Institute for Comparative Genomics Guest Lecture
Dr. Jesse McNichol (Oct 31st, 2025)
'omics-Driven Marine Microbial Ecology: Tracking Food Web Structure, Climate Change Impacts, and Ecosystem Resilience
- Part I: "Carl Sagan" intro
- Part II: Coastal Complexity
- Part III: Microbiome madness
- Part IV: How not to go mad: CAFCA
- Contextualized
- Affordable / Accessible
- FAIR
- Comprehensive
- Archived
- My thoughts on the future of coastal microbial ecology
- Questions / discussion
Talk outline
Part I: "Carl Sagan" intro

If you could step back to the edge of the solar system...

...and look at the light passing through our atmosphere...

...what would you see?
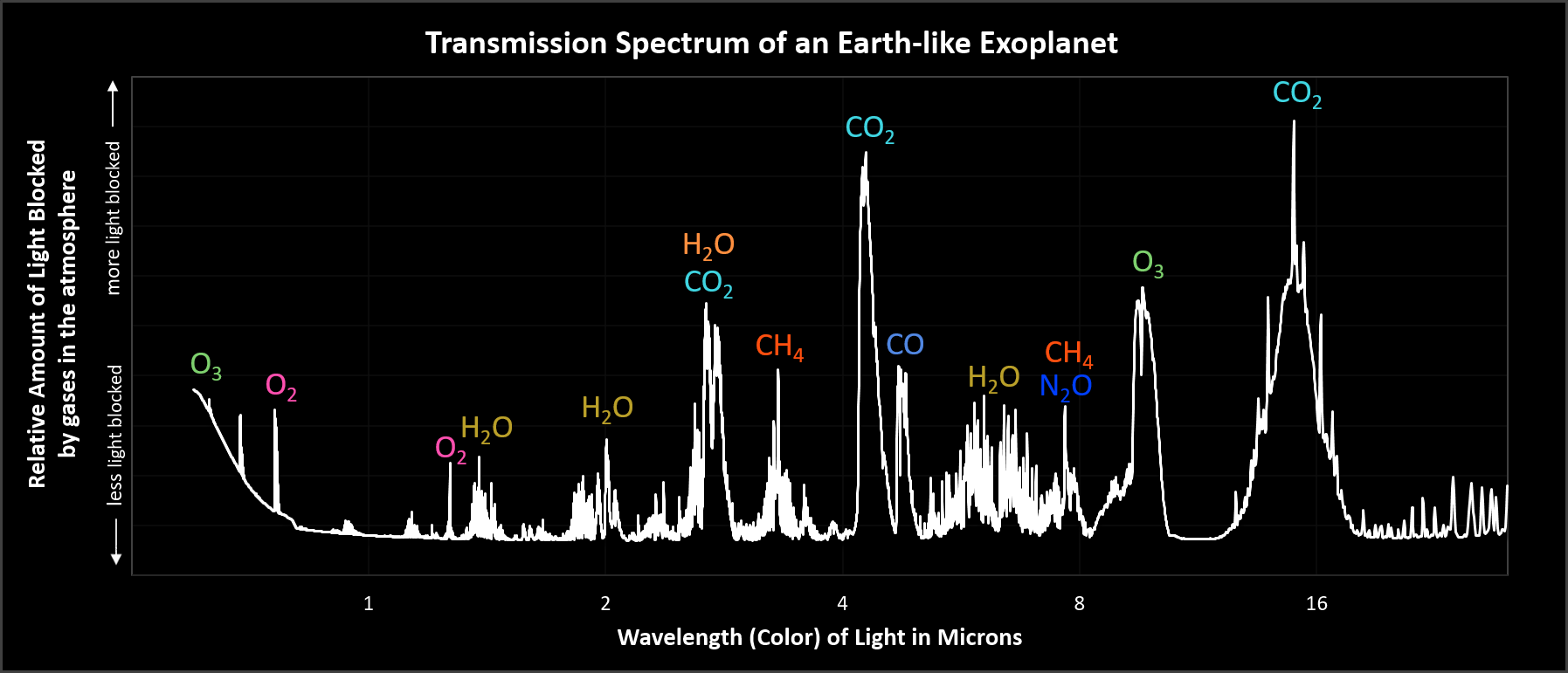
Chemical signatures of a living planet!
- Primary production
- Nutrient cycling
- GHG production / consumption
- Influence geology, climate, chemistry, and biology of Earth
Microbial life

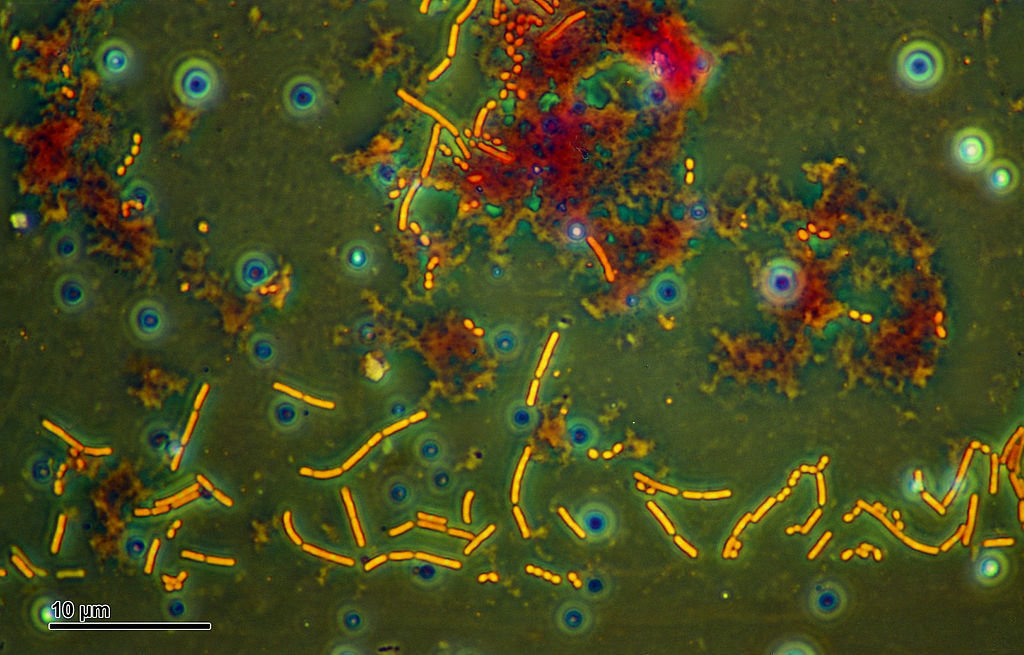
(Micro)
Kump, Kasting, and Crane, The Earth System
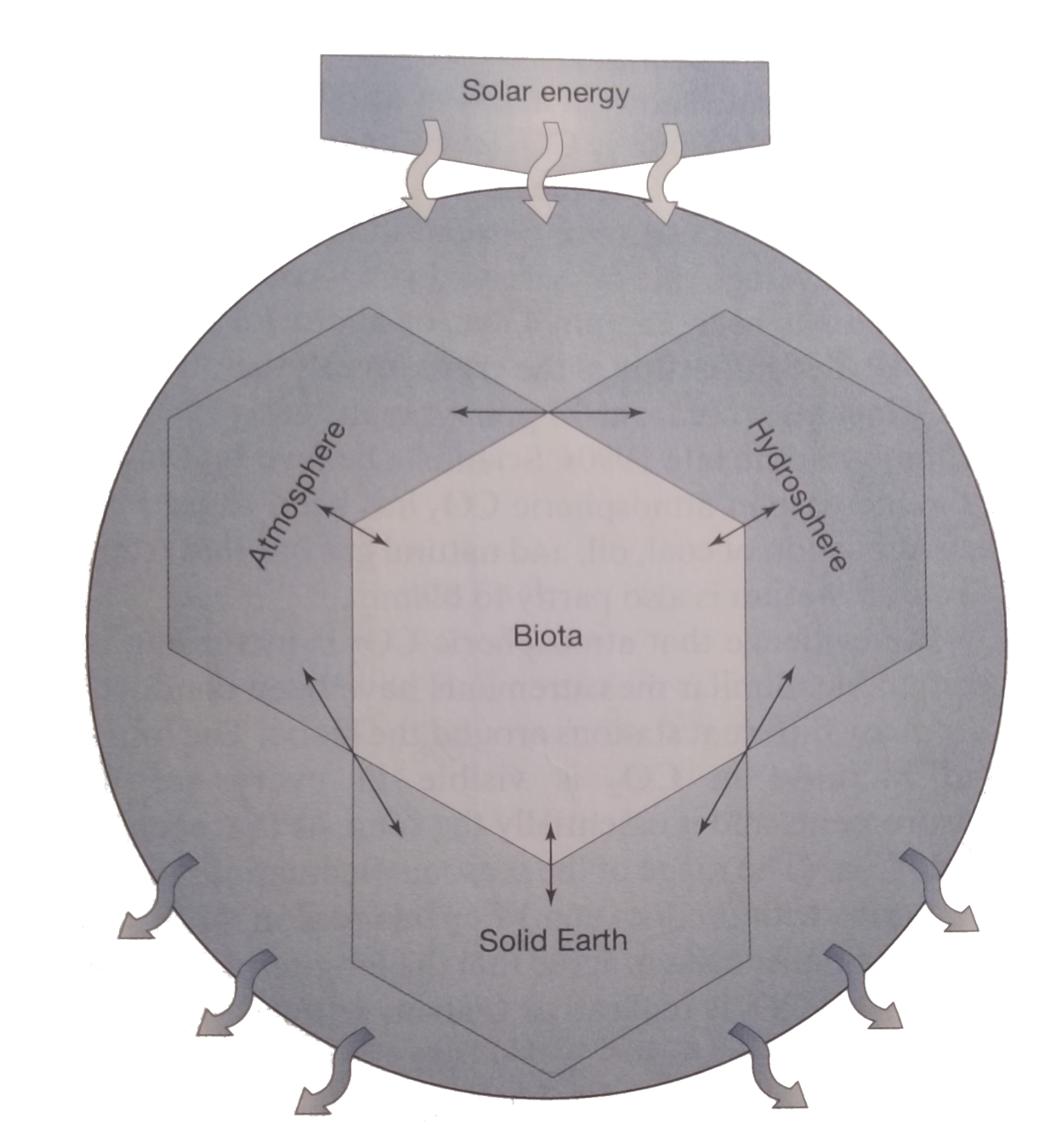
Microbial life keeps the big wheel turning...
Looking back:
Where did we come from?

