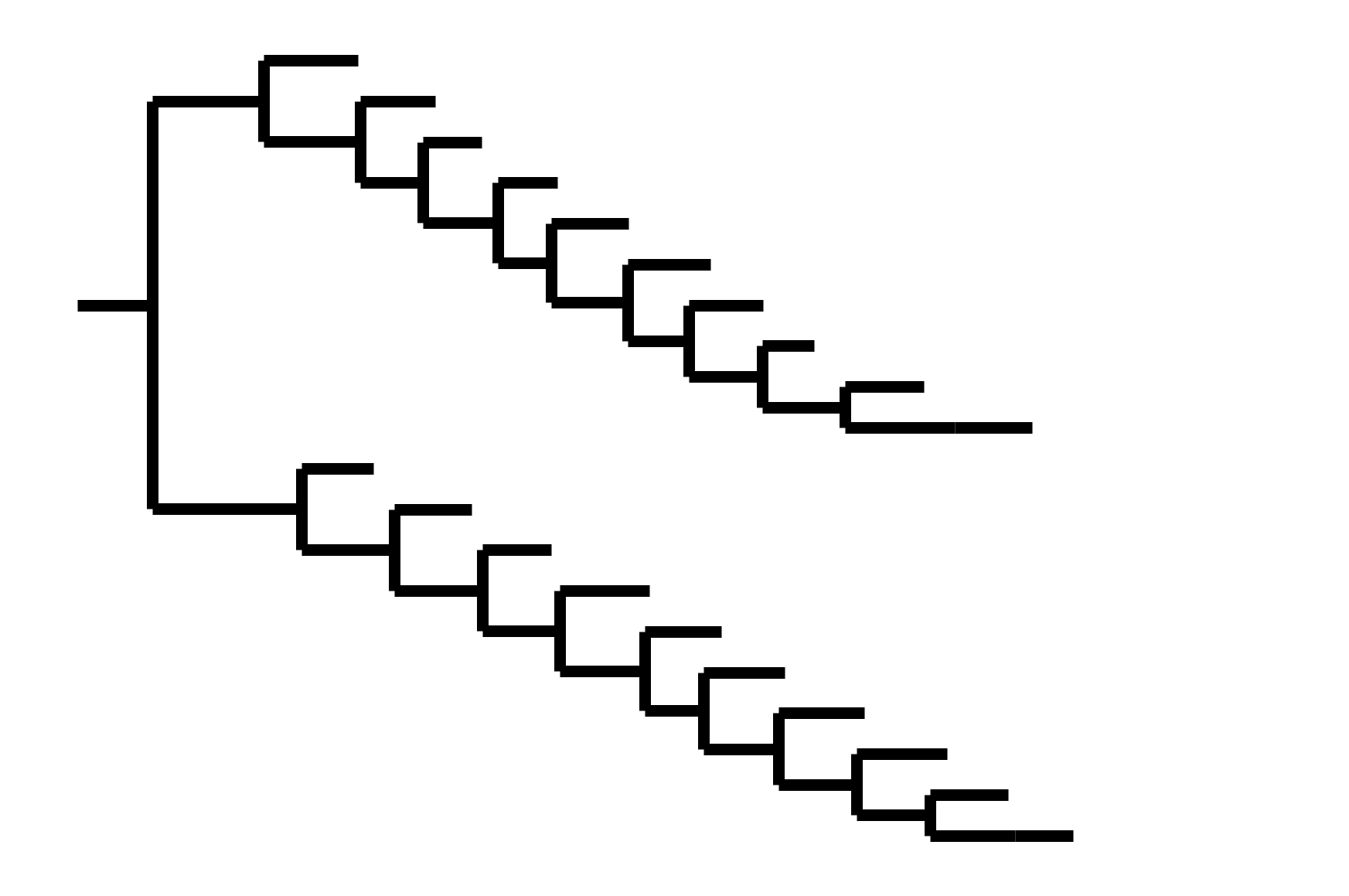

CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.
CoV-OC43 split into two ladder-like lineages. Influenza B evolves this way too. It's theoretically possible to pick well-matched bivalent vaccine.
