Interpreting the evolution
of SARS-CoV-2
Jesse Bloom
Fred Hutch Cancer Research Center / HHMI
Slides at https://slides.com/jbloom/sars-cov-2-evolution
Disclosures
- I am on the scientific advisory boards of Flagship Labs 77 and Oncorus
- I consult for Moderna
- I am an inventor on Fred Hutch licensed patents related to deep mutational scanning of viral proteins
- My lab has unfunded research collaborations with Vir Biotechnology
Evolution was historically studied by examining changes in phenotype
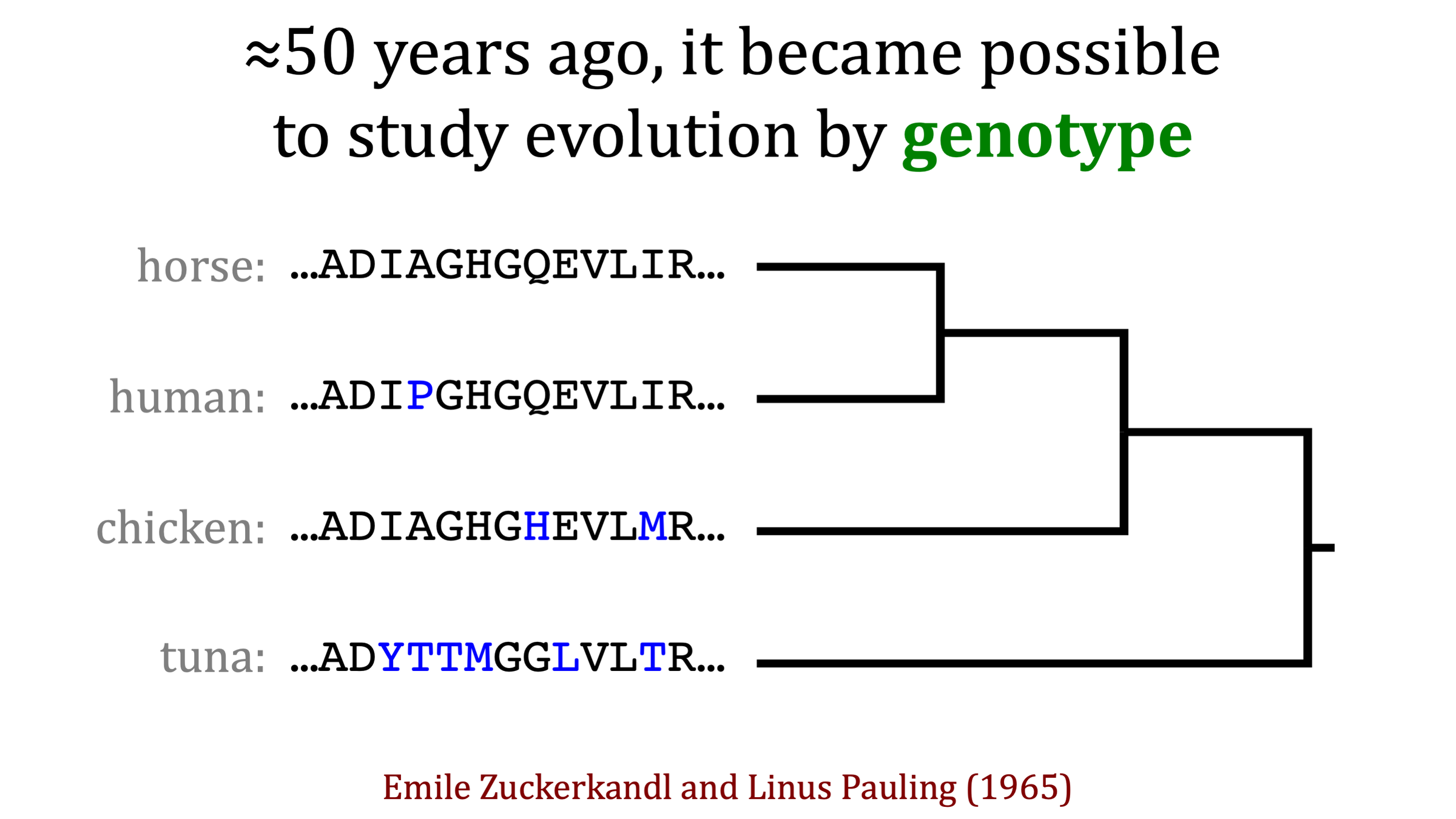
~55 years ago, it became possible to also study evolution in terms of genotype
How changes in genotype affect phenotype is complex
"There is no reason to expect that the extent of functional change in a polypeptide chain is proportional to the number of amino-acid substitutions... It is the type rather than the number of substitutions that is decisive."
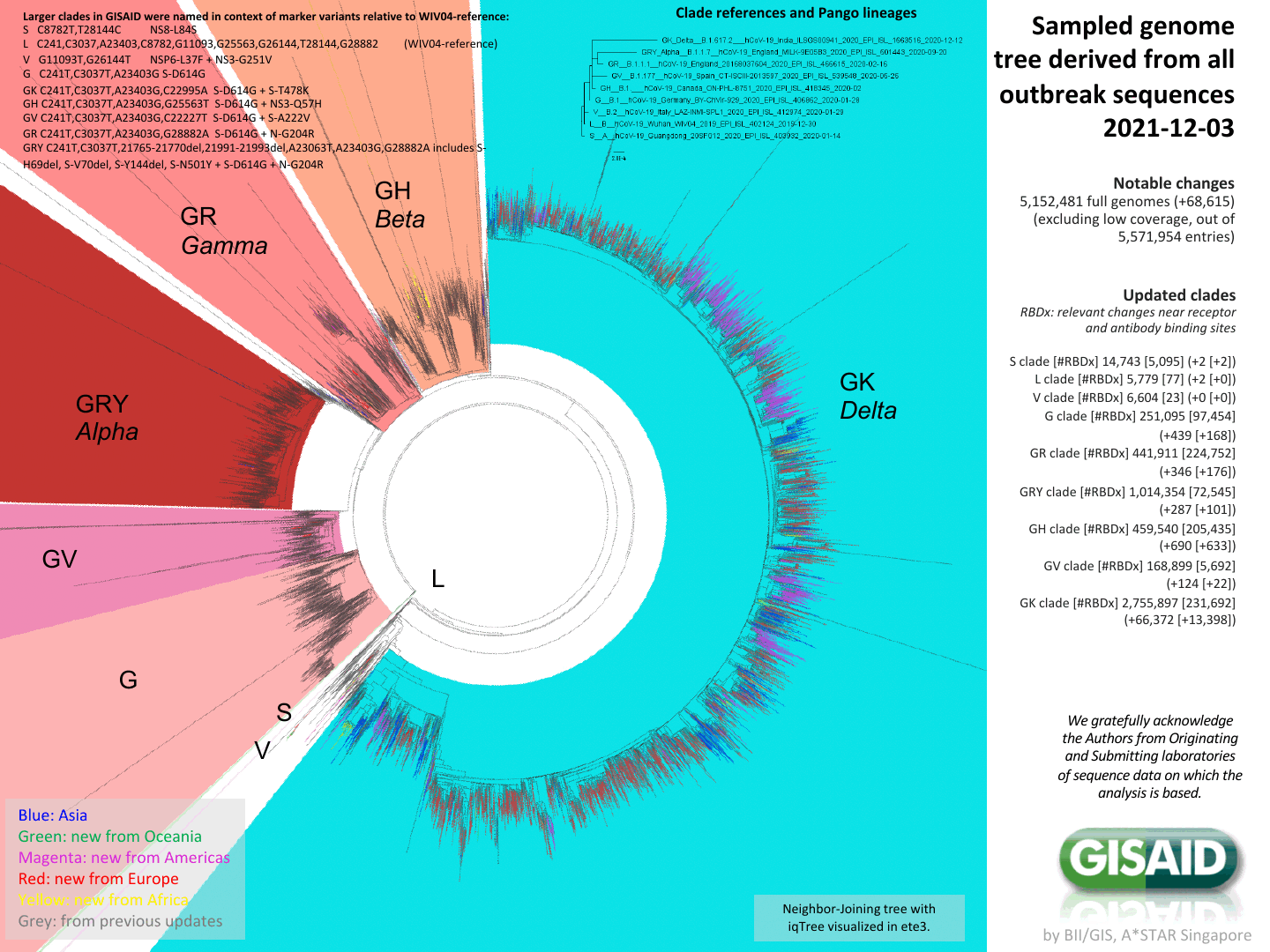
We can now characterize evolution of genotype at high resolution
Phylogenetic tree of 5,152,481 full-length SARS-CoV-2 genomes provided by GISAID
We want to know how changes in genotype affect phenotype


