SARS-CoV-2 evolution
Jesse Bloom
Fred Hutch Cancer Center / HHMI
These slides at https://slides.com/jbloom/sars2-variants-2022-10-17
Disclosures
I am on the scientific advisory boards of Apriori Bio and Oncorus
I have consulted on topics related to viral evolution for Moderna and Merck
I am an inventor on Fred Hutch licensed patents related to deep mutational scanning of viral proteins
Conceptual background on human coronavirus evolution
Current SARS-CoV-2 variants
Conceptual background on human coronavirus evolution
Current SARS-CoV-2 variants
Circulation patterns of other human CoVs
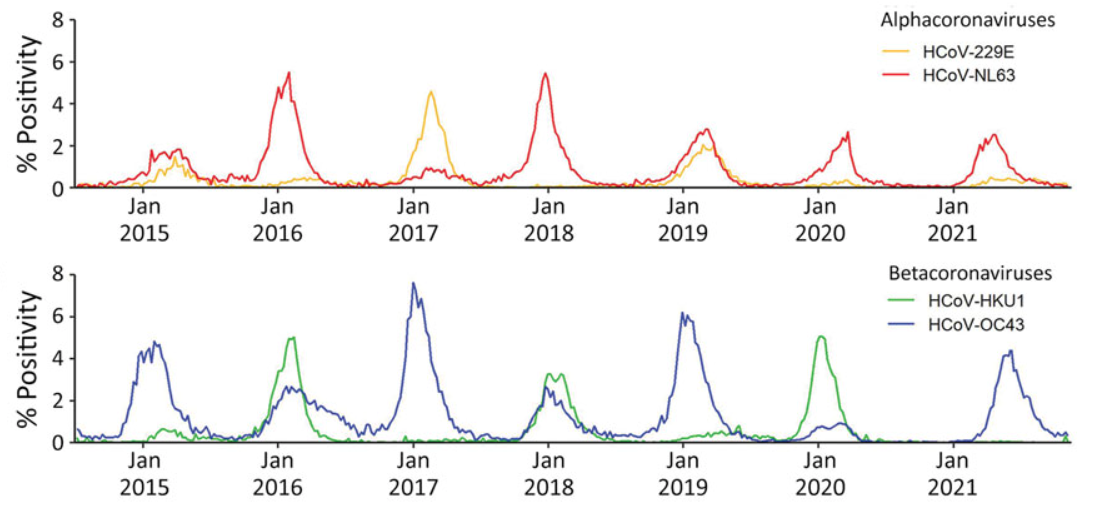
- Cause seasonal waves starting late Oct / early Nov, peaking Jan to mid Feb
- Usually only one each of the two alpha- and beta-coronaviruses causes wave
- The 2021 season was delayed, likely due to COVID-19 related behavior changes
Human CoV-229E undergoes rapid antigenic evolution