Phylogenetic tree shape and vaccine strategy


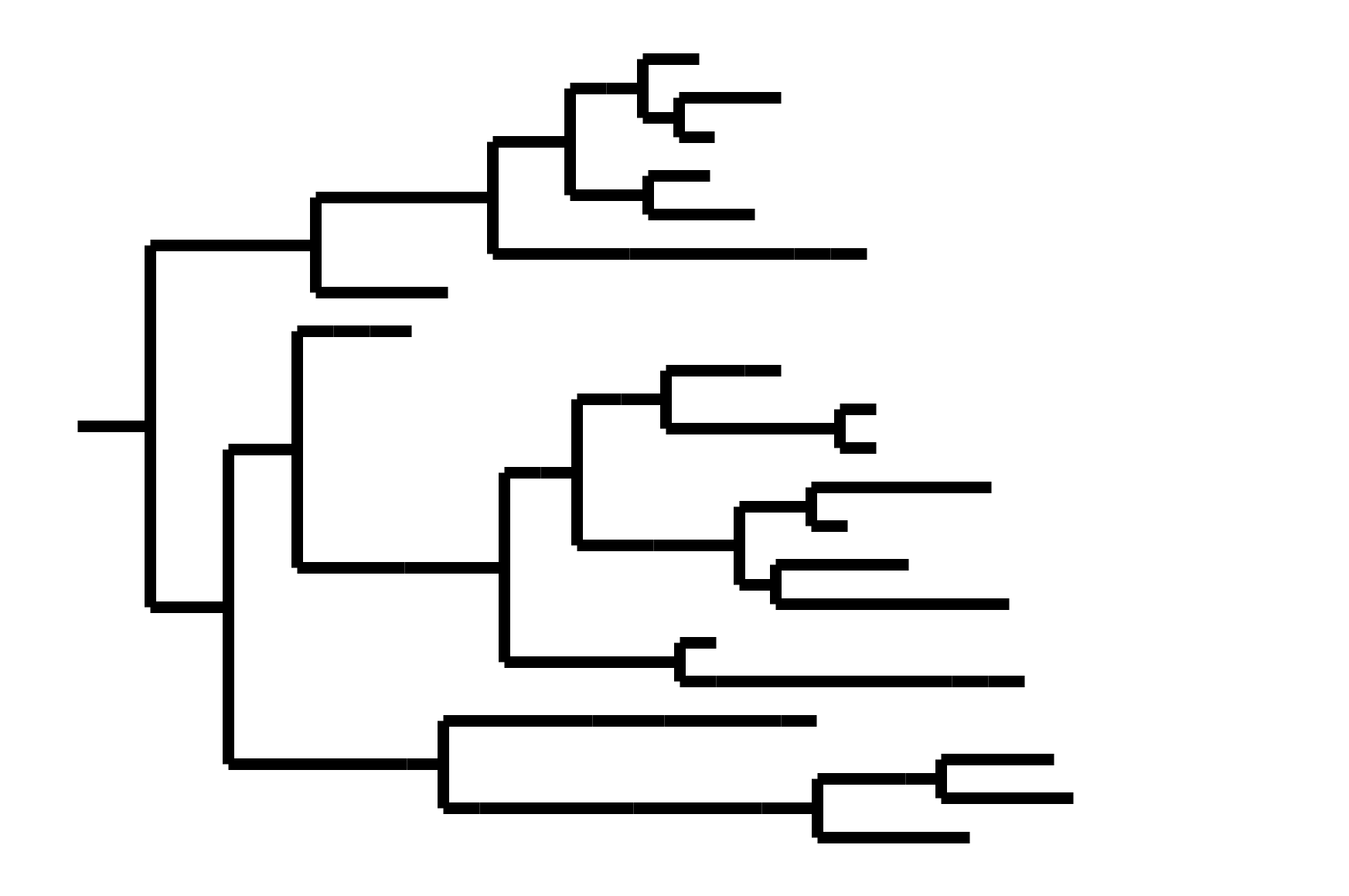
CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.
CoV-OC43 split into two ladder-like lineages. Influenza B evolves this way too. It's theoretically possible to pick well-matched bivalent vaccine.
In non-ladder-like tree, next variant not descended from recent successful one. Makes picking vaccine strains difficult.
After early fixation of D614G, the SARS-CoV-2 tree has not been ladder-like

Strongest evolutionary selection is in RBD
Sites of evolutionary change in the spike of CoV-229E over the last four decades
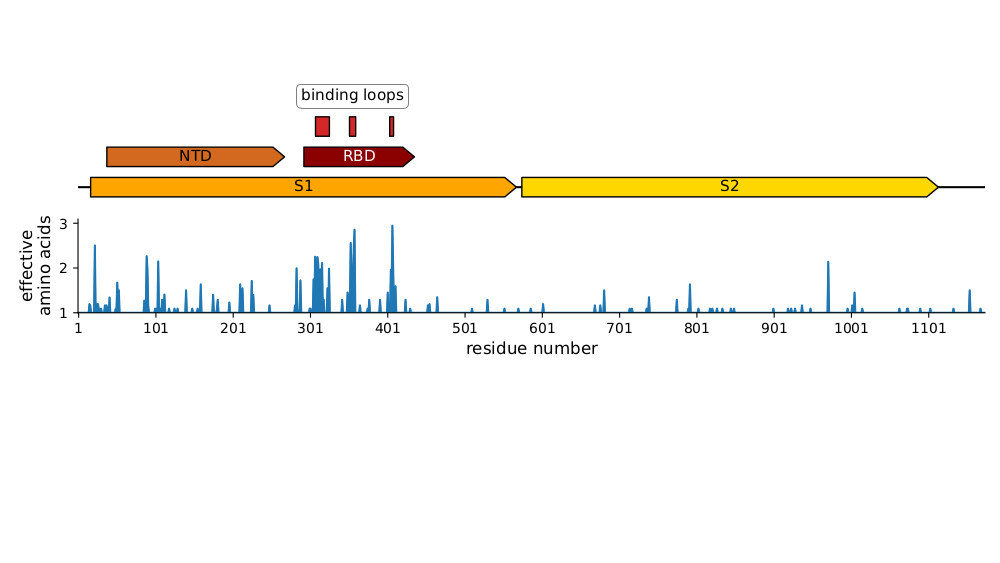
Strongest evolutionary selection is in RBD
Sites of evolutionary change in the spike of CoV-229E over the last four decades