The Coronavirus Is Mutating. What Does That Mean For Us?
The Coronavirus Is Mutating. What Does That Mean For Us?
"There is no reason to expect that the extent of functional change in a polypeptide chain is proportional to the number of amino-acid substitutions... It is the type rather than the number of substitutions that is decisive."

biochemical phenotypes
- protein folding
- affinity for ACE2
- binding by antibodies
Protein structure matters implicitly, as it largely determines mapping from mutation to phenotype.
-Asn-Ile-Thr-Asn-Leu-
-Asn-Ile-Thr-Glu-Leu-
-Asn-Lys-Thr-Asn-Leu-
My lab studies how changes in genotype affect phenotype for viral proteins
Outline
Outline
- SARS-CoV-2 spike and its receptor-binding domain (RBD)
Outline
- SARS-CoV-2 spike and its receptor-binding domain (RBD)
- How do coronavirus spikes evolve?
Outline
- SARS-CoV-2 spike and its receptor-binding domain (RBD)
- How do coronavirus spikes evolve?
- How mutations affect function of RBD
Outline
- SARS-CoV-2 spike and its receptor-binding domain (RBD)
- How do coronavirus spikes evolve?
- How mutations affect function of RBD
- How mutations affect antibody recognition of RBD
Outline
- SARS-CoV-2 spike and its receptor-binding domain (RBD)
- How do coronavirus spikes evolve?
- How mutations affect function of RBD
- How mutations affect antibody recognition of RBD
SARS-CoV-2 virions are decorated by spike proteins that mediate viral entry
virion image from https://phil.cdc.gov/Details.aspx?pid=23312

See Tortorici and Veesler (2019) for general review on coronavirus spikes.
See Walls et al (2020) and Wrapp et al (2020) for details on structure of SARS-CoV-2 spike.
A key portion of the spike is the receptor-binding domain (RBD)

The RBD binds ACE2 receptor on cells
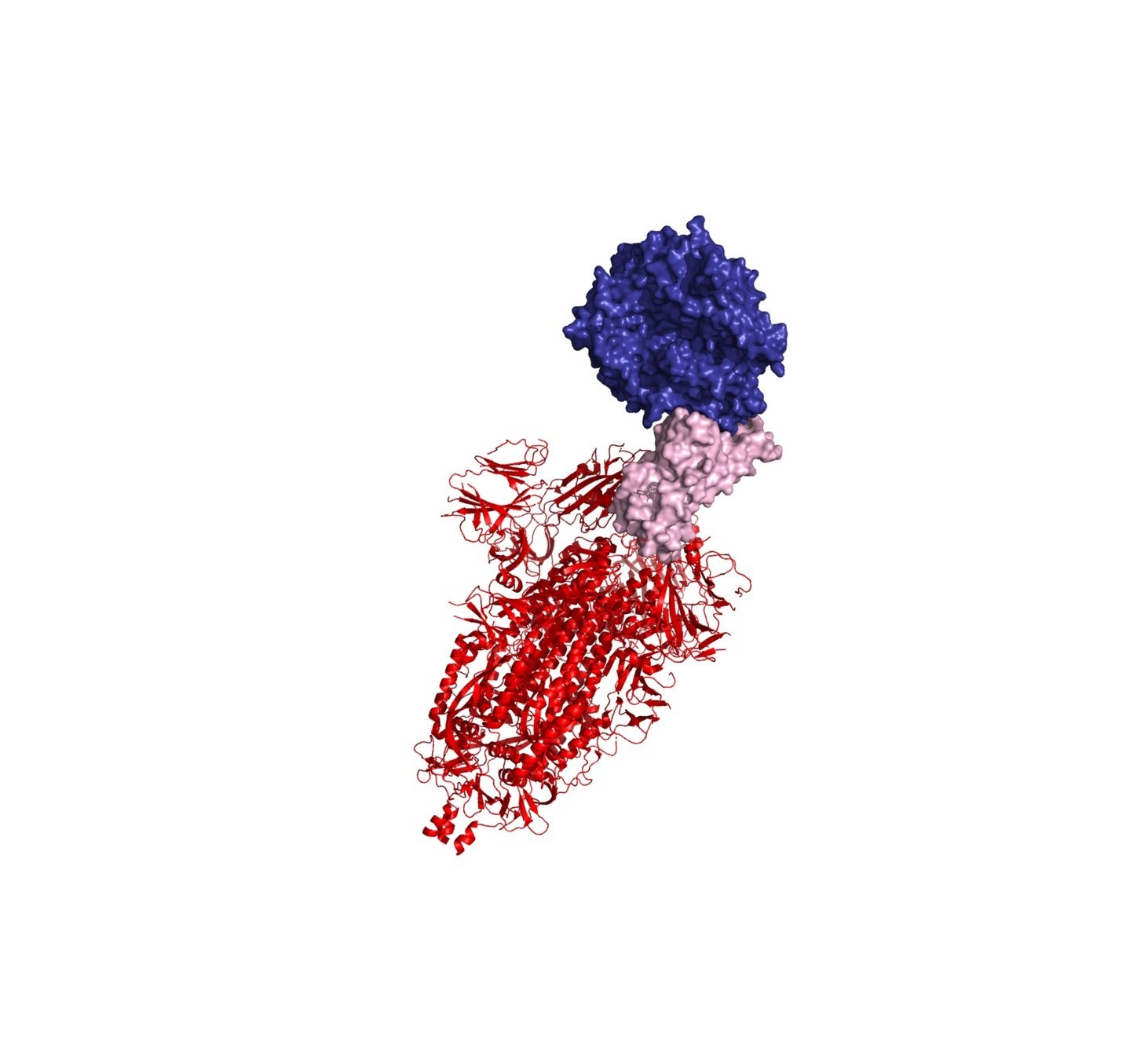
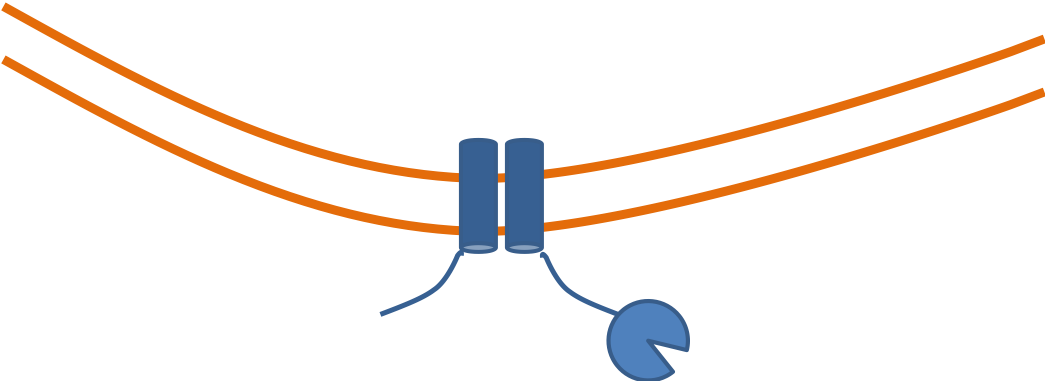
Neutralizing antibodies target spike; often bind RBD and block interaction with ACE2