Reconstructing evolution of CoV-229E spike

We experimentally generated CoV-229E spikes at ~8 year intervals so we could study them in the lab:
- 1984
- 1992
- 2001
- 2008
- 2016
Note "ladder-like" shape of tree
Evolution of CoV-229E spike erodes neutralization by human antibody immunity
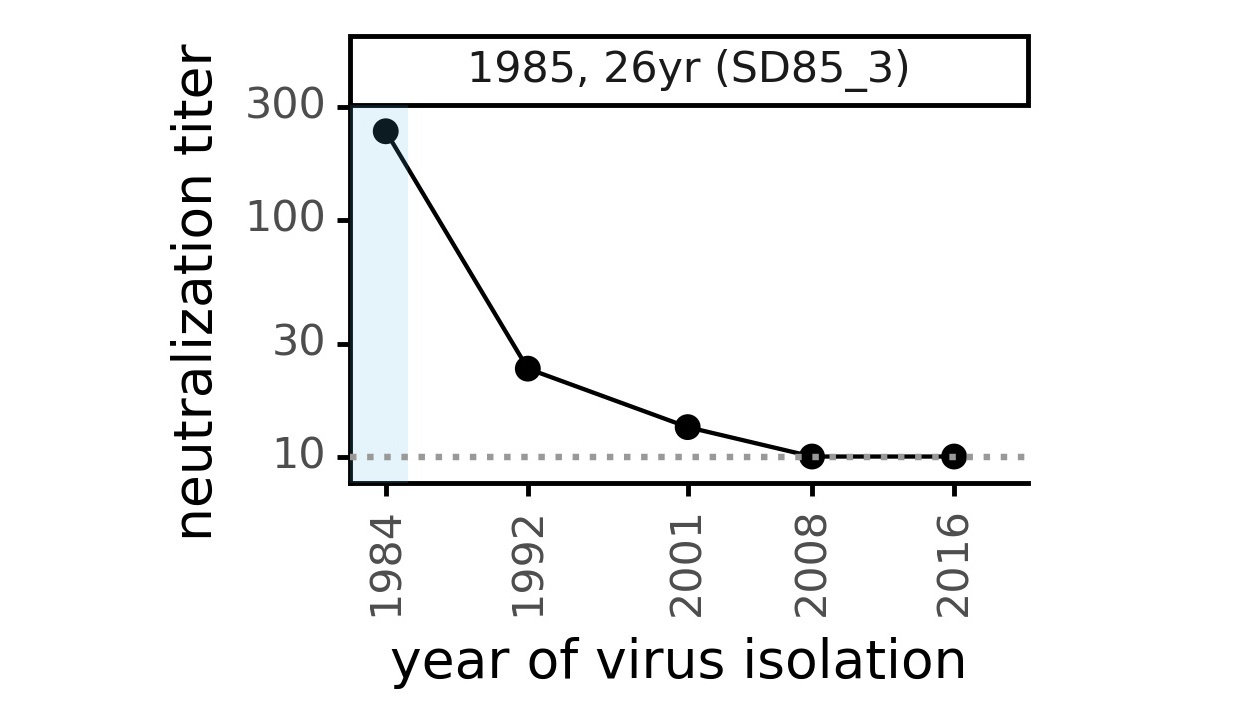
Serum collected in 1985 neutralizes virus with spike from 1984, but less effective against more recent viruses.
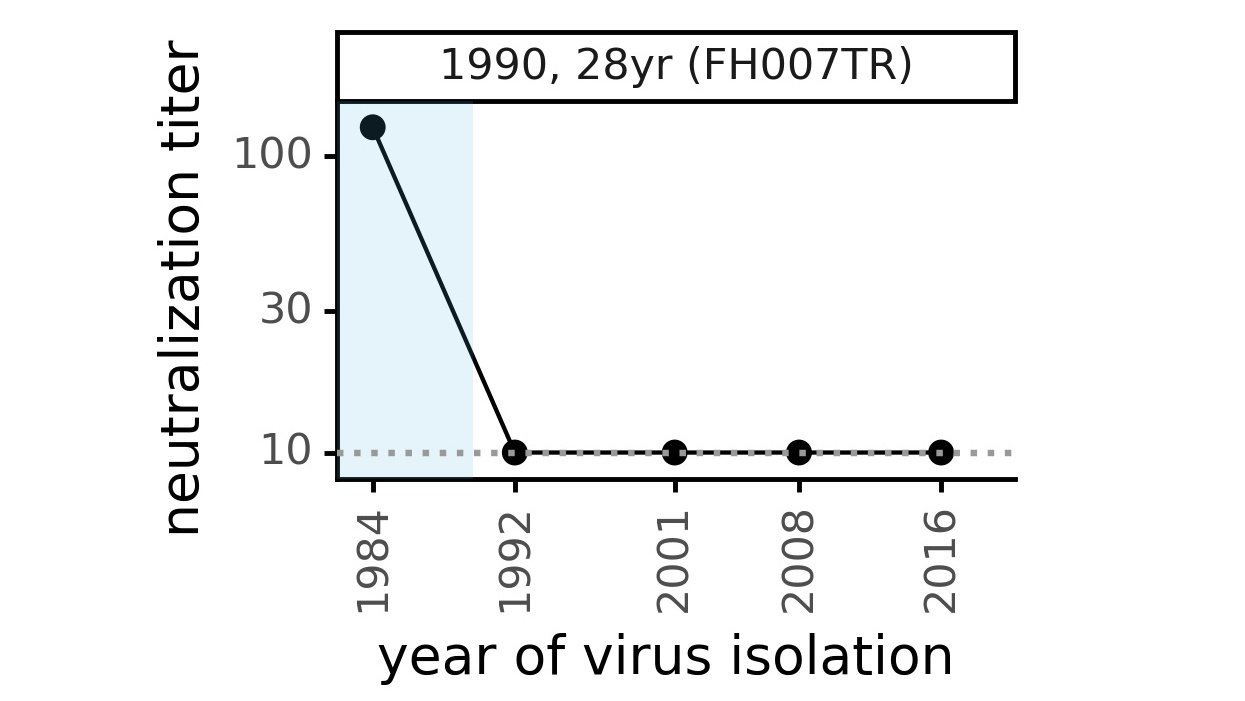
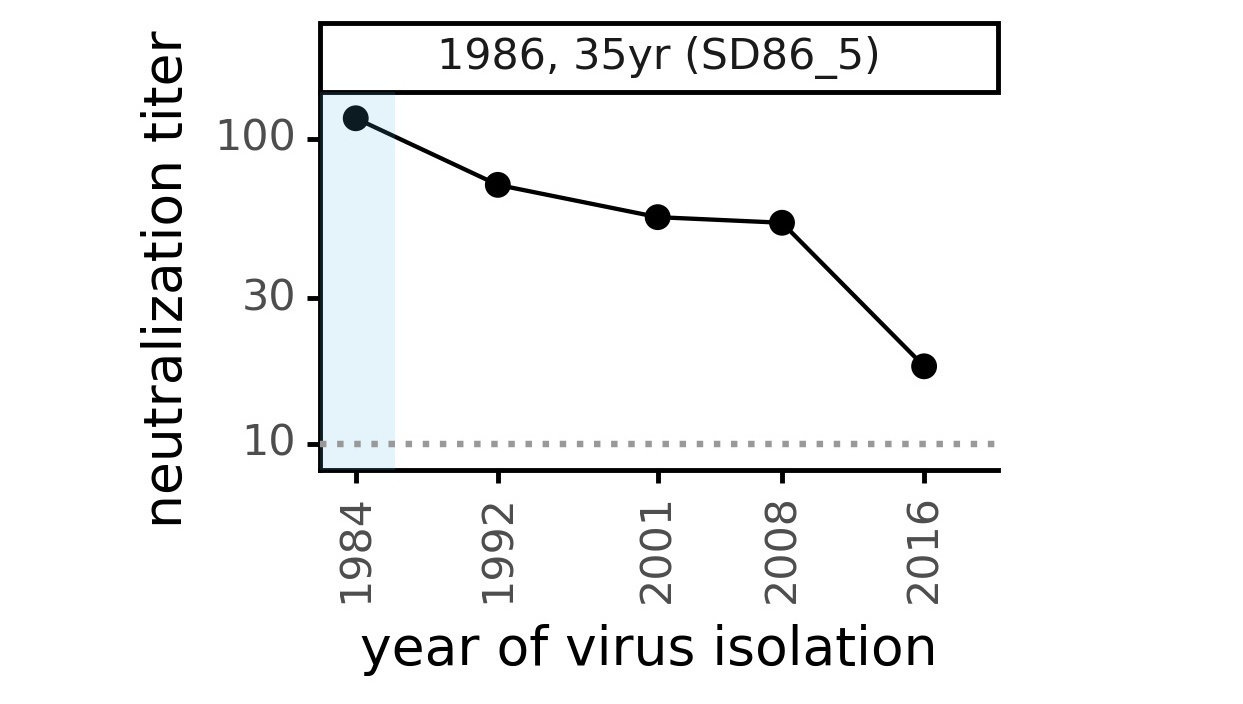
Viral evolution erodes antibody immunity of different people at different rates

Phylogenetic tree shape
CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.

CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.
CoV-OC43 split into two ladder-like lineages. Influenza B evolves this way too. It's theoretically possible to pick well-matched bivalent vaccine.
Phylogenetic tree shape

CoV-229E has ladder-like tree:
- new variants displace old ones
- new variants descend from recent successful ones
Human influenza A evolves this way too. It's theoretically possible to pick single well-matched vaccine strain.

CoV-OC43 split into two ladder-like lineages. Influenza B evolves this way too. It's theoretically possible to pick well-matched bivalent vaccine.
In non-ladder-like tree, next variant not descended from recent successful one. Makes picking vaccine strains difficult.
Phylogenetic tree shape
Omicron emerged on a branch with excess amino-acid mutations only in spike

For each Nextstrain clade, number of mutations plotted versus date of clade origin, with trend line fit only to non-Omicron clades. Omicron clades are circled in red.
Data extracted from Neher (2022) and re-plotted.
This pattern is consistent with antibody selection in chronic human infection

See here for more details on why Omicron probably evolved in a chronic human infection.