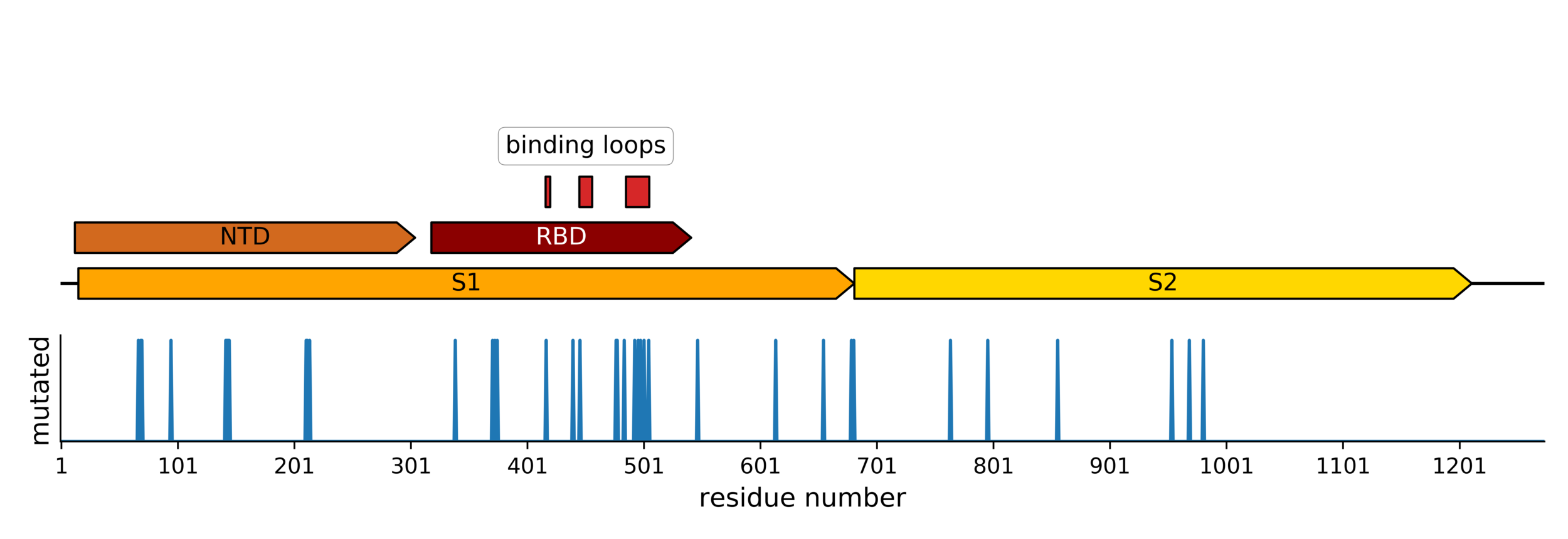
Sites of mutations in SARS-CoV-2 Omicron (BA.1) spike relative to Wuhan-Hu-1
Main difference is SARS-CoV-2 also fixing transmissibility-enhancing spike mutations that affect proteolytic processing and stabilize defects cause by furin-cleavage site
Majority of neutralizing antibody response in vaccinated/infected humans targets RBD
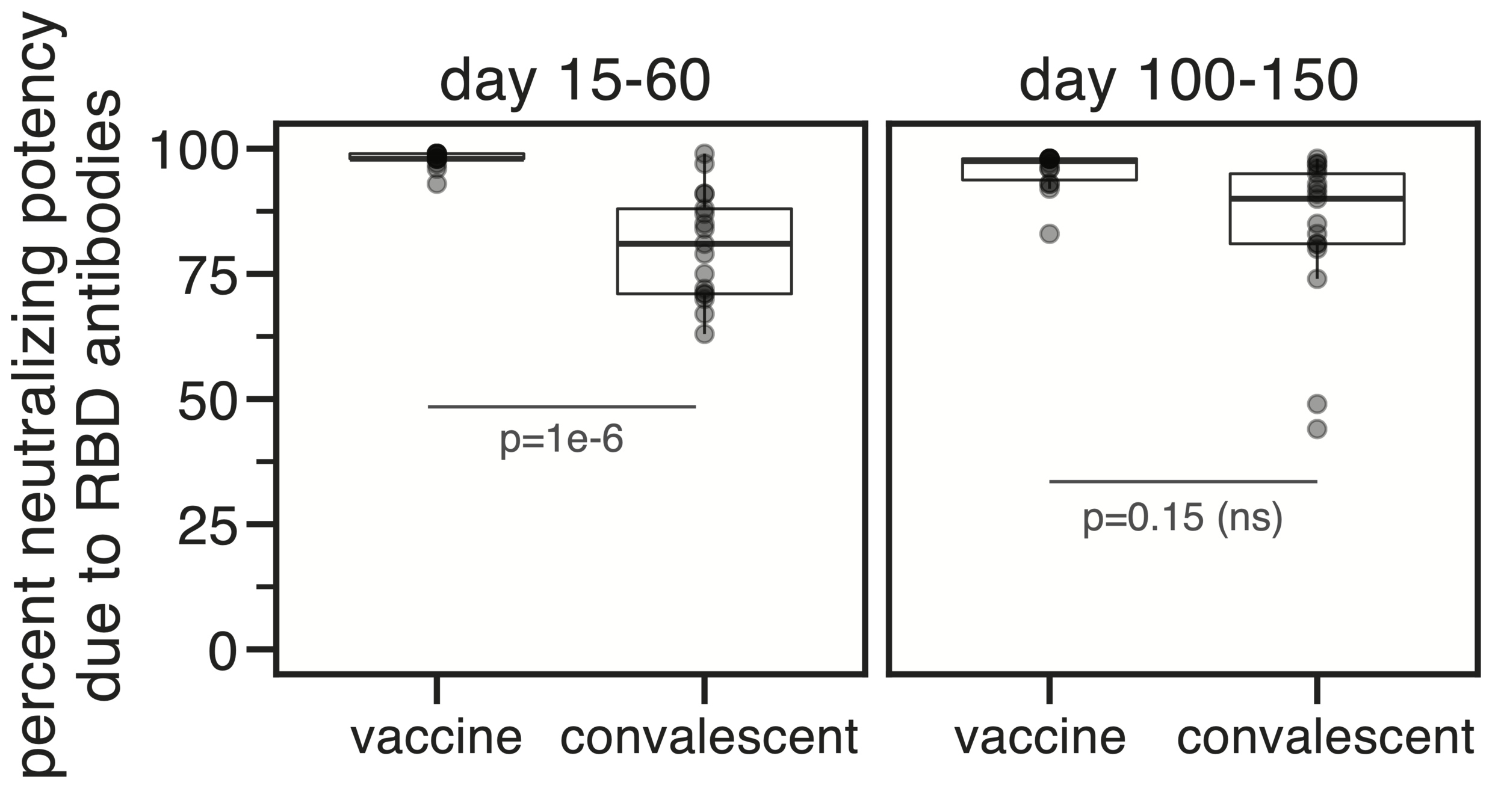
Importance of RBD
Human CoVs, which evolve to escape transmission-blocking immunity, show strongest selection in RBD. So virus is telling us RBD antibodies matter most for blocking transmission. But non-RBD antibodies and T-cells still matter, especially for reducing disease severity while putting less selection on virus.