It is a wholesome and necessary thing for us to turn again to the earth and in the contemplation of her beauties to know the sense of wonder and humility. ”
― Rachel Carson
Why study microbiology?
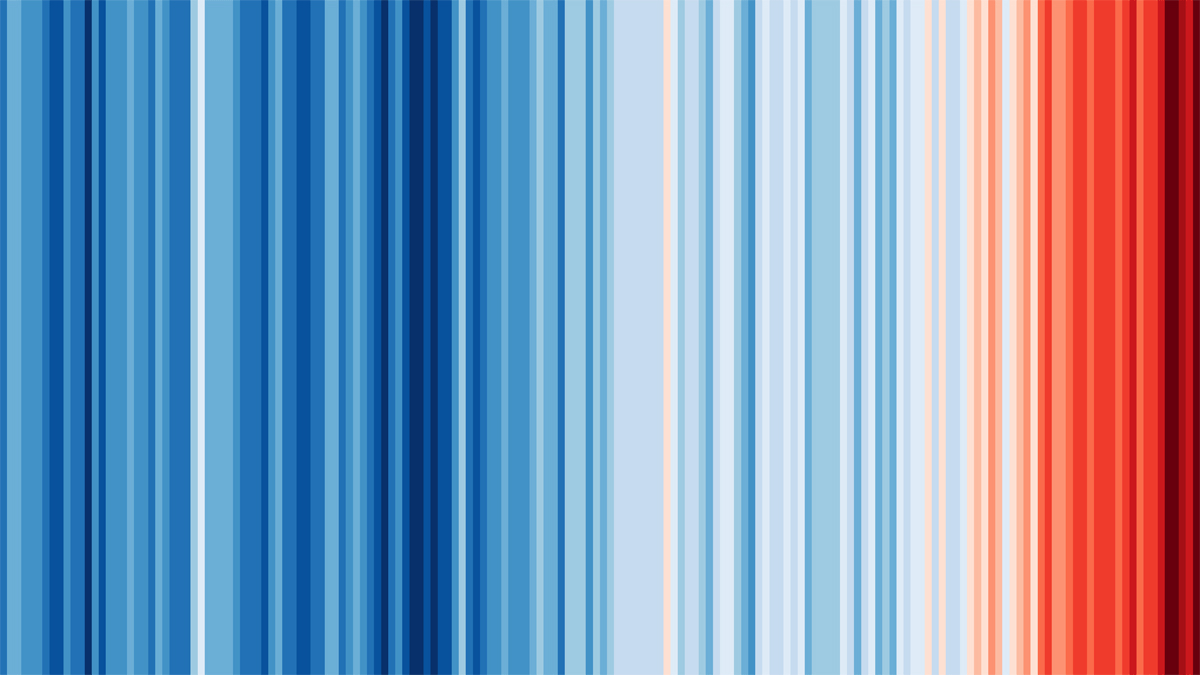
But what about the future?

Microbial life


But what about the future?

(Micro)
Kump, Kasting, and Crane, The Earth System
What effect will anthropogenic pressures such as climate change and coastal eutrophication have on microbial "ecosystem services"?

Microbial life


But what about the future?

(Micro)
Kump, Kasting, and Crane, The Earth System
What is the role of microbial ecology / microbiome science in an age of crisis?
As we witness our planet transforming around us we watch, listen, measure … respond."
― Alise Singer
Looking ahead:
Where are we going?