Strongest evolutionary selection is in RBD
Sites of evolutionary change in spike of CoV-229E over last four decades
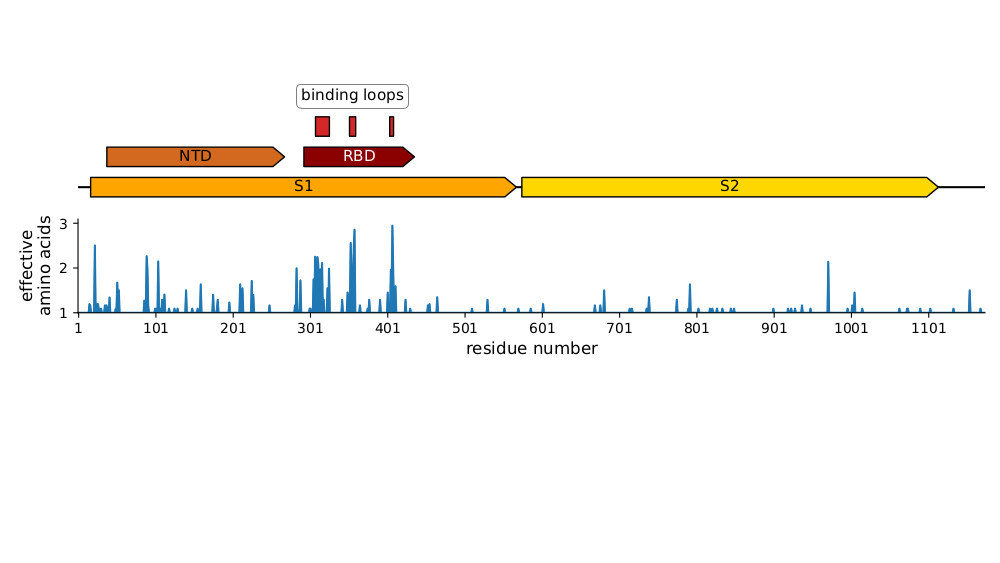
Strongest evolutionary selection is in RBD
Sites of evolutionary change in spike of CoV-229E over last four decades

Sites of mutations in SARS-CoV-2 Omicron BQ.1.1 spike relative to Wuhan-Hu-1
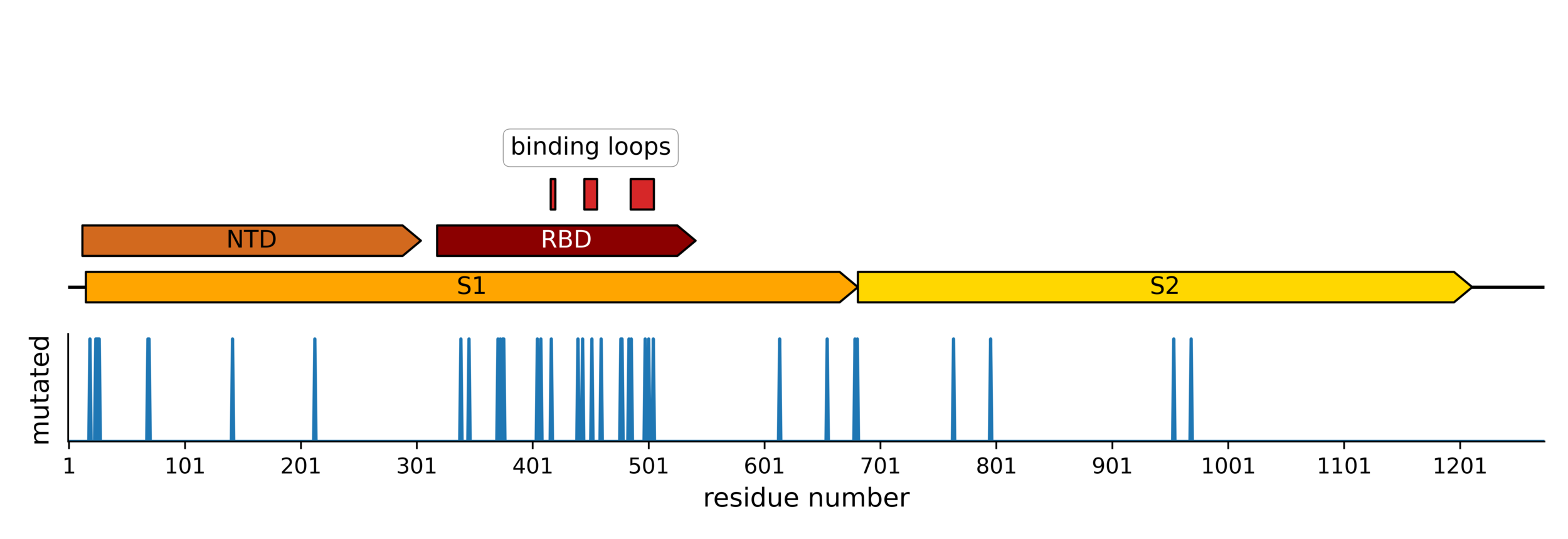
Majority of neutralizing antibody response targets RBD
RBD will not run out of evolutionary space
25 of 31 residues in CoV-229E RBD that contact receptor varied during virus's evolution in humans over last ~50 years (Li et al, 2019)