RBD-binding antibodies are responsible for most neutralizing activity
Data from Greaney et al (2021a, 2021b) using lentiviral pseudotypes on ACE2-overexpressing cells. Similar results seen by Piccoli et al (2020). Neutralizing antibodies can target other spike regions such as NTD (e.g., McCallum et al, 2021).
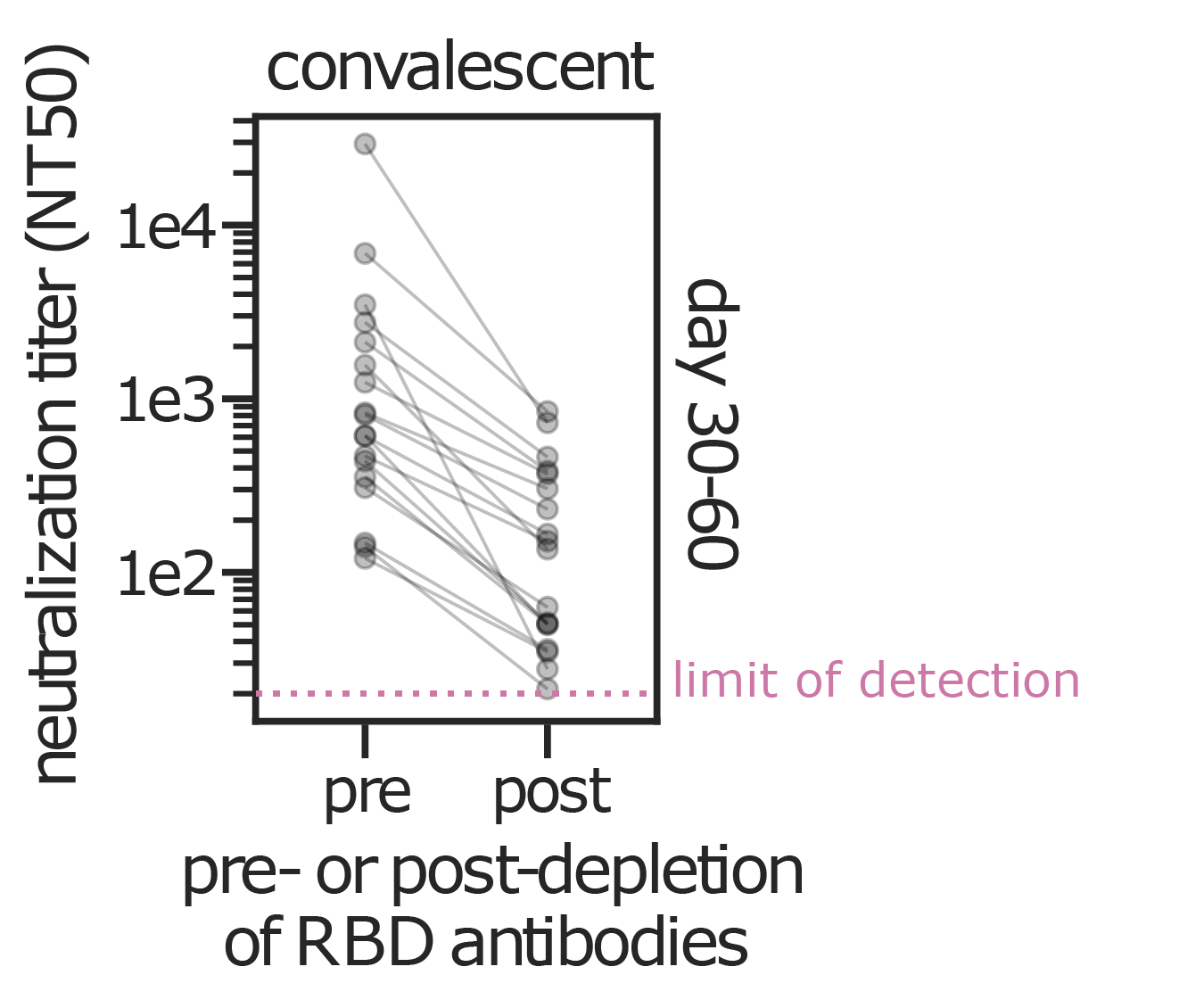
RBD-binding antibodies are responsible for most neutralizing activity
Data from Greaney et al (2021a, 2021b) using lentiviral pseudotypes on ACE2-overexpressing cells. Similar results seen by Piccoli et al (2020). Neutralizing antibodies can target other spike regions such as NTD (e.g., McCallum et al, 2021).