Being an ecologist in the Anthropocene
Part II: Coastal Complexity

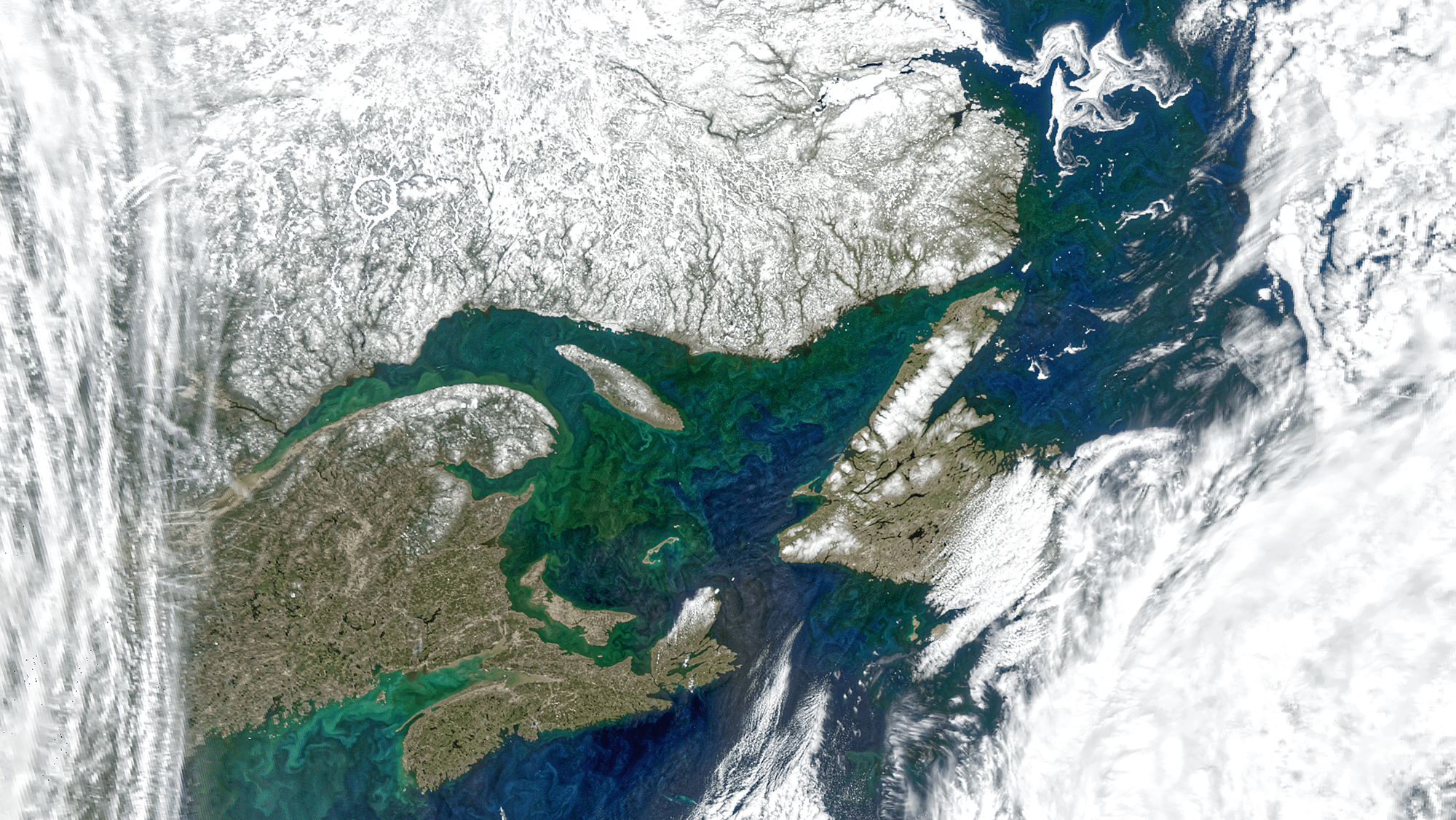
NASA PACE
Coastal ecosystems cover huge spatial scales
Coastal ecosystems are incredibly dynamic
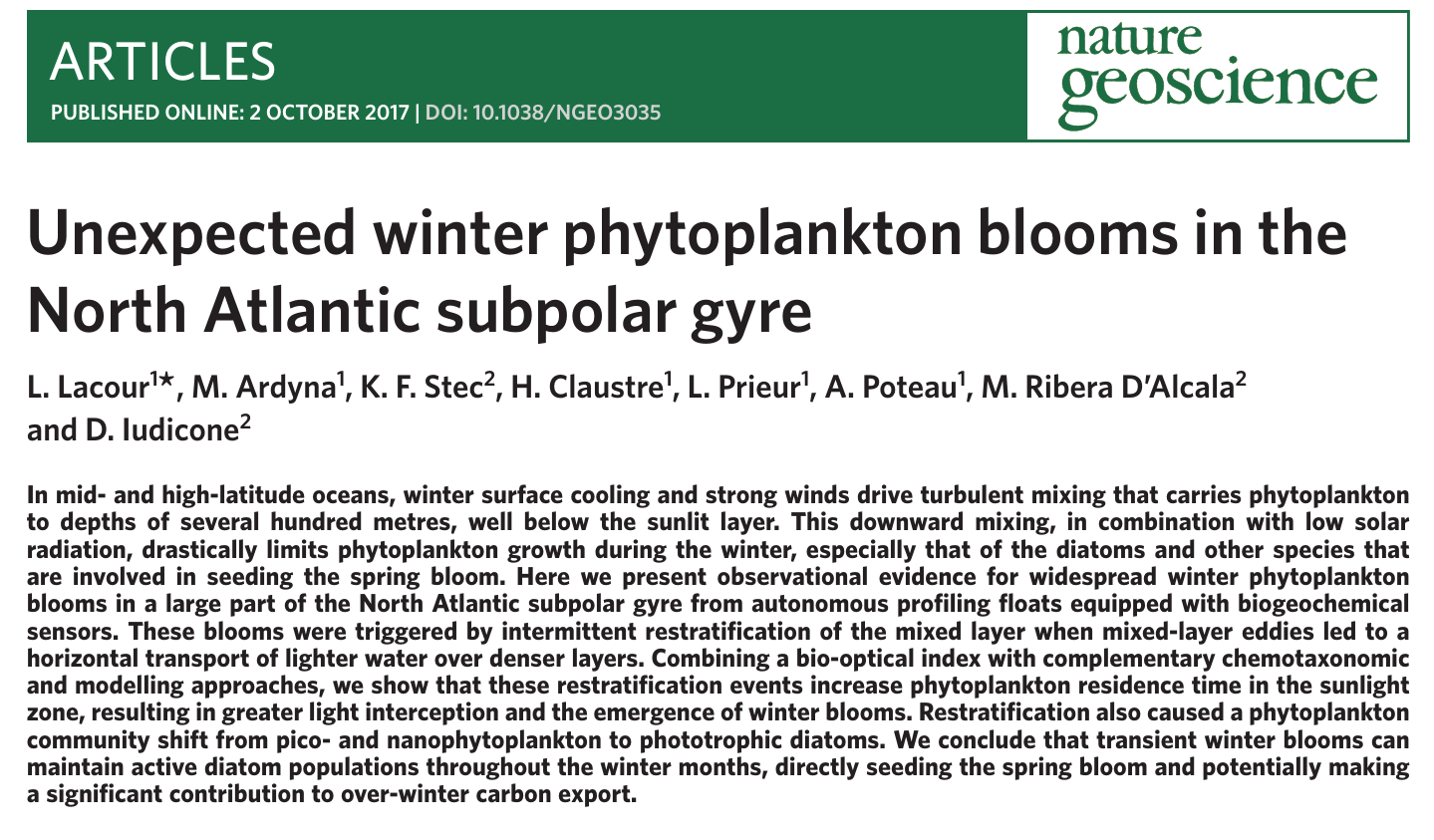
Coastal ecosystems are incredibly dynamic
Coastal ecosystems are incredibly dynamic
Land-sea interactions and temporal variability
Data: Environment Canada
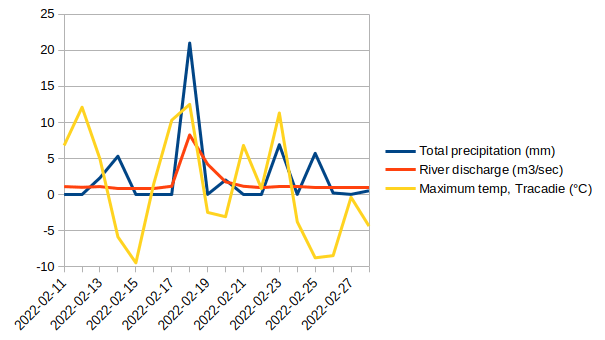
Land-sea interactions and temporal variability
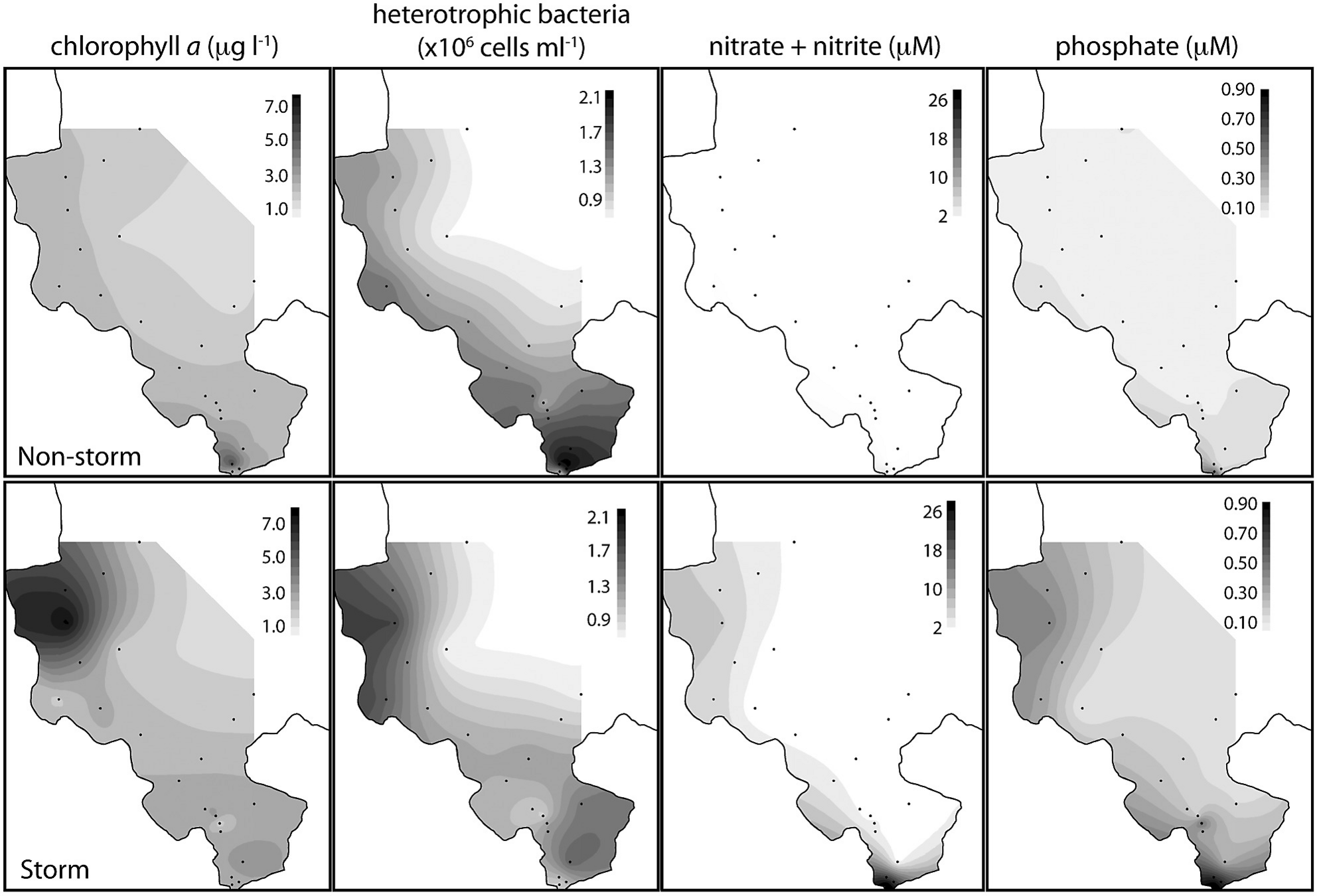
Yeo et al., 2013
Normal
Land-sea interactions and temporal variability
Post-storm disturbance