There are lots of mutations to SARS-CoV-2 RBD that retain (and sometimes even enhance) ACE2 affinity (Starr et al, 2020; Starr et al, 2022)
Conceptual background on human coronavirus evolution
Current SARS-CoV-2 variants
Fastest growing in USA are BQ.1 and BQ.1.1
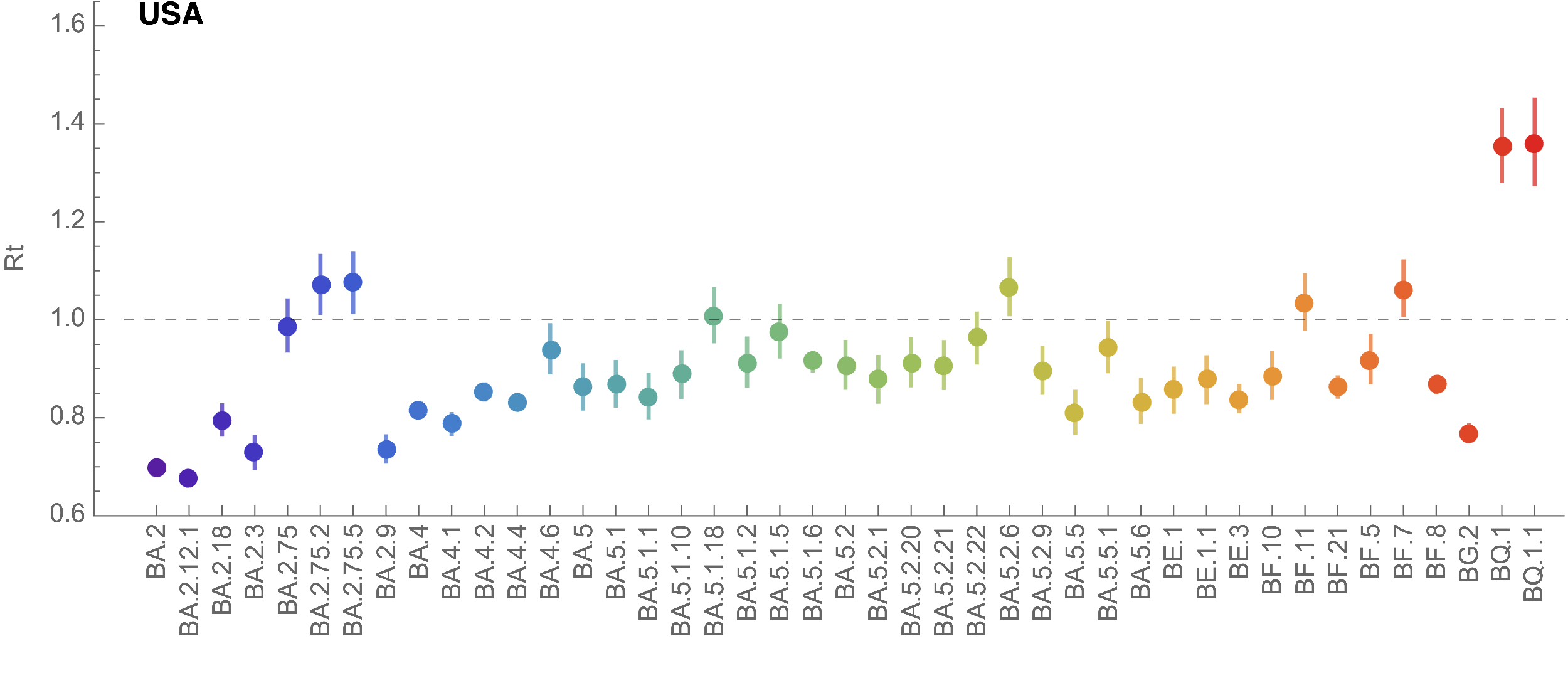
Figure taken from this analysis by Trevor Bedford
BQ.1 is descendant of BA.5 with additional RBD mutations K444T and N460K. BQ.1.1 also has RBD mutation R346T. Both variants evolved by accumulating mutations sequentially.