RBD-binding antibodies are responsible for most neutralizing activity
Data from Greaney et al (2021a, 2021b) using lentiviral pseudotypes on ACE2-overexpressing cells. Similar results seen by Piccoli et al (2020). Neutralizing antibodies can target other spike regions such as NTD (e.g., McCallum et al, 2021).
Outline
- SARS-CoV-2 spike and its receptor-binding domain (RBD)
- How do coronavirus spikes evolve?
- How mutations affect function of RBD
- How mutations affect antibody recognition of RBD
Only sometimes does virus evolution lead to changes in antigenic phenotype
- Measles virus: Does not evolve to escape immunity. People are infected at most once in their lives. A vaccine developed in the 1960s still works today.
- Influenza virus: Evolves to escape immunity. People are infected every ~5 years. The vaccine needs to be updated annually.
Initially, many people suggested coronaviruses didn't evolve antigenically

Coronaviruses are the only RNA viruses with a proofreading polymerase (Denison et al, 2011), meaning they have a lower mutation rate than influenza or measles virus
Not a good argument
Evolution isn't just mutation. Rather, it involves three related factors:
- The rate at which mutations arise.
- The phenotypic impact of these mutations.
- How selection (and drift) act on these mutations.
We studied CoV-229E, a coronavirus that causes common colds and has been circulating in humans since at least the 1960s.
Reconstructing evolution of CoV-229E spike

We experimentally generated CoV-229E spikes at ~8 year intervals so we could study them in the lab:
- 1984
- 1992
- 2001
- 2008
- 2016
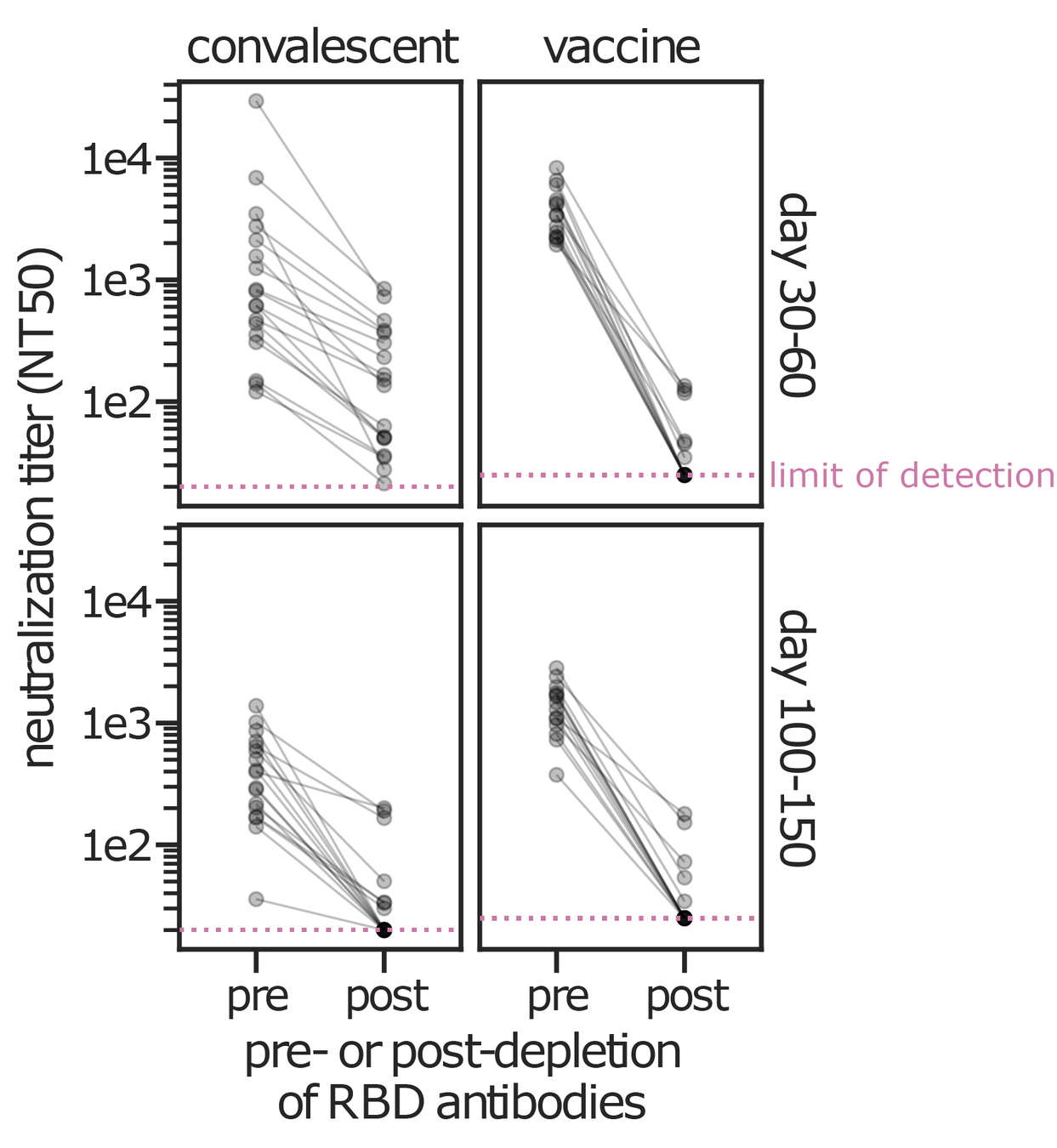
Evolution of CoV-229E spike erodes neutralization by human antibody immunity
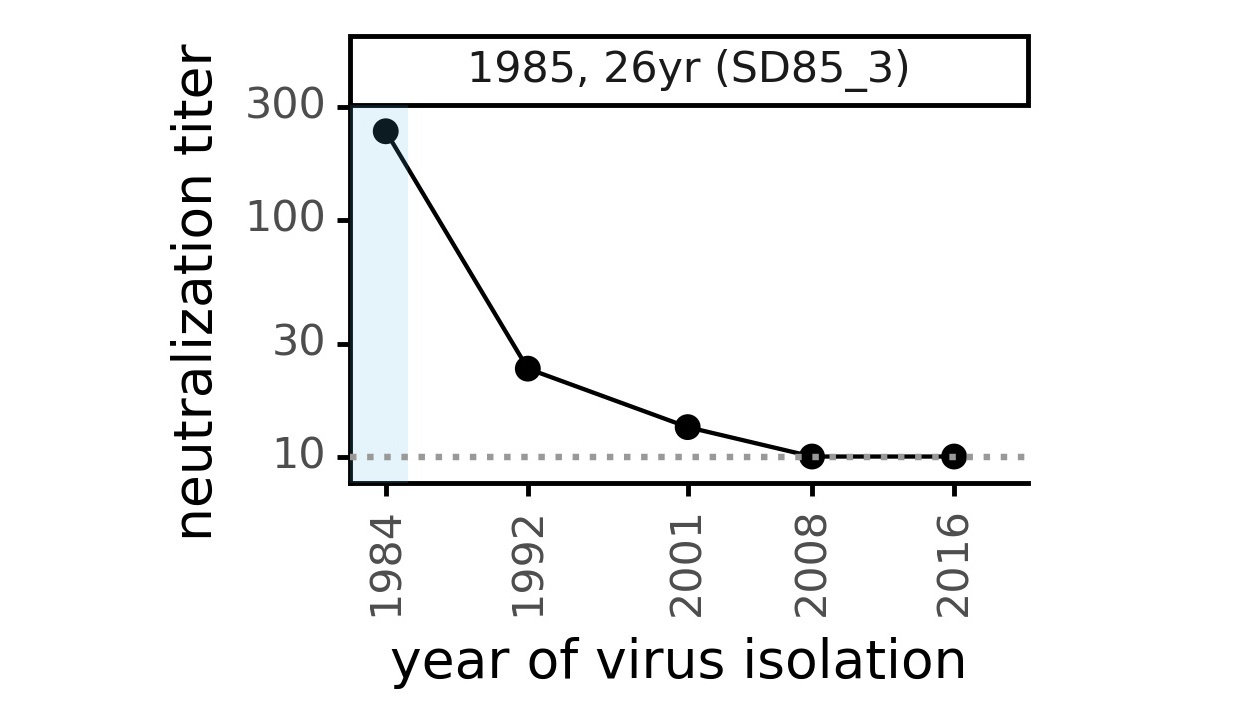
Serum collected in 1985 neutralizes virus with spike from 1984, but less effective against more recent viruses.
Viral evolution erodes antibody immunity of different people at different rates