Most neutralizing activity from RBD antibodies (although antibodies to other domains such as NTD can also be neutralizing).
However, a minority of all anti-spike antibodies elicited by current vaccines target RBD. Given these facts, we should continue to weigh pros / cons of RBD-only vaccines.
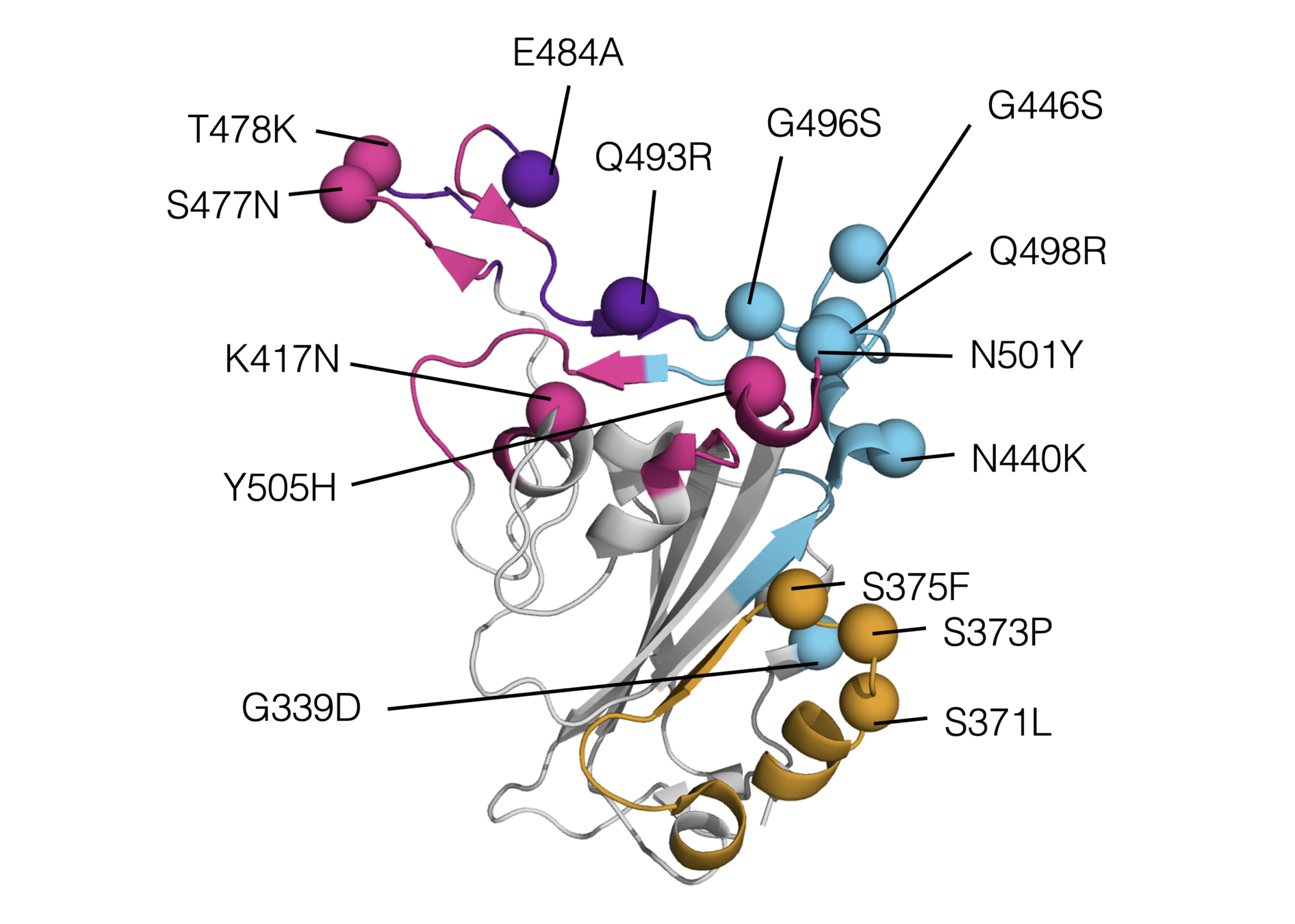
Omicron has a lot of mutations, especially in the receptor binding domain (RBD)
RBD will not run out of evolutionary space
25 of 31 residues in CoV-229E RBD that contact receptor varied during virus's evolution in humans over last ~50 years (Li et al, eLife, 2019)
There are lots of mutations to SARS-CoV-2 RBD that retain (and sometimes even enhance) ACE2 affinity (Starr et al, 2020)
