Deep mutational scanning to interpret viral evolution
Jesse Bloom
Fred Hutch Cancer Center / HHMI
Disclosures
I am on the scientific advisory boards of Apriori Bio and Oncorus
I have consulted on topics related to viral evolution for Moderna and Merck
I am an inventor on Fred Hutch licensed patents related to deep mutational scanning of viral proteins
We now have lots of viral sequences
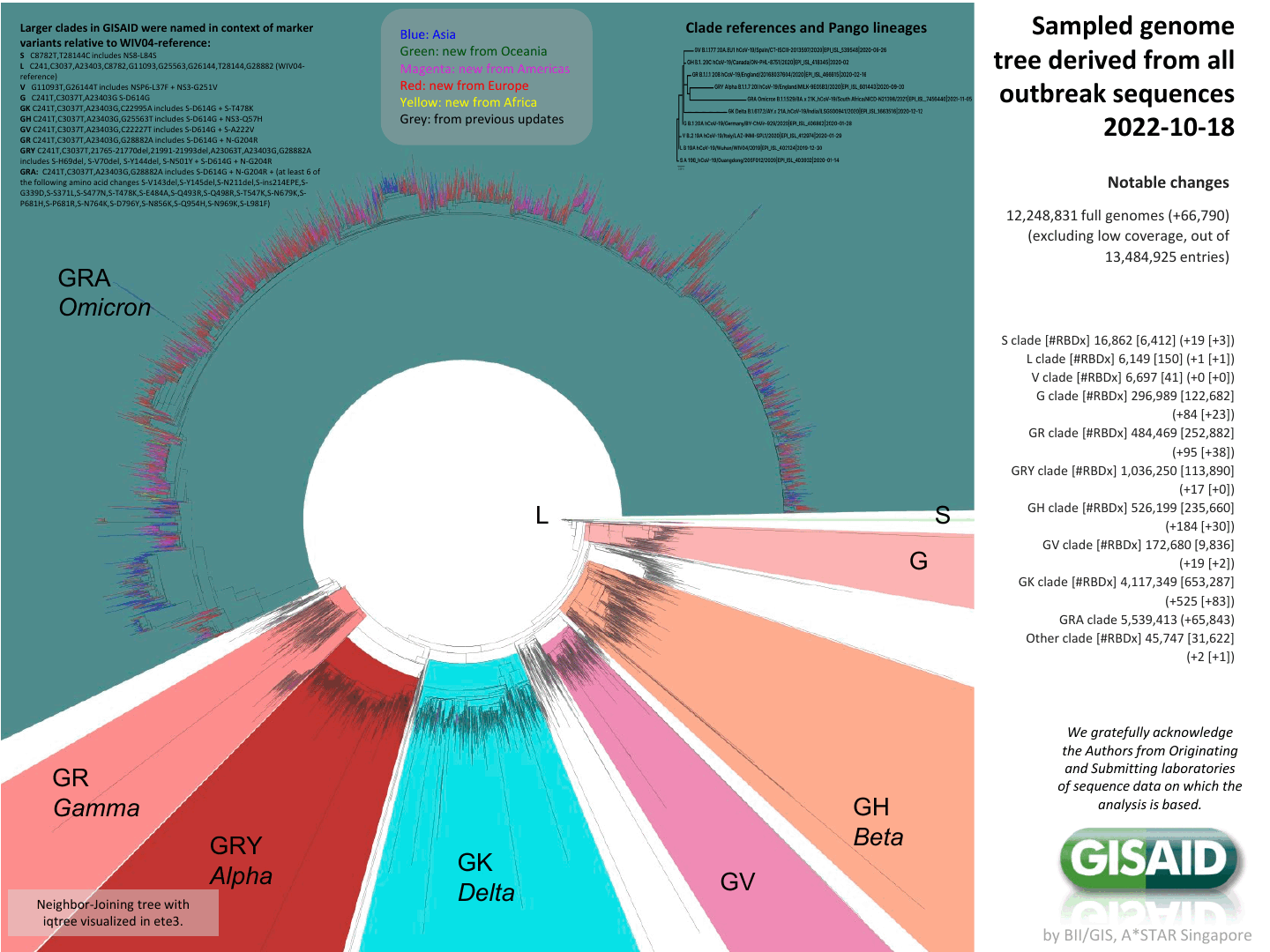
Tree of the ~12-million SARS-CoV-2 sequences in GISAID
What are the implications of the mutations in these sequences?