Internationally, XBB growing similarly fast
Figure taken from this analysis by Trevor Bedford
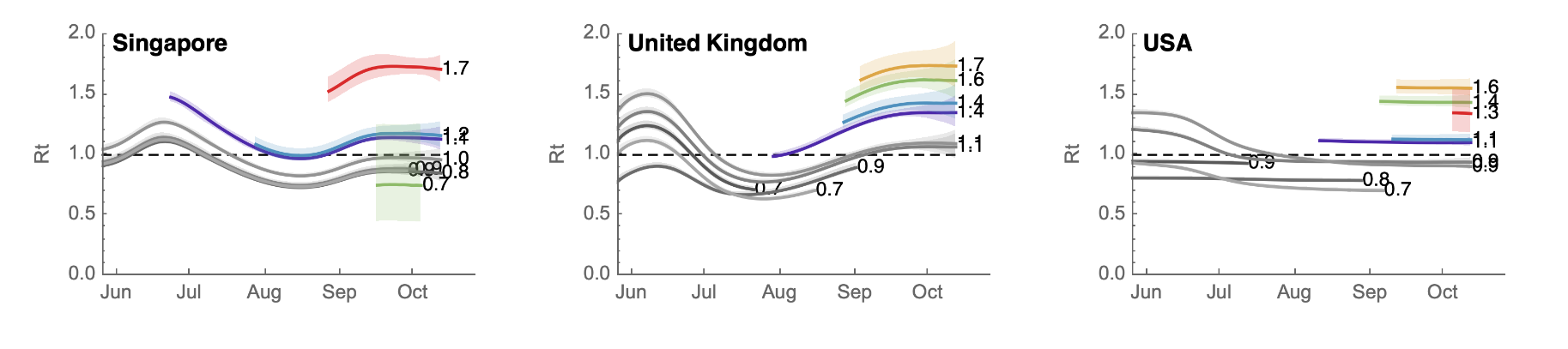

XBB descended from BA.2 as a recombinant of two BA.2 sublineages. Relative to BA.2 it has RBD mutations: R346T, L368I, V445P, G446S, N460K, F486S, F490S, & R493Q reversion.
BQ.1, BQ.1.1 and XBB all bad news for therapeutic antibodies
Data taken from Cao et al (2022)
-
XBB
- fully escapes neutralization by COV2-2130 + COV2-2196 (EvuShield)
- fully escapes neutralization by LY-CoV1404 (bebtelovimab)
- weak (BA.2-level) neutralization by S309 (sotrovimab)
-
BQ.1
- fully escapes neutralization by COV2-2130 + COV2-2196 (EvuShield)
- mostly escapes neutralization by LY-CoV1404 (bebtelovimab)