We map antibody escape mutations using yeast display deep mutational scanning
RBD
fluorescently labeled antibody
yeast
fluorescent tag on RBD
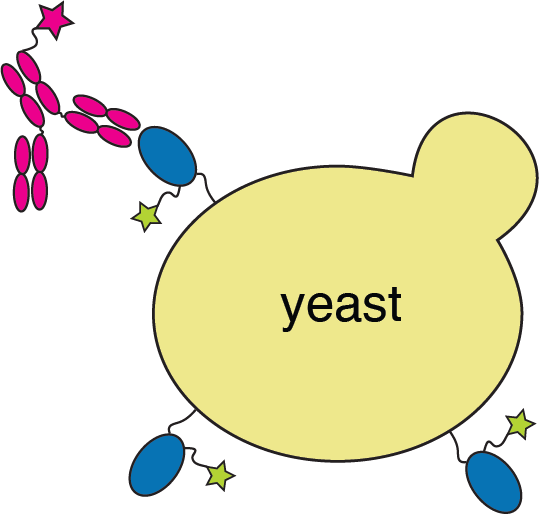
Escape map from a single antibody
Escape maps from lots of antibodies
Infection / vaccination elicit polyclonal antibodies that can bind many epitopes
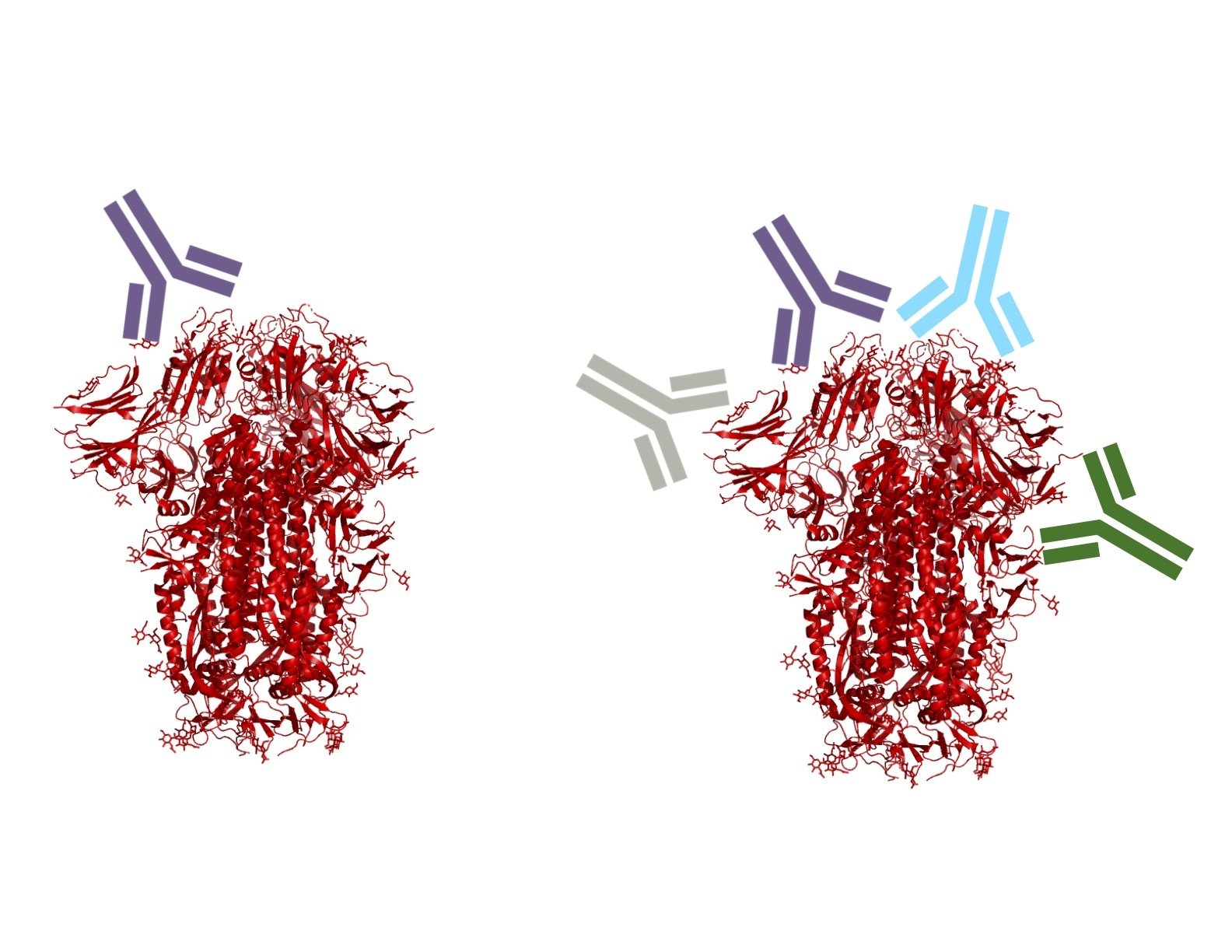
Monoclonal antibodies bind one epitope, so can usually be escaped by single mutation
Polyclonal antibodies can bind many epitopes, so often more resistant to escape
We know where in RBD antibodies elicited by current Wuhan-Hu-1-like vaccines bind