Deep mutational scanning is method to measure effects of many mutations
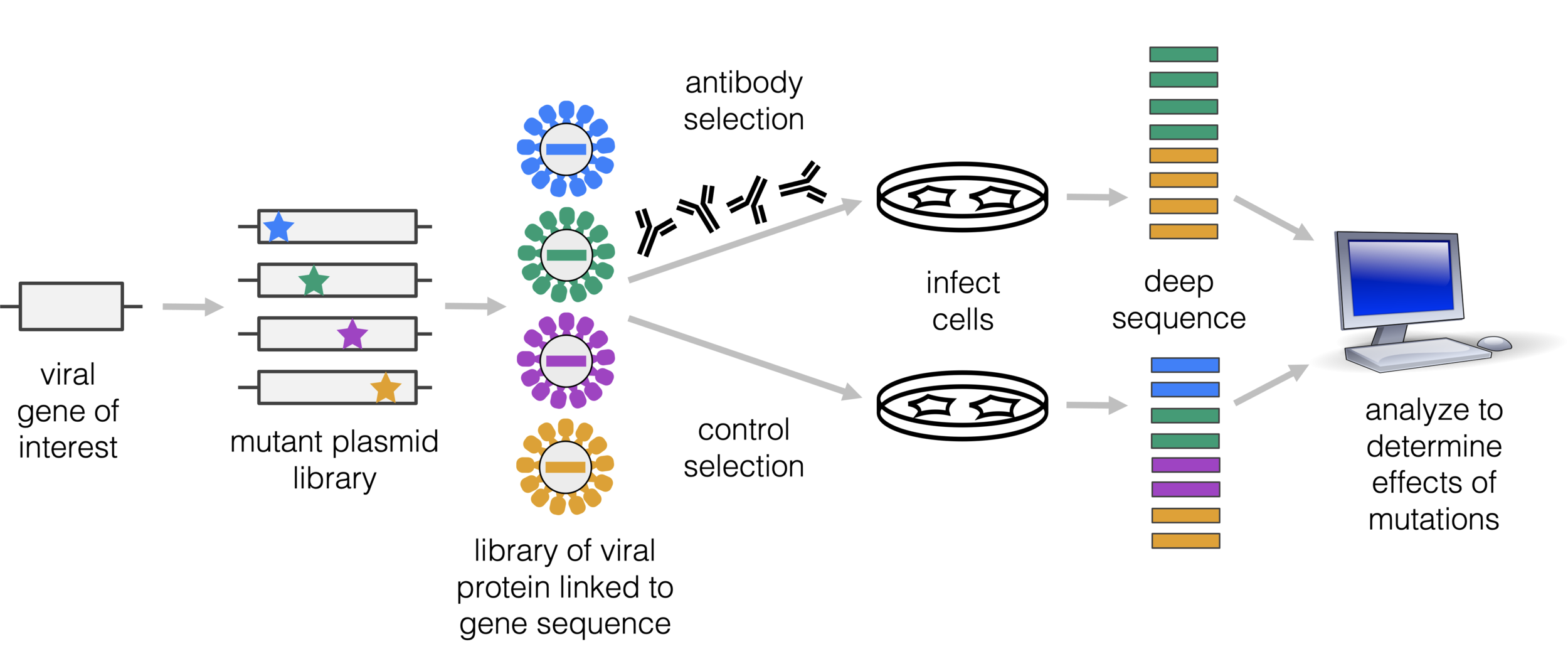
Amenable to multiple ways to link viral protein to gene sequence, and different selections (not just antibodies)
Before COVID-19, we were developing deep mutational scanning to study influenza
When SARS-CoV-2 emerged, we started studying it's receptor binding domain (RBD)
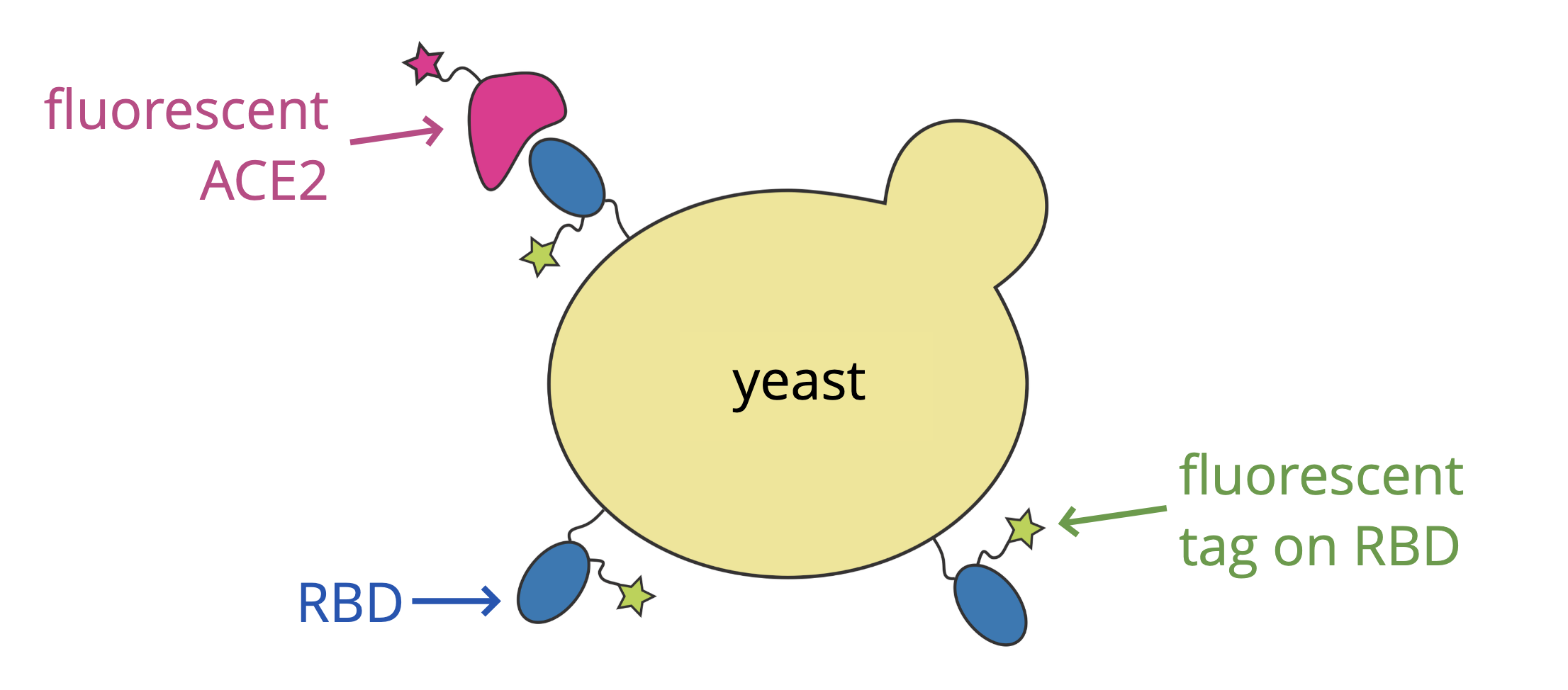
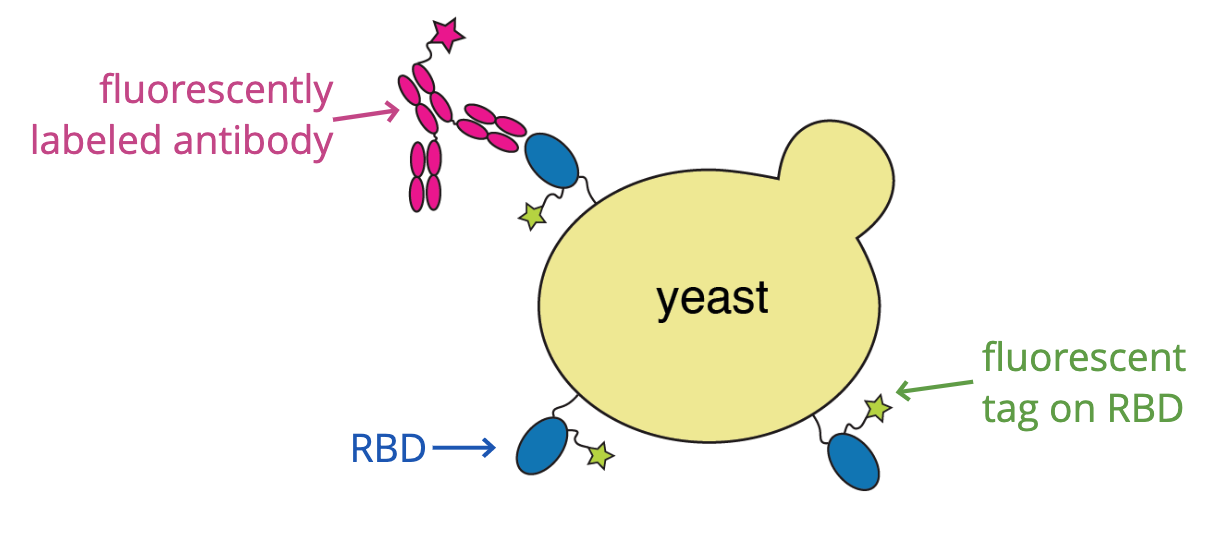

For these studies, we displayed the RBD on the surface of yeast, so we could measure how mutations affects its affinity for ACE2 or ability to escape antibody binding.
Antibody-escape calculator extends this principle to deep mutational scanning data for ~1,500 different human antibodies
Escape calculator is described in Greaney et al (2022), and is available at https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/escape-calc/
36 antibodies mapped by Tyler Starr & Allie Greaney in Bloom lab, from early SARS-CoV-2 strains
1,522 (!) antibodies mapped by Sunney Xie, Richard Cao, Fanchong Jian, et al at Peking University. From early strains, BA.1, & patients with prior SARS-CoV-1 infection. See here.
Limitations of RBD yeast-display deep mutational scanning
- Only examines mutations in RBD, which is just part of spike
- Measures antibody binding, not neutralization. They are unequal in polyclonal serum.
- Only works for viral entry proteins with domains amenable to yeast display.
Alternative: lentiviral pseudotyping
- Many viruses have entry proteins amenable to lentiviral pseudotyping.
- However, traditional pseudotyping does not create genotype-phenotype link.