- weak (two-fold worse than BA.2) neutralization by S309 (sotrovimab)
-
BQ.1.1
- fully escapes neutralization by COV2-2130 + COV2-2196 (EvuShield)
- fully escapes neutralization by LY-CoV1404 (bebtelovimab)
- very weak (7-fold worse than BA.2) neutralilzation by S309 (sotrovimab)
Infection / vaccination elicit polyclonal antibodies
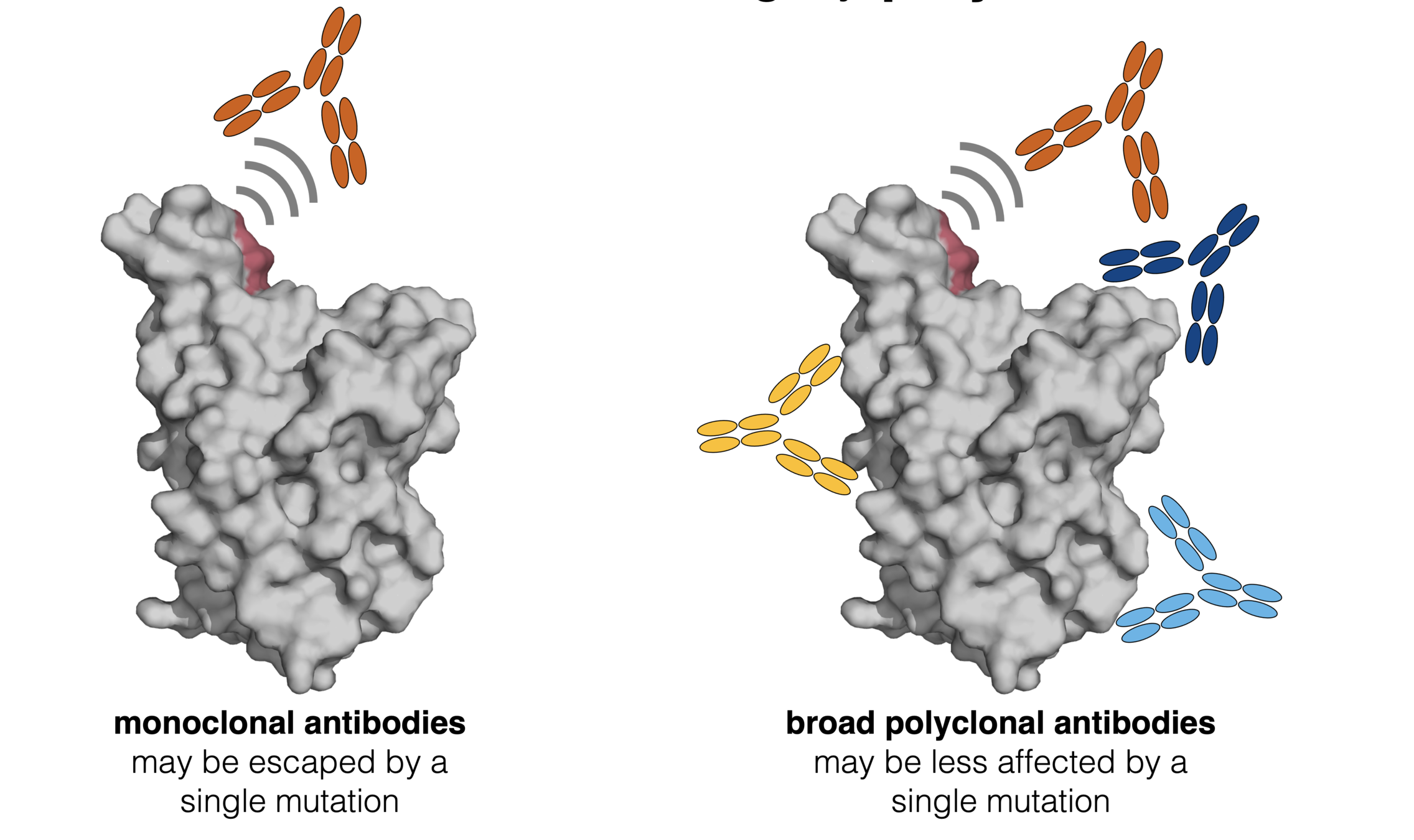
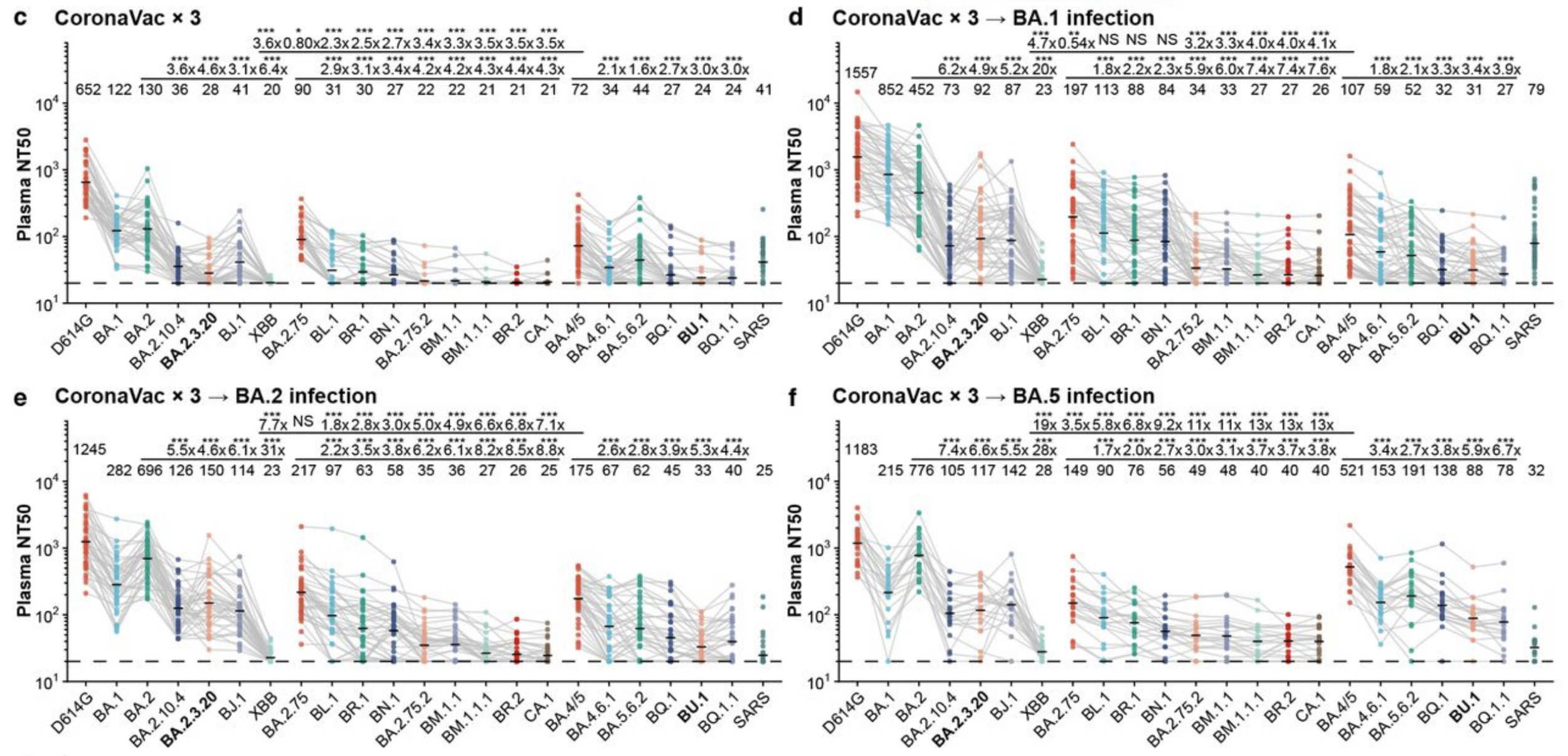
BQ.1.1 and XBB neutralized 4-5 fold worse than BA.5 by polyclonal serum
Although BQ.1.1 escape all histories, BA.5 breakthrough has best neutralization. Suggests BA.5 booster was still good choice. Data taken from Cao et al (2022)
Why do these new variants have so much antibody escape?