Plot from escape calculator described in Greaney et al (2021)

Plot from escape calculator described in Greaney et al (2021)

Omicron (BA.1) is extensively mutated at important RBD antigenic sites
Plot from escape calculator described in Greaney et al (2021)

Omicron (BA.2) shares most but not all key RBD antigenic mutations with BA.1
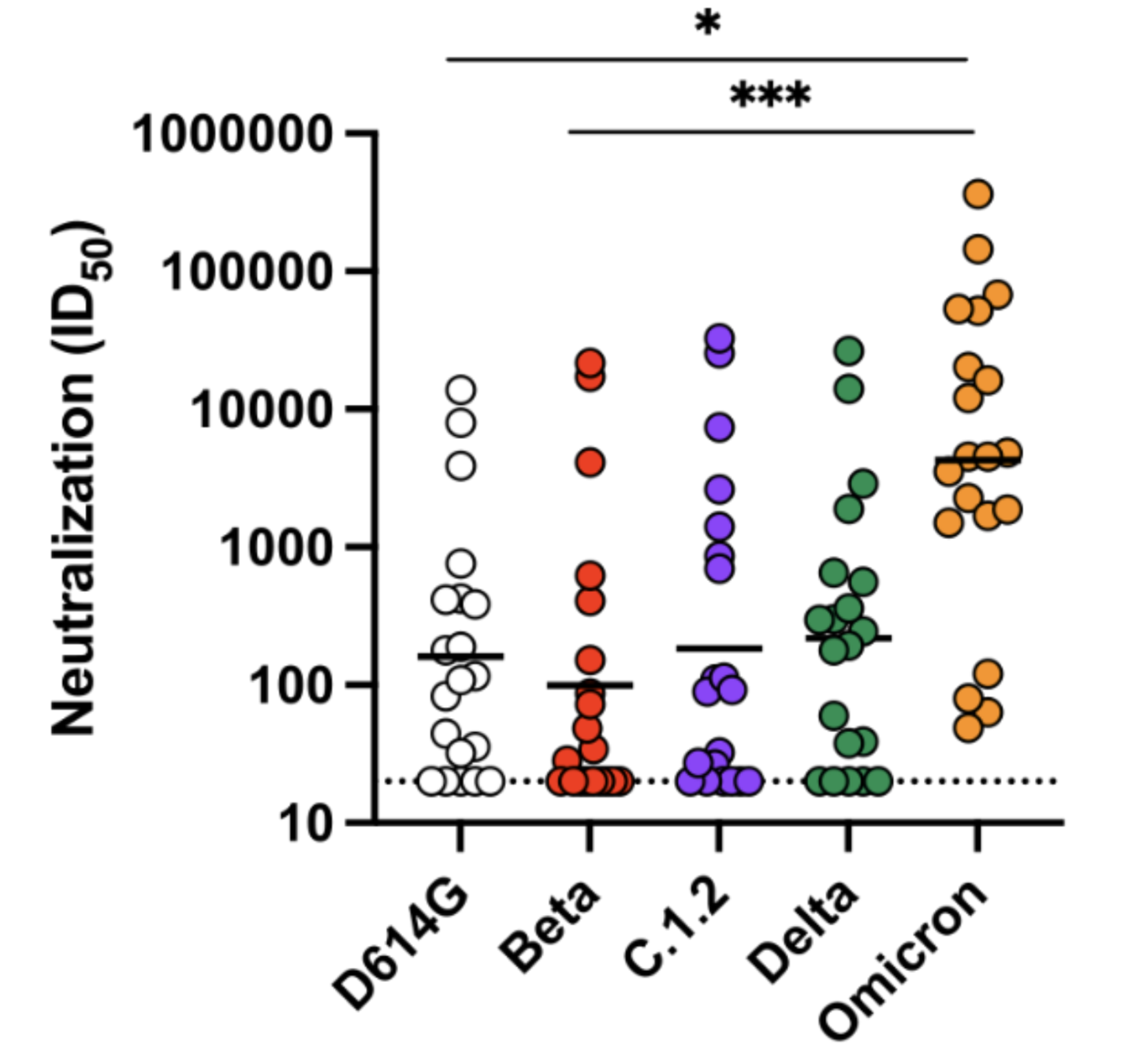
Current vaccines elicit substantially worse titers to Omicron than Delta

Delta
Omicron (BA.1)
Most other studies find similar ~20-fold drop in Omicron neutralization for sera after two doses of current vaccine.
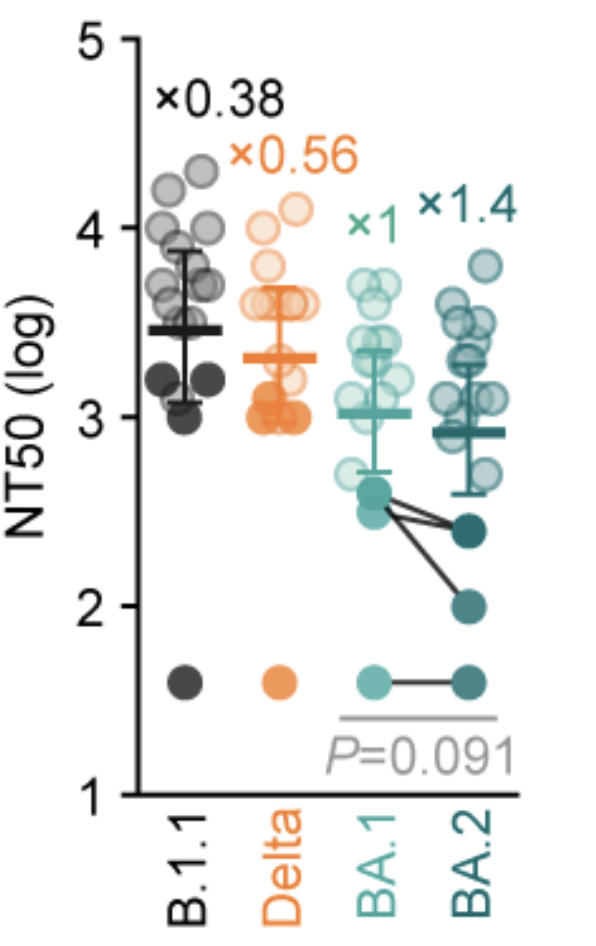
Similarly, Omicron infection without prior immunity elicits poor titers to other variants