Tveness, WMC
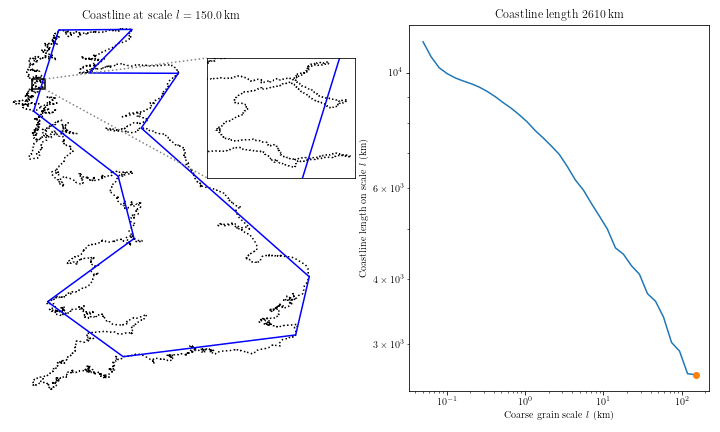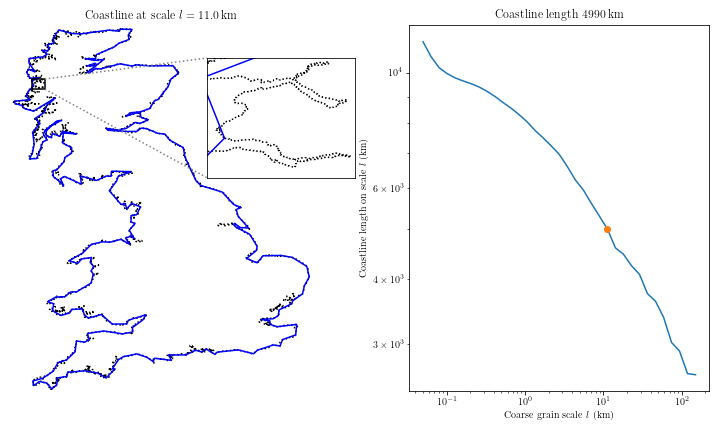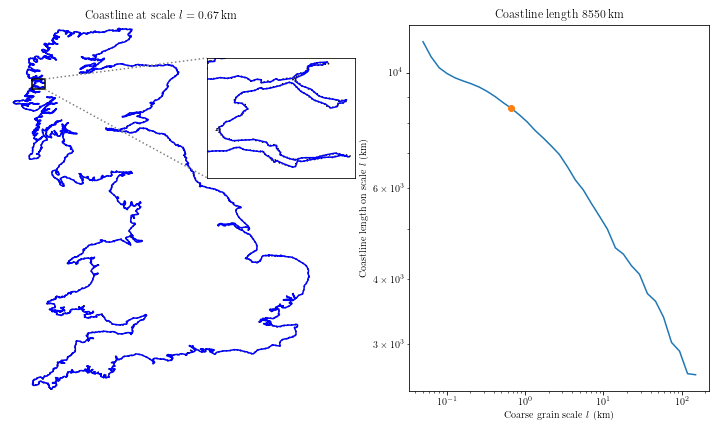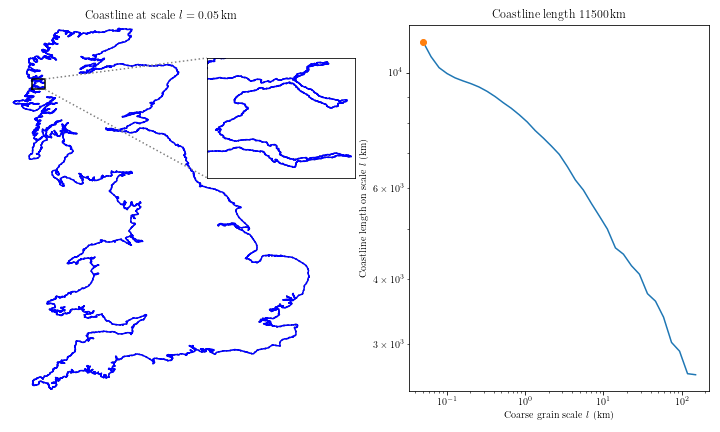
Complex coastal geometry → spatial heterogeneity
Complex coastal geometry → spatial heterogeneity