We can use deep mutational scanning to map antibody escape mutations:
- Deep mutational scanning of RBD described here
- Large deep mutational datasets described here
- Antibody escape calculator described here and here
- See https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/escape-calc/
Antibodies elicited by early SARS-CoV-2 that neutralize Wuhan-Hu-1 heavily focus on sites like 484, 417, 446
417
446
484

417
446
484
Omicron BA.1 is mutated at many of these sites, which is why it was neutralized substantially less well by current vaccines

417
446
484
Omicron BA.2 has some different RBD mutations than BA.1, but similar overall escape from antibodies from early SARS-CoV-2

Can identify mutations that mediate further escape. In Dec 2021 we predicted 486 as likely site of future evolution--in April 2022, mutation F486V was identified in Omicron BA.4 and BA.5.
486 is largest site of escape for antibodies not already escaped by mutations in BA.2

The reason we are seeing convergent evolution is because virus is getting mutations at key escape sites targeted by vaccine/infection induced immunity. Unfortunately, current antibody drugs target these same sites.
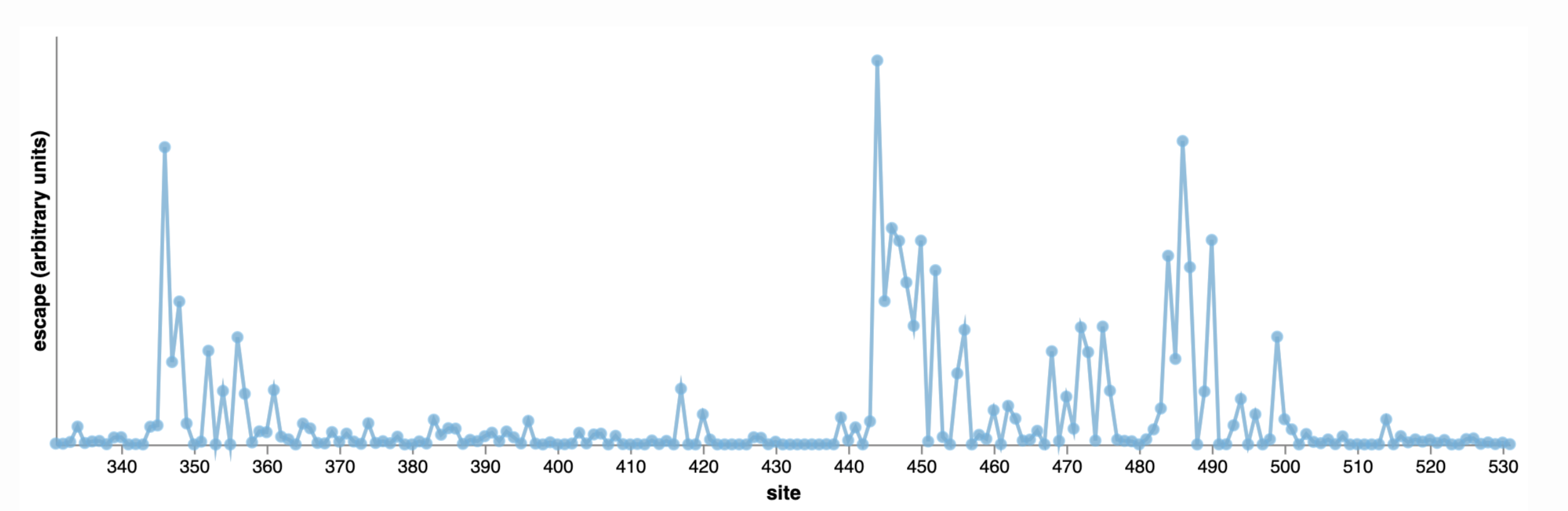
444
346
486
Explore yourself: https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/escape-calc/
Conclusions
- Human coronaviruses evolve to escape immunity
- Any given coronavirus causes a wave of infection some winters
- Current leading variants include BQ.1, BQ.1.1, and XBB
- They escape monoclonal antibody drugs
- They have reduced serum neutralization
- This is because of convergent antibody escape mutations
- Need antibody drugs that target sites different than natural immunity
- Updated boosters should help: BA.5 remains a good choice