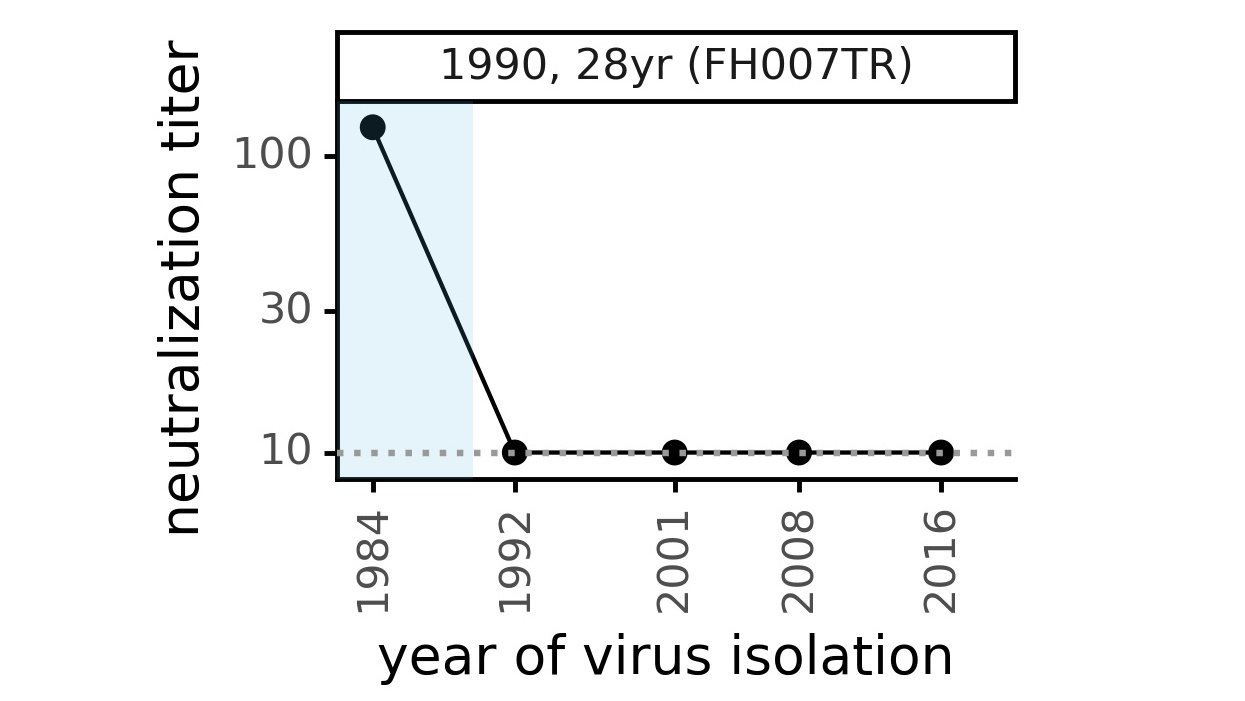
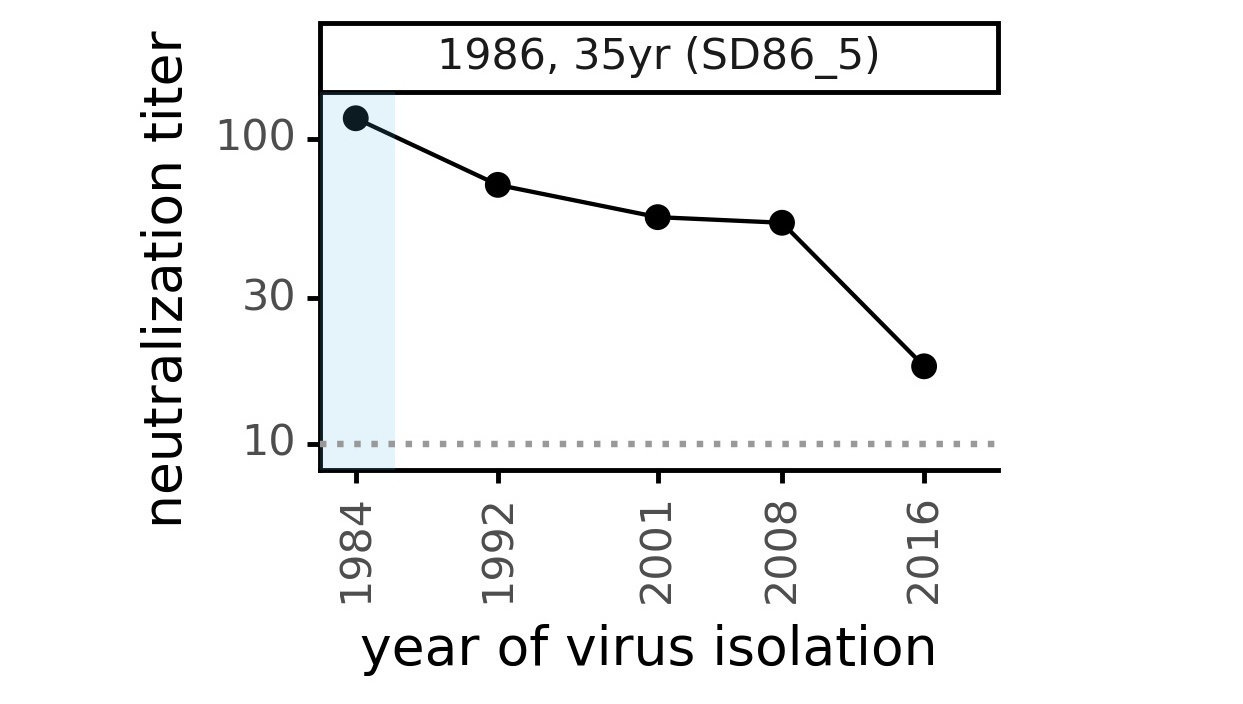
We are studying basis of these differences, as ideally vaccines would elicit more evolution-resistant sera as on the right.
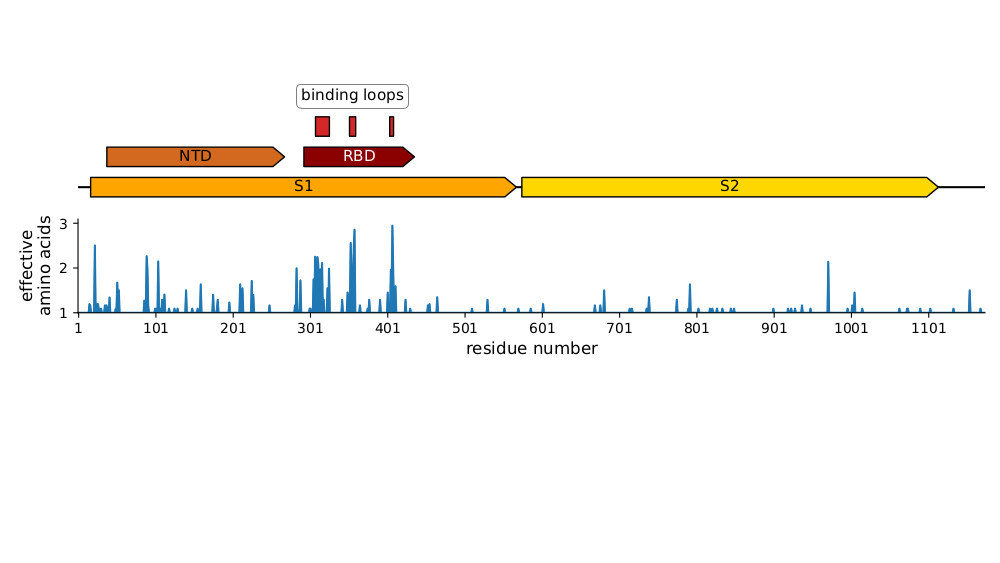
Most mutations in RBD & NTD
Plot of sequence variability across CoV-229E spike taken from Eguia, ..., Bloom, PLoS Pathogens (2021) . See also Wong, ..., Rini, Nature Communications (2017) and Li, ..., Rini, eLife (2019) for detailed structural studies of evolution in receptor-binding loops.
Outline
- SARS-CoV-2 spike and its receptor-binding domain (RBD)
- How do coronavirus spikes evolve?
- How mutations affect function of RBD
- How mutations affect antibody recognition of RBD
For SARS-CoV-2 RBD, want to prospectively map how each mutation affects phenotype
Impossible to measure true viral fitness in the lab, so we focus on three biochemical phenotypes that contribute to fitness:
1) Does RBD fold properly?
2) Does RBD bind ACE2 with high affinity?
3) Is RBD bound by anti-viral antibodies?
For SARS-CoV-2 RBD, want to prospectively map how each mutation affects phenotype
Impossible to measure true viral fitness in the lab, so we focus on three biochemical phenotypes that contribute to fitness:
1) Does RBD fold properly?
2) Does RBD bind ACE2 with high affinity?
3) Is RBD bound by anti-viral antibodies?
Evolutionary pressure is to maintain these two phenotypes...
... while changing this phenotype.
We use yeast display to enable high-throughput experiments
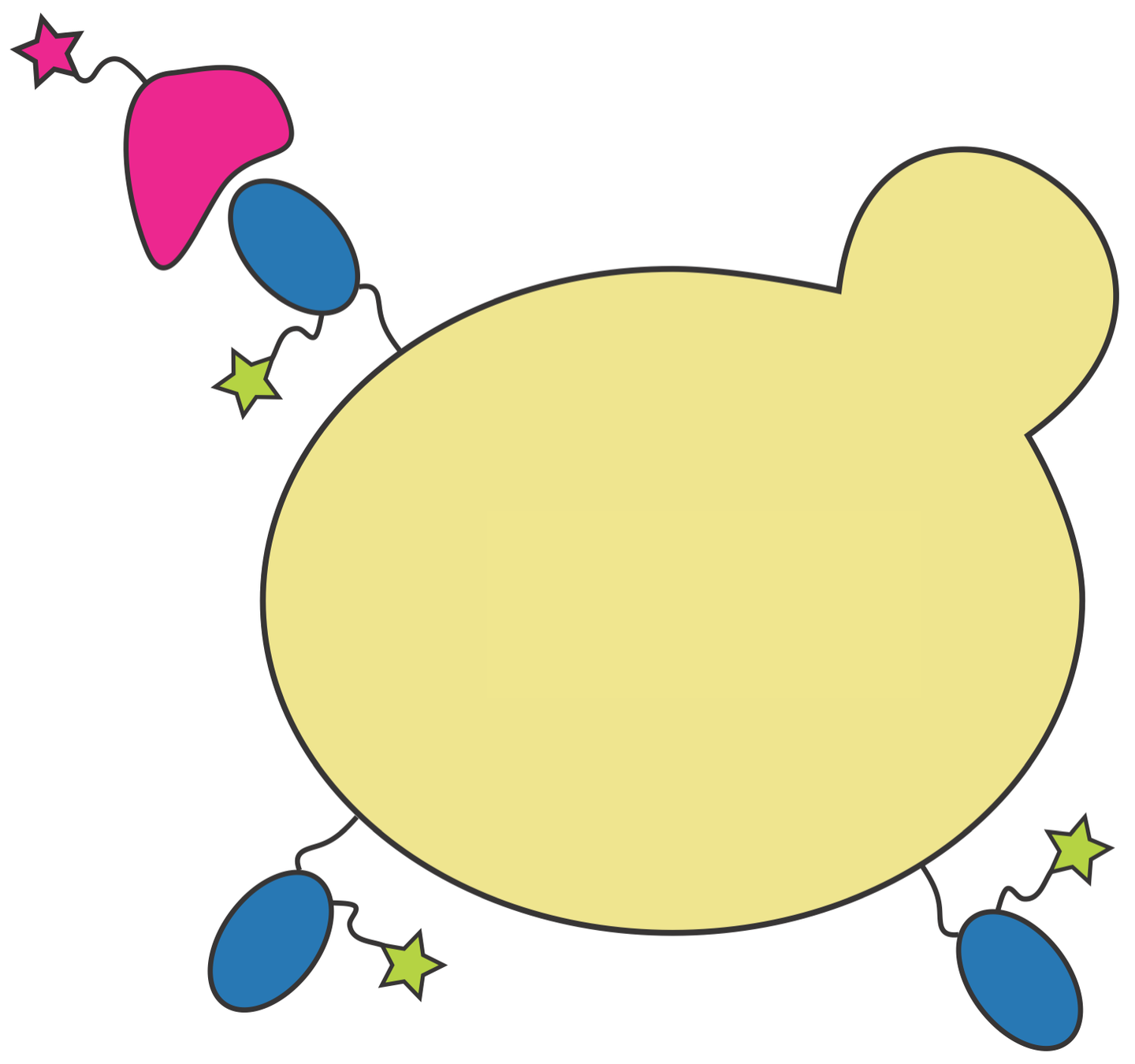
RBD
fluorescent ACE2
yeast
fluorescent tag on RBD
Importantly, we use ACE2 titrations to measure true affinities, not just relative FACS binding signal; see here for details.
Library of yeast, each expressing different RBD mutant with identifying 16 nt barcode