Image: Google Maps
Complex coastal geometry → spatial heterogeneity
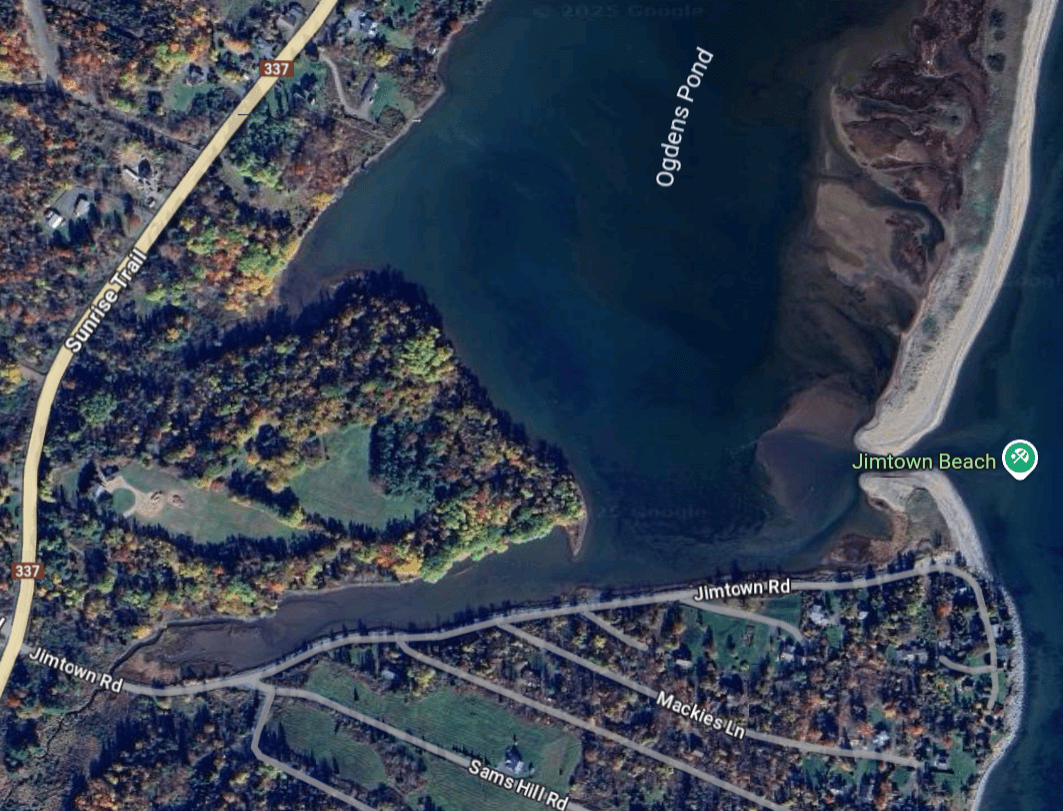
Image: J. McNichol
Complex coastal geometry → spatial heterogeneity

Image: J. McNichol
Complex coastal geometry → spatial heterogeneity

Image: J. McNichol
Complex coastal geometry → spatial heterogeneity

Image: J. McNichol
Complex coastal geometry → spatial heterogeneity

Image: J. McNichol
Complex coastal geometry → spatial heterogeneity

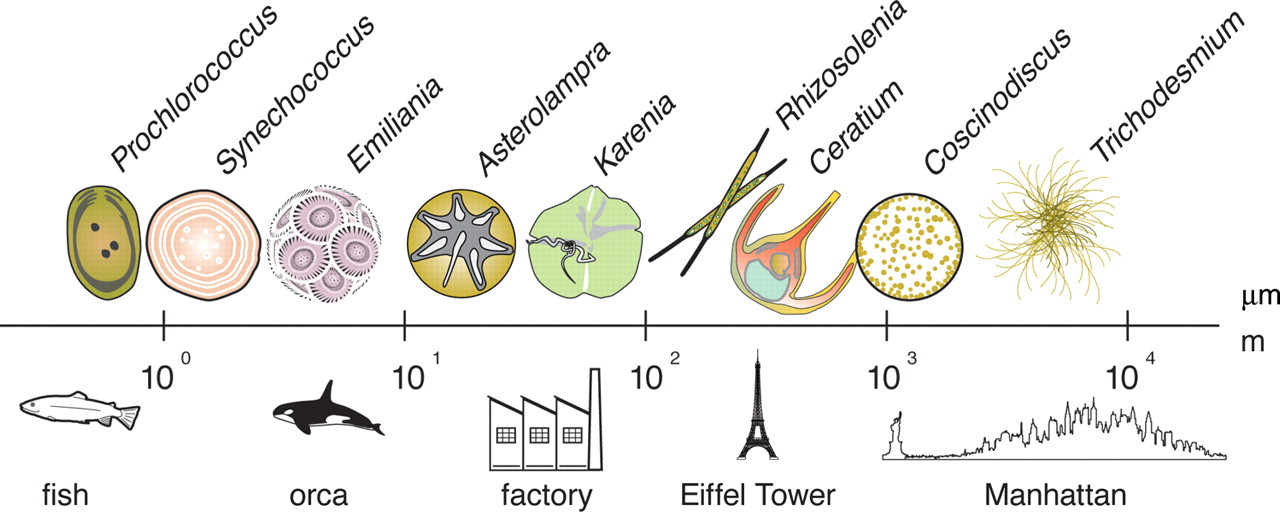
For microbes, spatial heterogeneity extends to the microscale
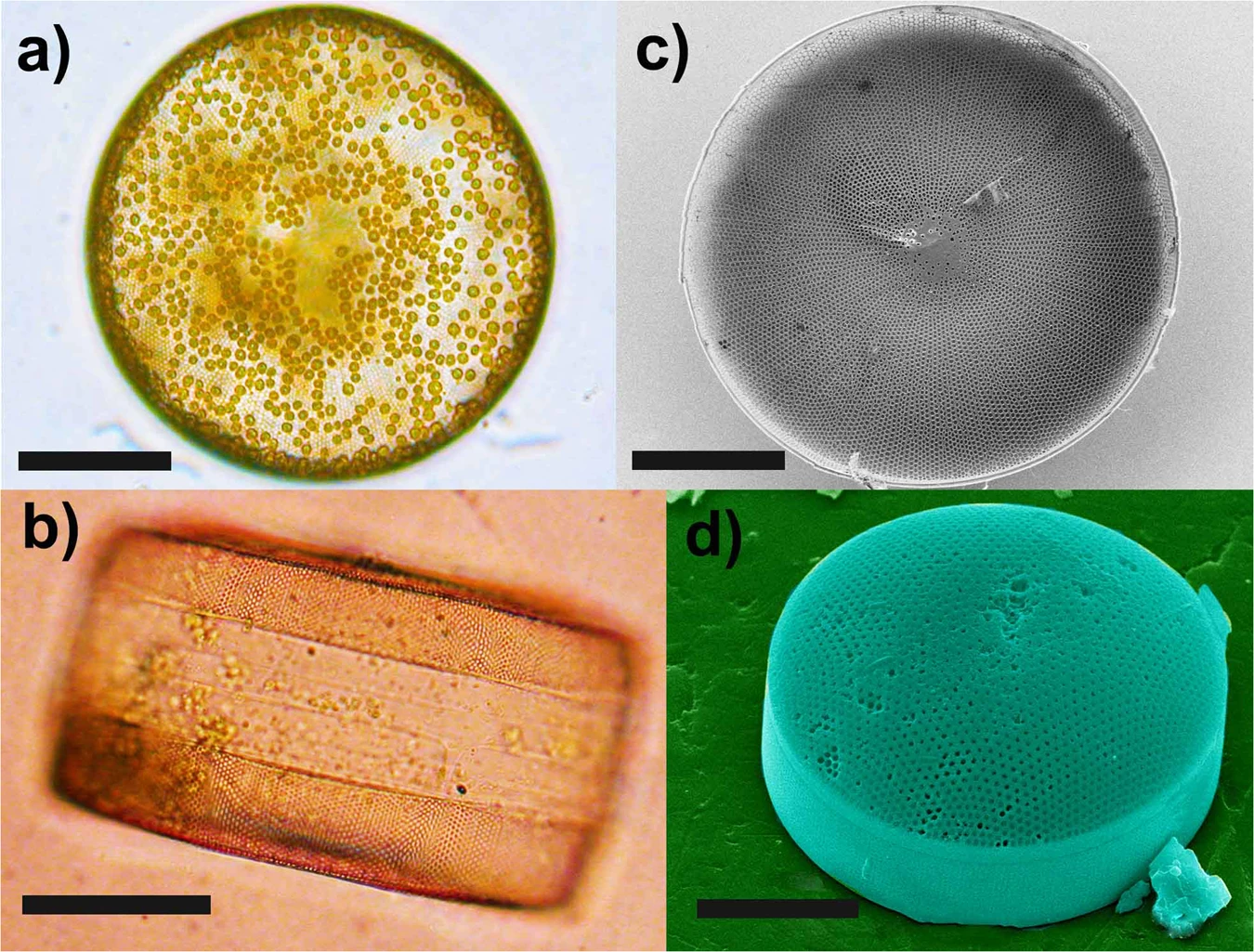
Coscinodiscus
Davis Laundon / De Tommasi et al
Living at the microscale - plankton