Serum neutralization titers after primary human infections with Omicron
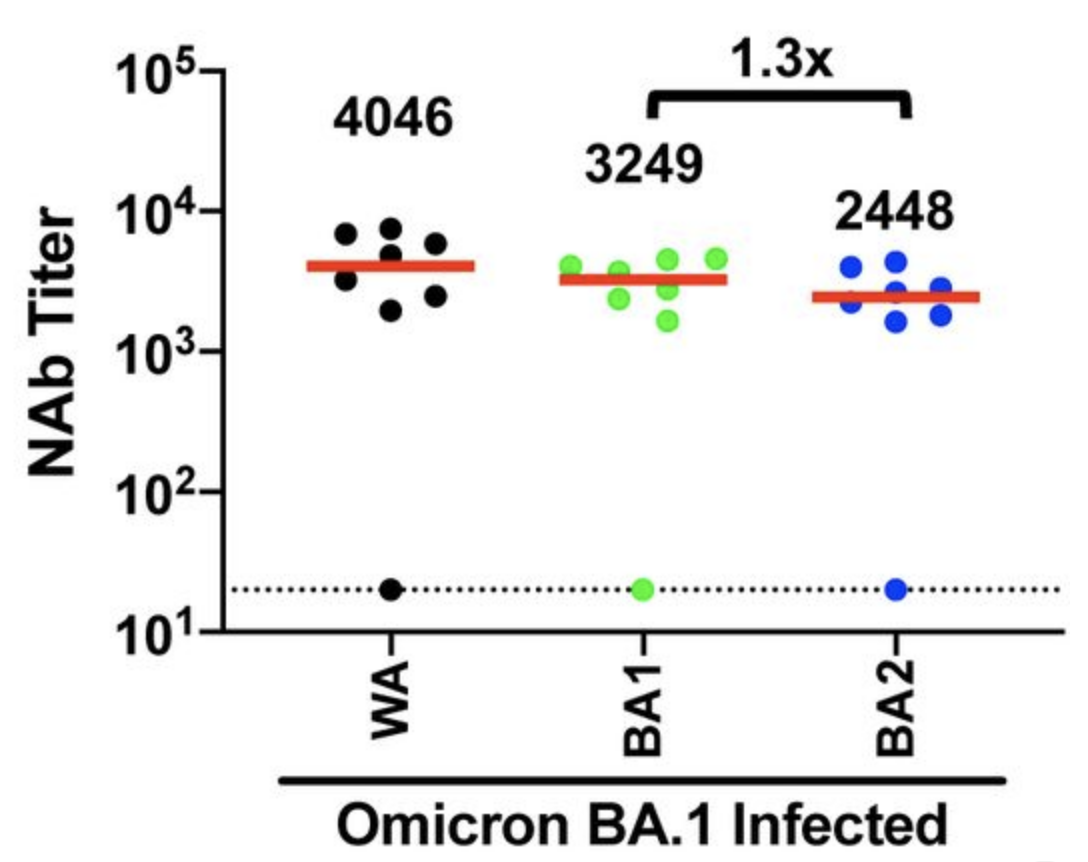
Omicron infection after current vaccine elicits good neutralization of Omicron
Humans vaccinated with current vaccines then infected with Omicron BA.1 neutralize Omicron BA.1 and BA.2 comparably to Delta and older Wuhan-Hu-1-like viruses
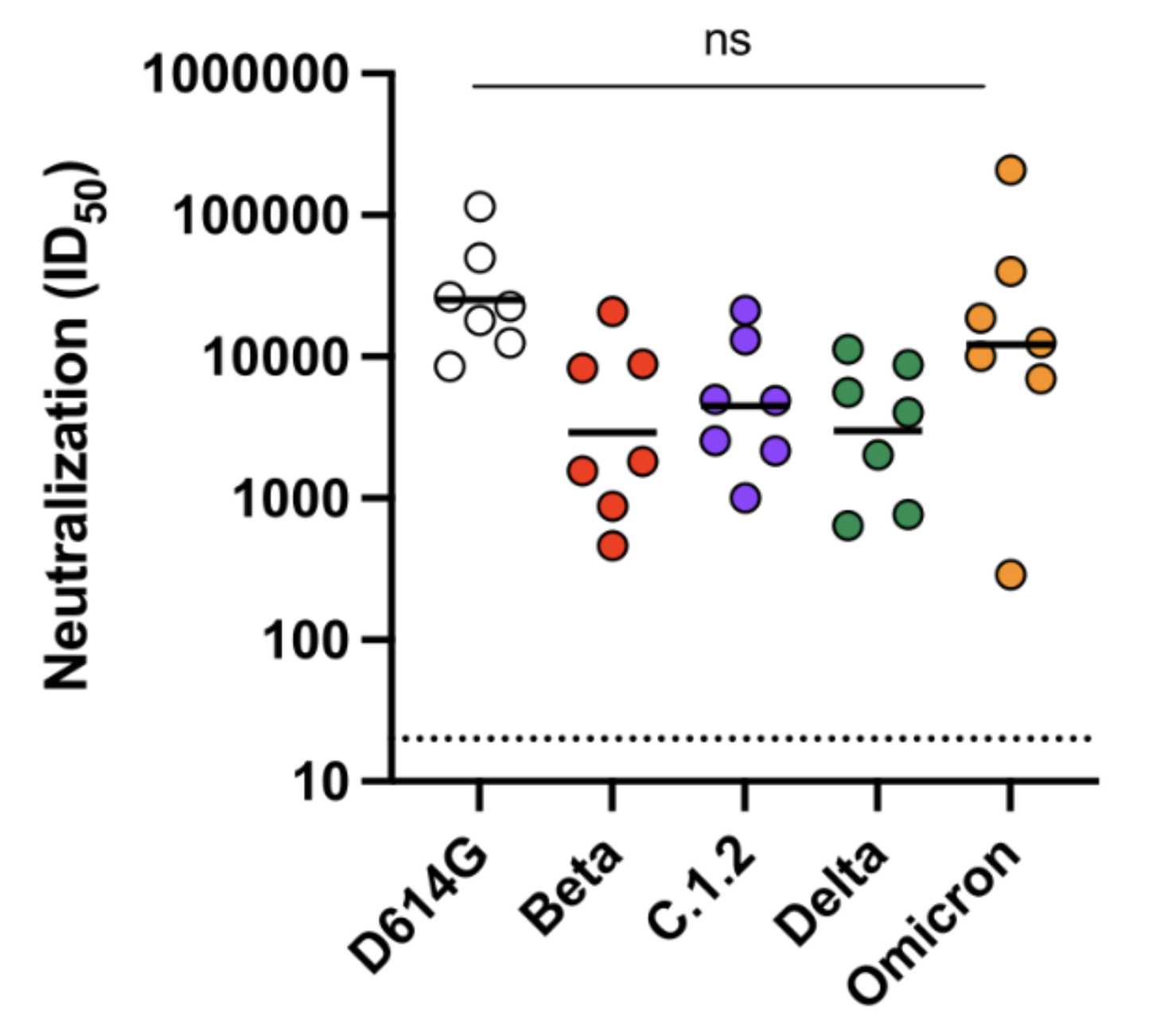
Even with an update to Omicron, there will still be a strong effect from immune imprinting with earlier vaccines / variants
If Omicron remains dominant in future, we should probably update vaccine to Omicron
What if Omicron isn't dominant in future?