Click here for details on how library is made.
Phenotypic maps of how mutations affect RBD folding & ACE2 affinity
Outline
- SARS-CoV-2 spike and its receptor-binding domain (RBD)
- How do coronavirus spikes evolve?
- How mutations affect function of RBD
- How mutations affect antibody recognition of RBD
We map escape mutations by sorting for RBD variants that don't bind antibody
RBD
fluorescently labeled antibody
yeast
fluorescent tag on RBD
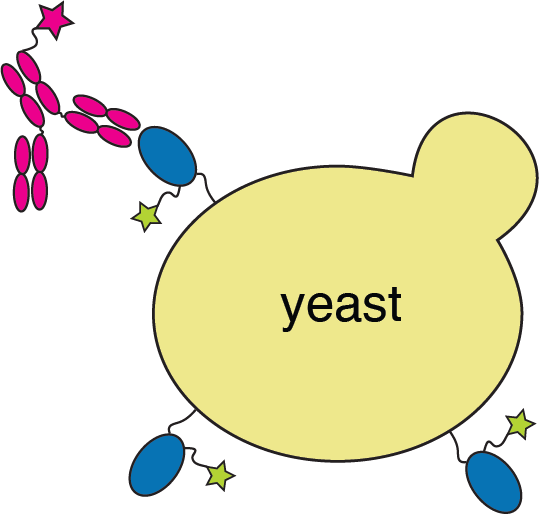
Escape map from a single antibody
Escape maps from lots of antibodies
Infection / vaccination elicit polyclonal antibodies that can bind many epitopes
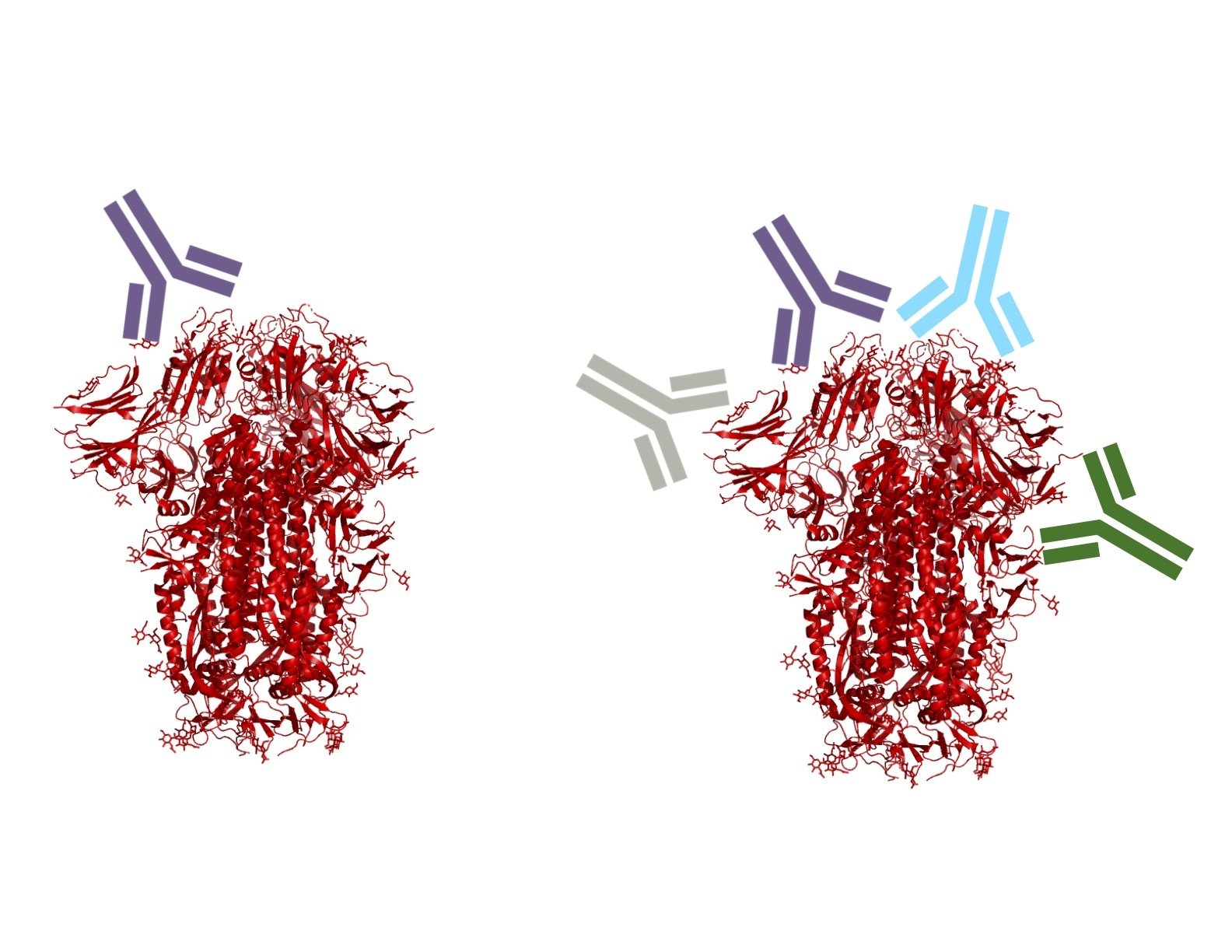
Monoclonal antibodies bind one epitope, so can usually be escaped by single mutation
Polyclonal antibodies can bind many epitopes, so often more resistant to escape
If polyclonal antibodies bind multiple epitopes, escape needs multiple mutations
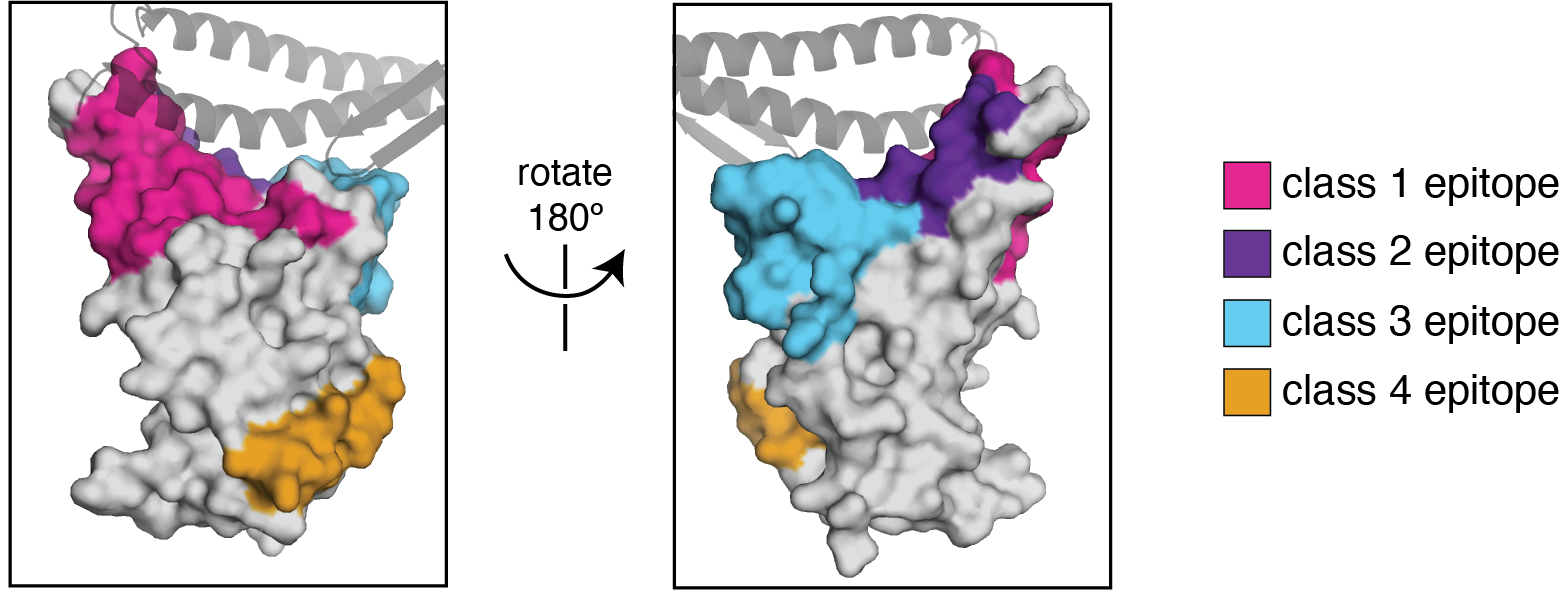
We use the antibody classification scheme from Barnes et al Nature (2020). The extension of these antibody classes to escape mapping is in Greaney et al (2021), and escape maps are available here. Class 1, 2, and 3 antibodies are often potently neutralizing, while class 4 antibodies are less neutralizing (see: Piccoli et al (2020), Dejnirattisai et al (2021), Liu et al (2020), Zost, et al (2020)).
We can visualize escape from different antibody classes
We can calculate how combinations of mutations affect polyclonal binding
Toy example of calculating escape from polyclonal mix:
https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/mini-example-escape-calc/
Escape-calculator with deep mutational scanning data from many antibodies:
https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/escape-calc/
Predictions correlate well with neutralization assays:
https://jbloomlab.github.io/RBD_escape_calculator_paper/neut_studies.html
See Greaney et al (2021) for paper on escape-calculator, and here for more examples on usage.
Omicron has extensive antigenic change
Omicron has a lot more escape than other variants.
Sites to watch for future antigenic mutations include 346.