Stocker, 2012: "Marine Microbes See a Sea of Gradients"
Living at the microscale - plankton
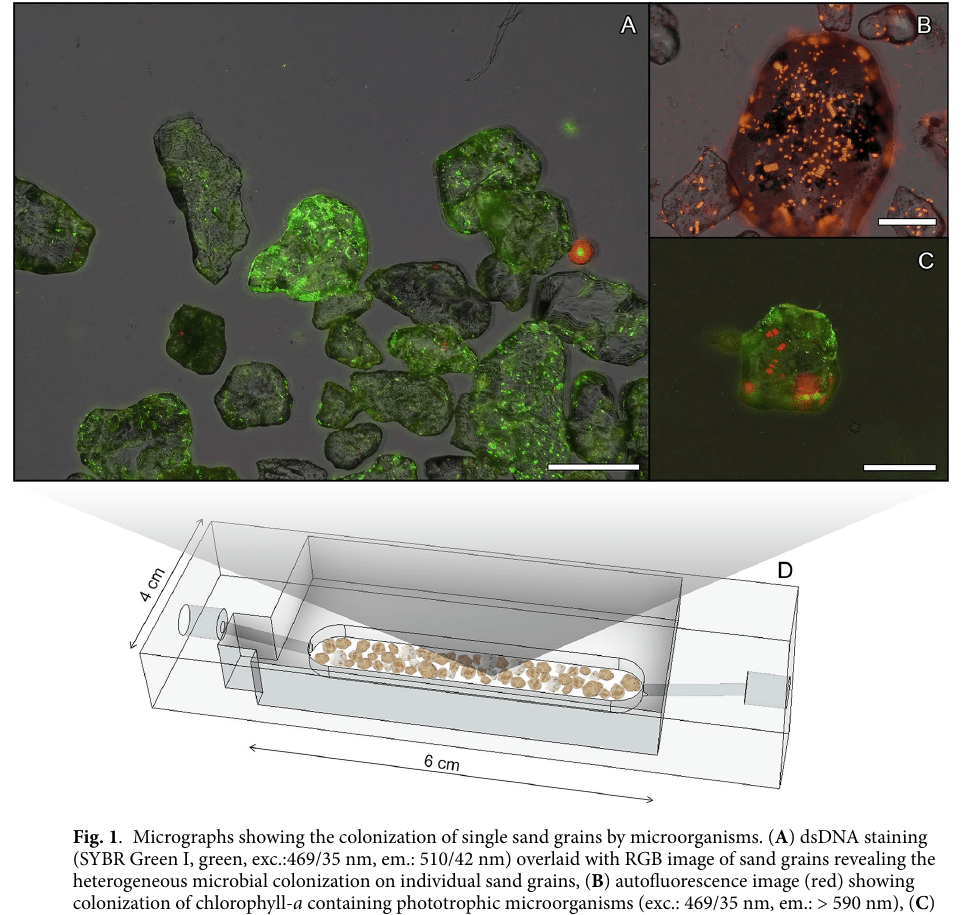
Living at the microscale - sand grains
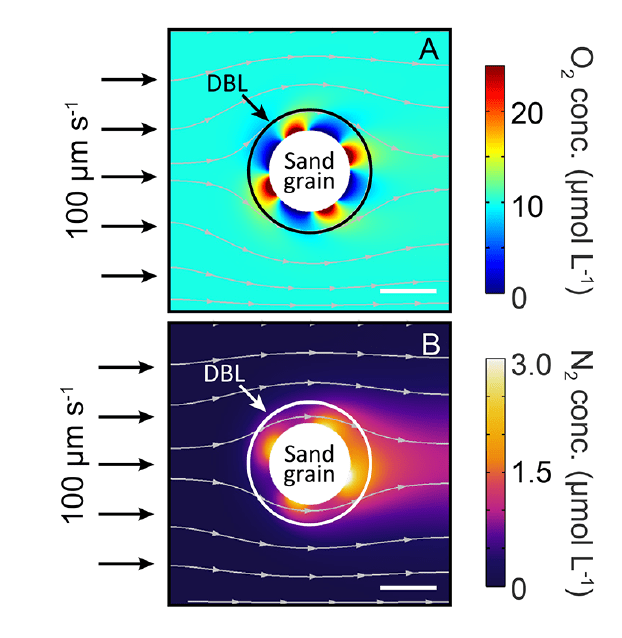

Microscale biogeochemical partitioning (sand grains)
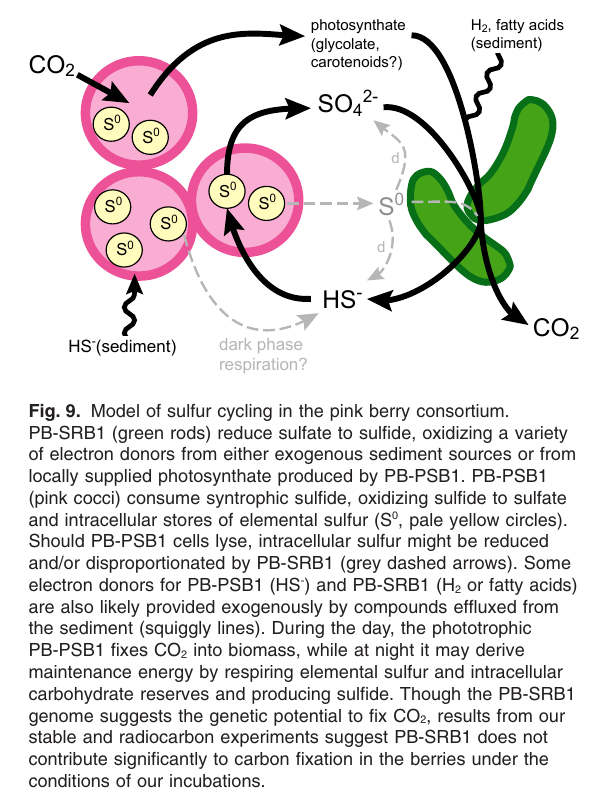

found in salt marshes near Woods Hole, but I've yet to see them here!
Microscale biogeochemical partitioning ("pink berries")
Why do I like studying coastal systems?