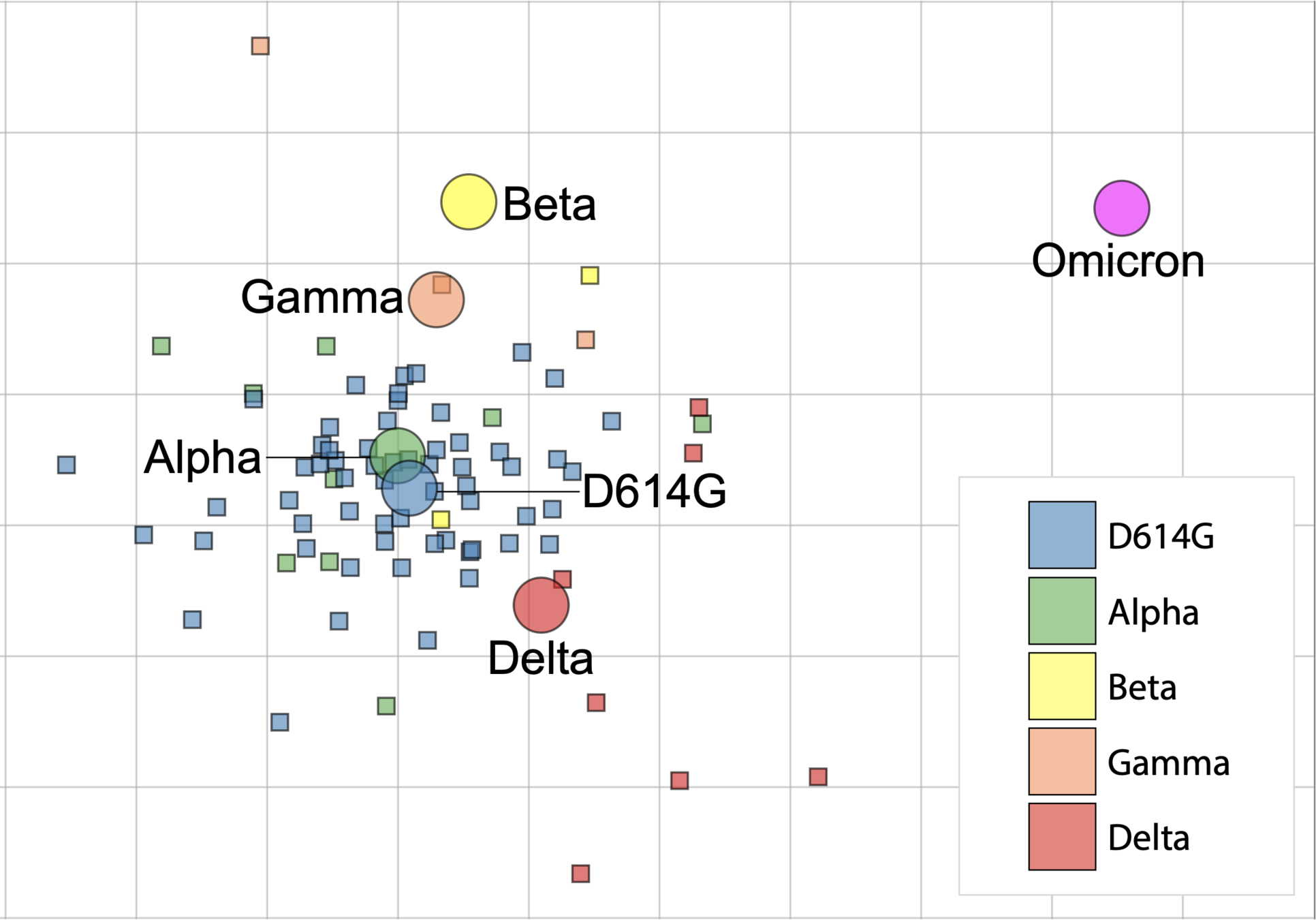
Data from van der Straten et al (2022); similar results in Wilks et al (2022)
All pre-Omicron variants are more antigenically similar
Continued Omicron circulation will make vaccine strain update more urgent
Effect of any update will be somewhat muted by immune imprinting to current vaccine
Consider cocktail vaccines, which might induce broader response and buffer uncertainty in evolutionary forecasts
Crowe lab (Vanderbilt)
Chu lab (Univ Wash)
Veesler lab (Univ Wash)
King lab (Univ Wash)
Li lab (Brigham & Women's)
Boeckh lab (Fred Hutch)
Alex Greninger (Univ Wash)
Nussenzweig lab (Rockefeller)
Bjorkman lab (Caltech)






Tyler Starr
Allie Greaney
Rachel Eguia
Bloom lab (Fred Hutch)


Sarah Hilton
Kate Crawford


Andrea Loes