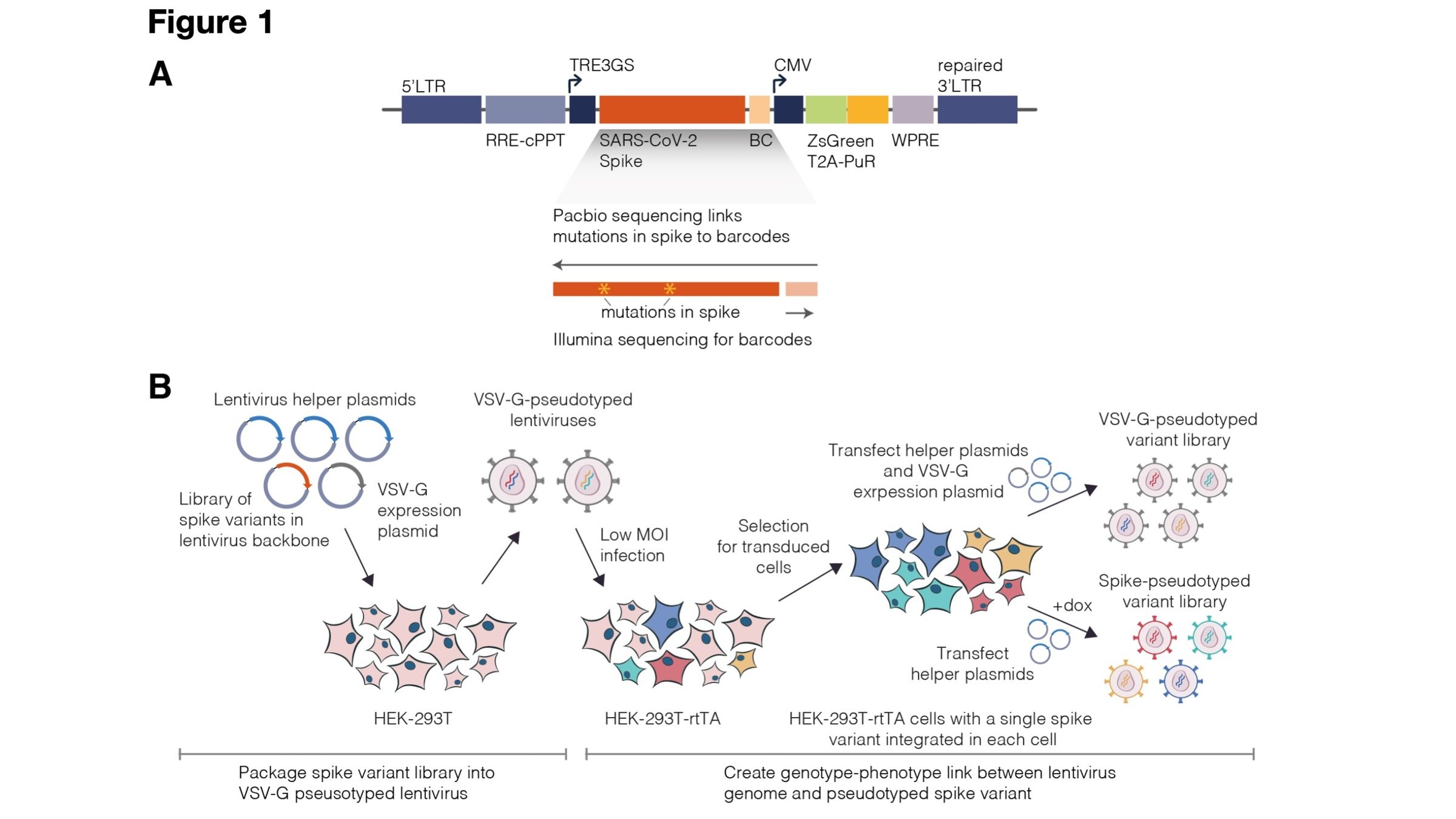
Two-step method to create genotype-phenotype linked spike-pseudotypes
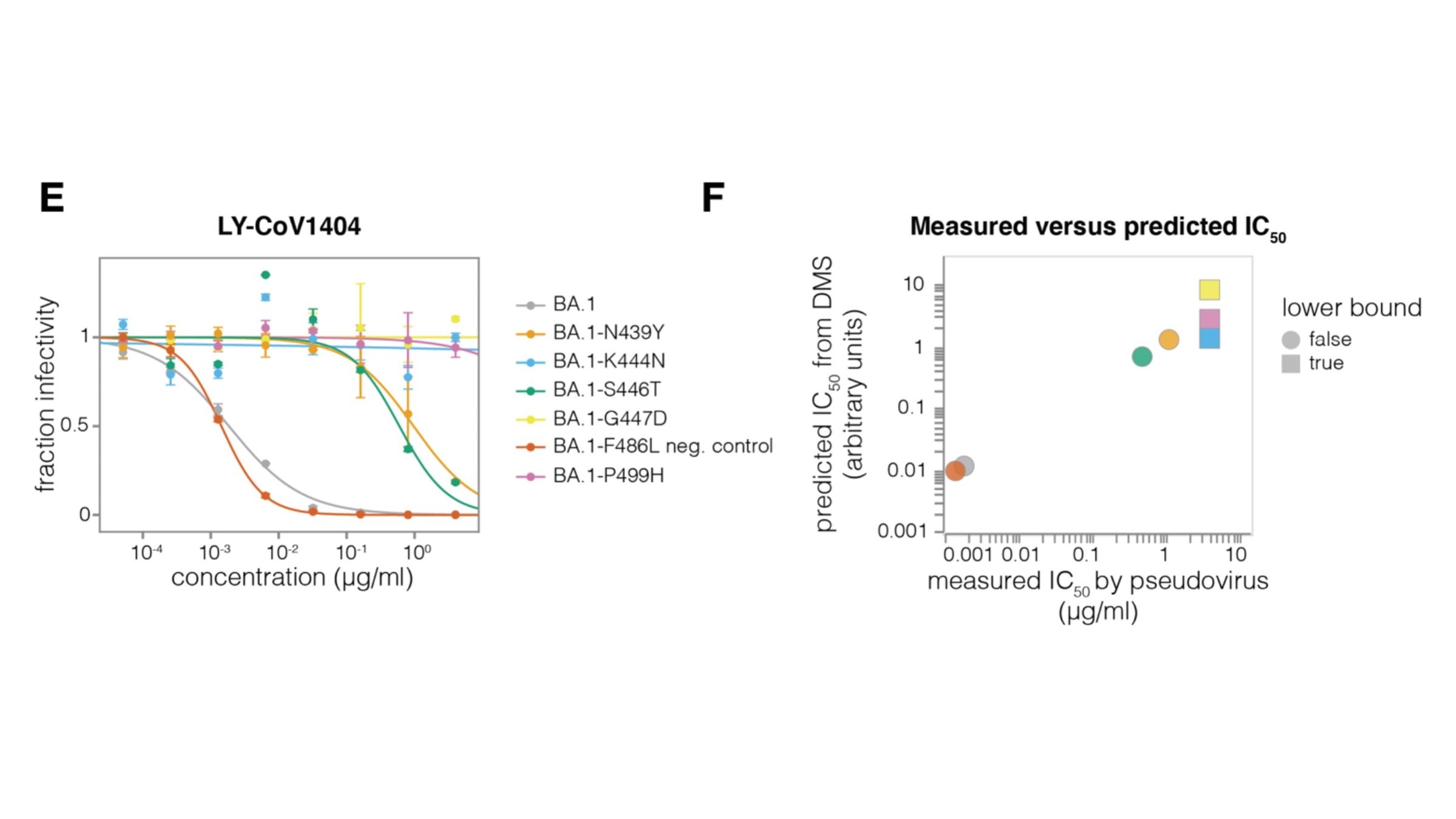
Sequencing only measures relative amounts, so include "absolute standard"