NASA PACE
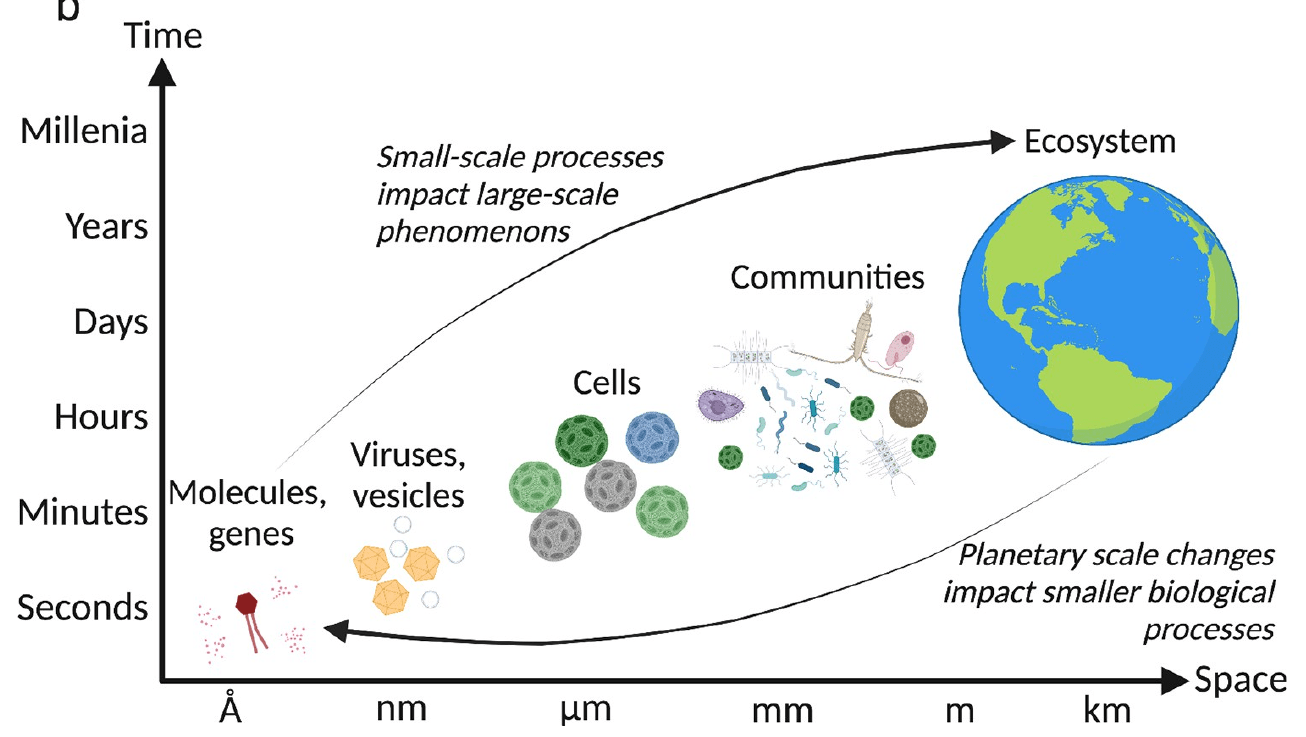
Vincent and Vardi, 2023
Can experiment across scales, depending on question
Vincent and Vardi, 2023

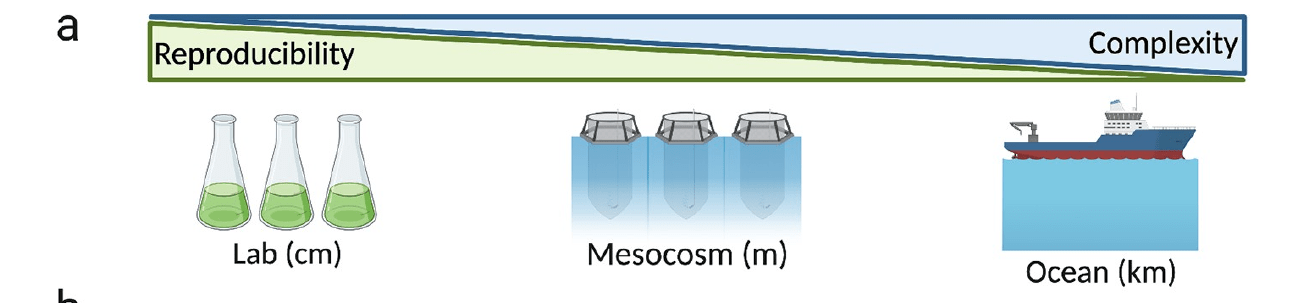
Can experiment across scales, depending on question
"Model ecosystems"
Bedford basin


Saanich inlet

SPOT (Los Angeles)


Minas basin
A "model ecosystem" in Cape Breton
- Stratified basin with anoxic, sulfidic waters ~ 17 m
- Using metabarcoding, metagenomics to understand taxa present, their dynamics, and functional potential
- Ultimate goal: use experiments to understand how resilient microbial "ecosystem services" are to disturbance (i.e. climate change, eutrophication)

Whycocomagh Bay (WB)
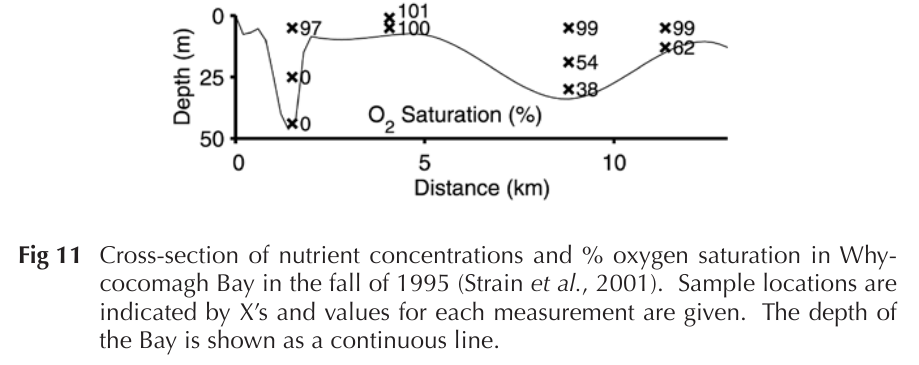
A "model ecosystem" in Cape Breton
Part III: Microbiome Madness
Early sequencing
"Next Generation Sequencing" (NGS)
"3rd generation" sequencing
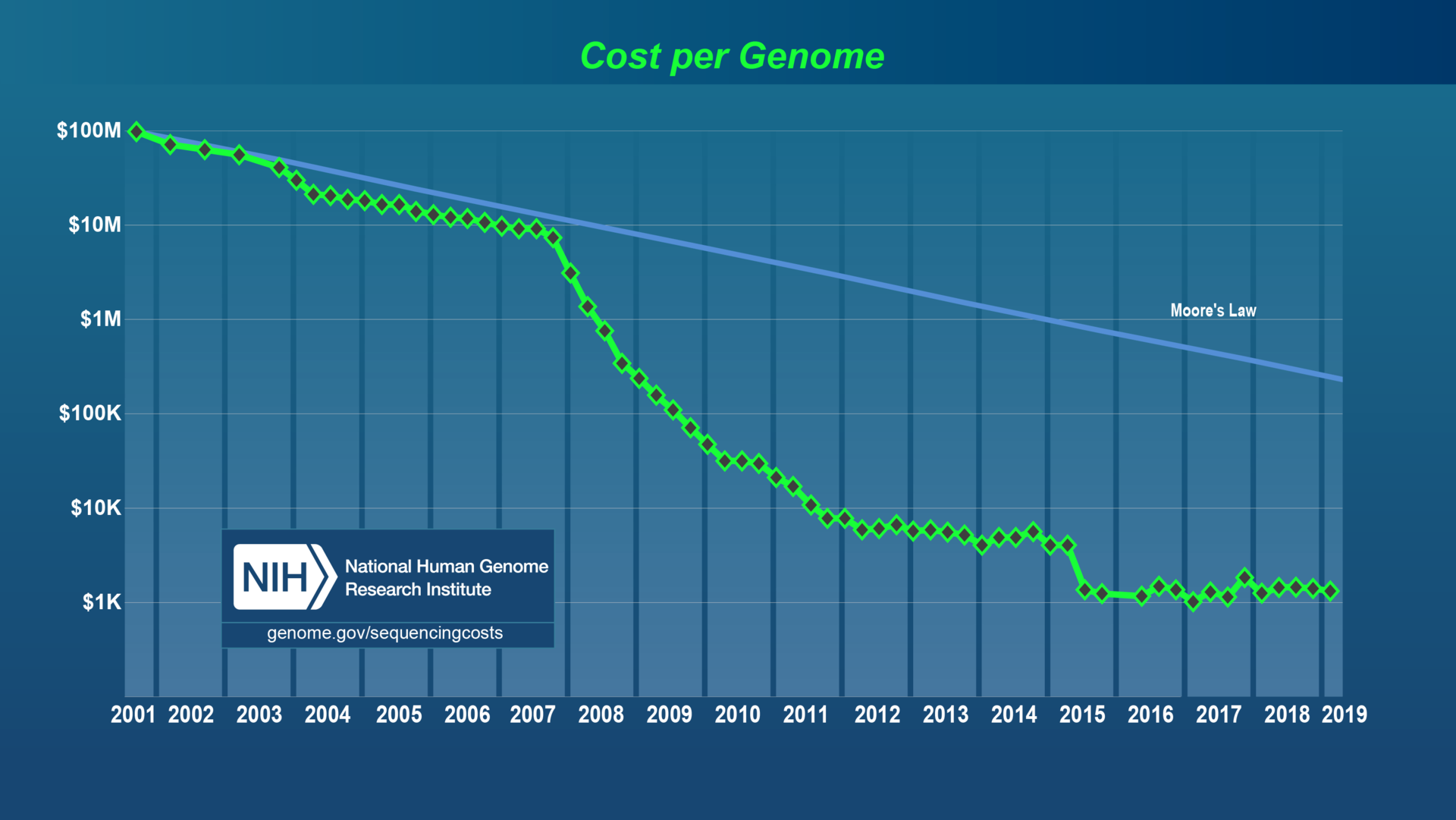
The era of cheap DNA sequencing
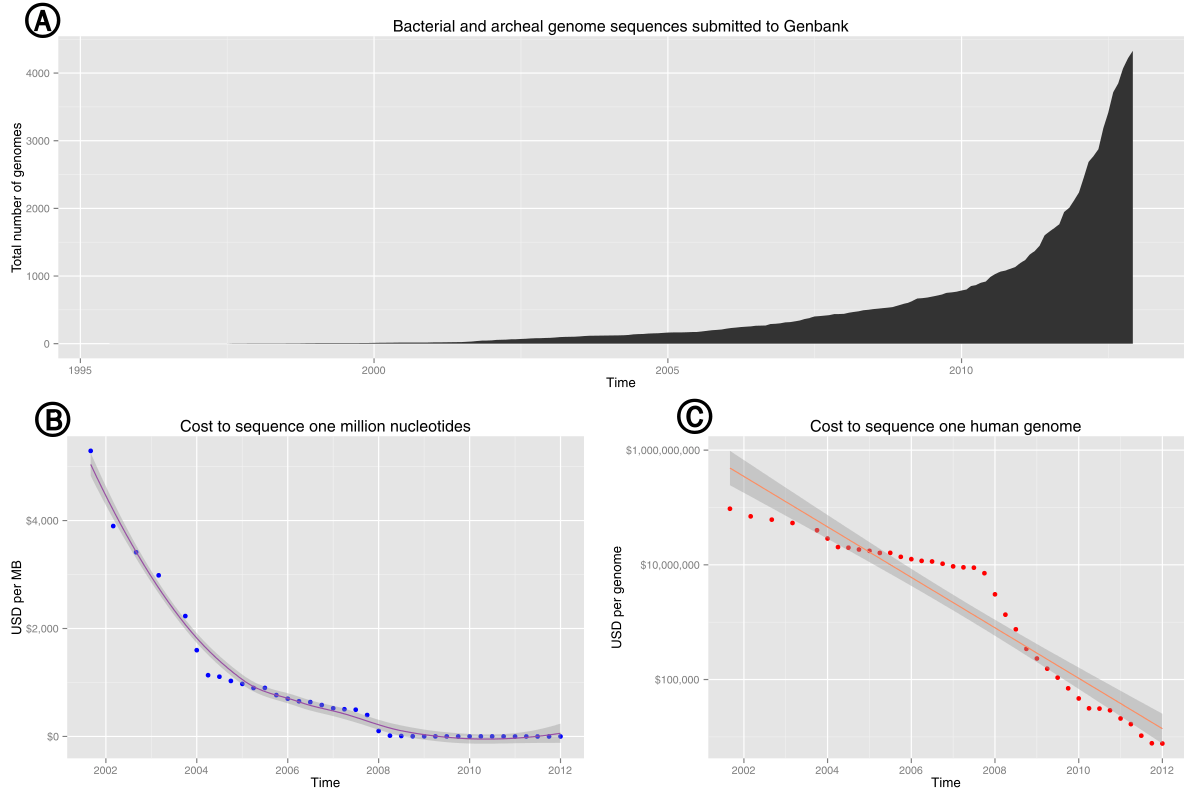
For a few thousand dollars, can get billions of "reads"