Conclusions
Relating change in genotype to phenotype is an old challenge in evolutionary biology
"There is no reason to expect that the extent of functional change in a polypeptide chain is proportional to the number of amino-acid substitutions... It is the type rather than the number of substitutions that is decisive."
Relating change in genotype to phenotype is an old challenge in evolutionary biology
The Coronavirus Is Mutating. What Does That Mean For Us?
Relating change in genotype to phenotype is an old challenge in evolutionary biology
"There is no reason to expect that the extent of functional change in a polypeptide chain is proportional to the number of amino-acid substitutions... It is the type rather than the number of substitutions that is decisive."
It's increasingly the bottleneck, since we've gotten good at obtaining genotypes
Phylogenetic tree of 5,152,481 full-length SARS-CoV-2 genomes provided by GISAID

We need to be able to interpret changes in phenotype at comparable scale
Crowe lab (Vanderbilt)
Chu lab (Univ Wash)
Veesler lab (Univ Wash)
King lab (Univ Wash)
Li lab (Brigham & Women's)
Boeckh lab (Fred Hutch)
Alex Greninger (Univ Wash)
Nussenzweig lab (Rockefeller)
Bjorkman lab (Caltech)




These slides: https://slides.com/jbloom/sars-cov-2-evolution
-
Adam Dingens
-
Will Hannon
-
Amin Addetia
-
Keara Malone


Tyler Starr
Allie Greaney
Rachel Eguia
Bloom lab (Fred Hutch)


Sarah Hilton
Kate Crawford


Andrea Loes