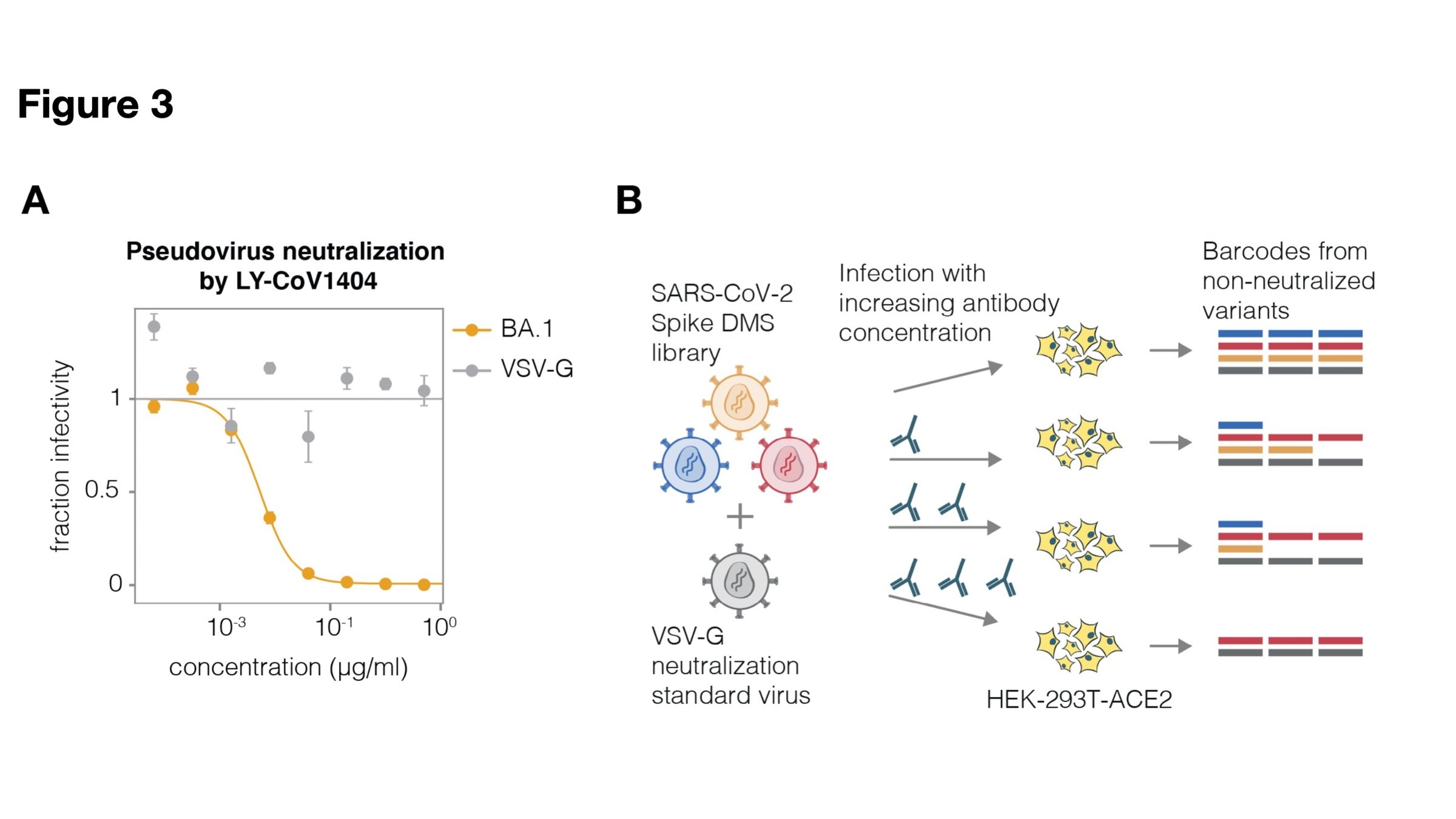
Validates very well
Method should be extensible to many other viral entry proteins
- System can provide safe way to do high-throughput mutational studies
- We need to be aware of information hazard
- We think hazard low for SARS-CoV-2 and seasonal influenza (already human adapted)
- But is potential hazard of human adaptive mutations in potential pandemic viruses
- For such viruses, will use target cells with natural host rather than human receptors