The era of cheap DNA sequencing
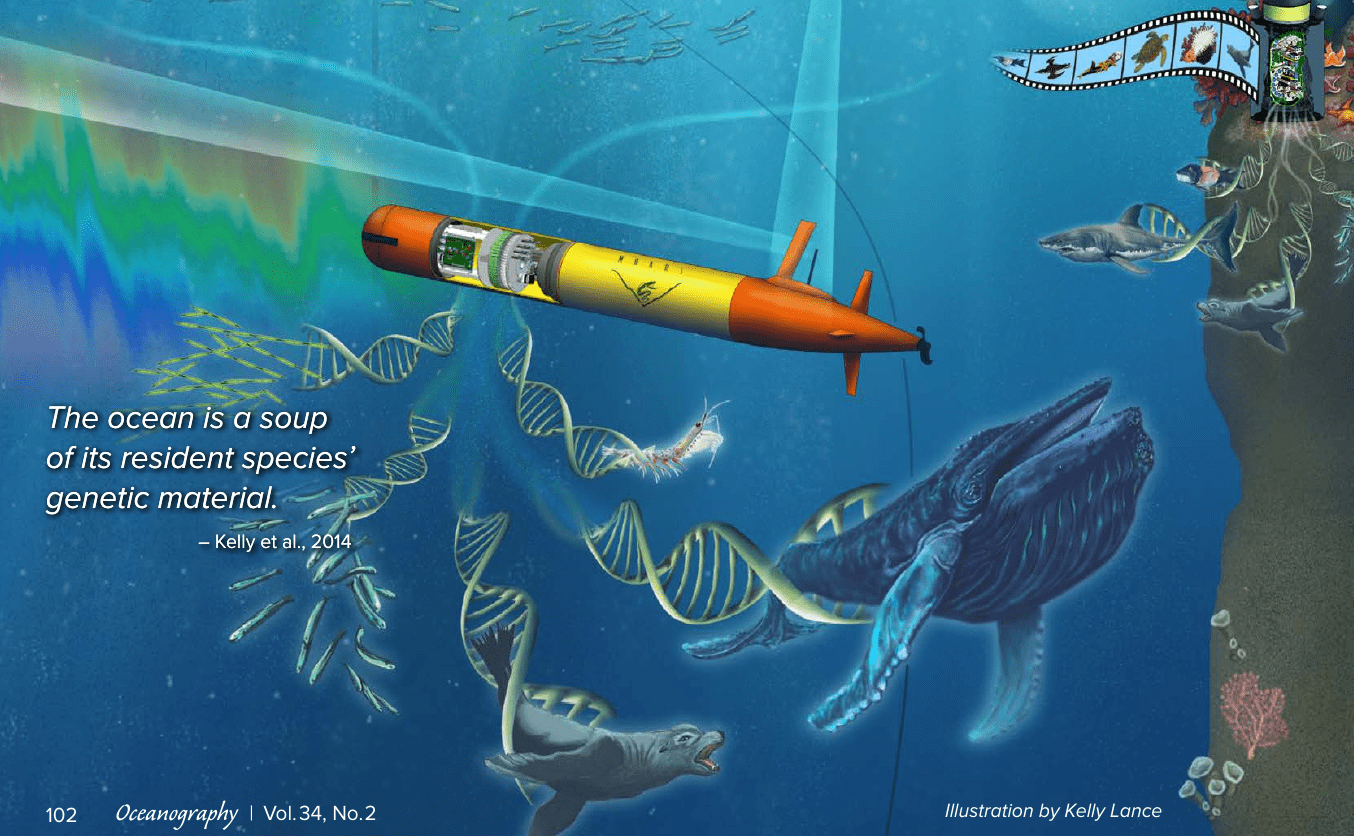
Image credit: Chavez et al., 2021
Cheap DNA sequencing → "eDNA assays" ( = amplicon sequencing = metabarcoding)
Image credit: Chavez et al., 2021
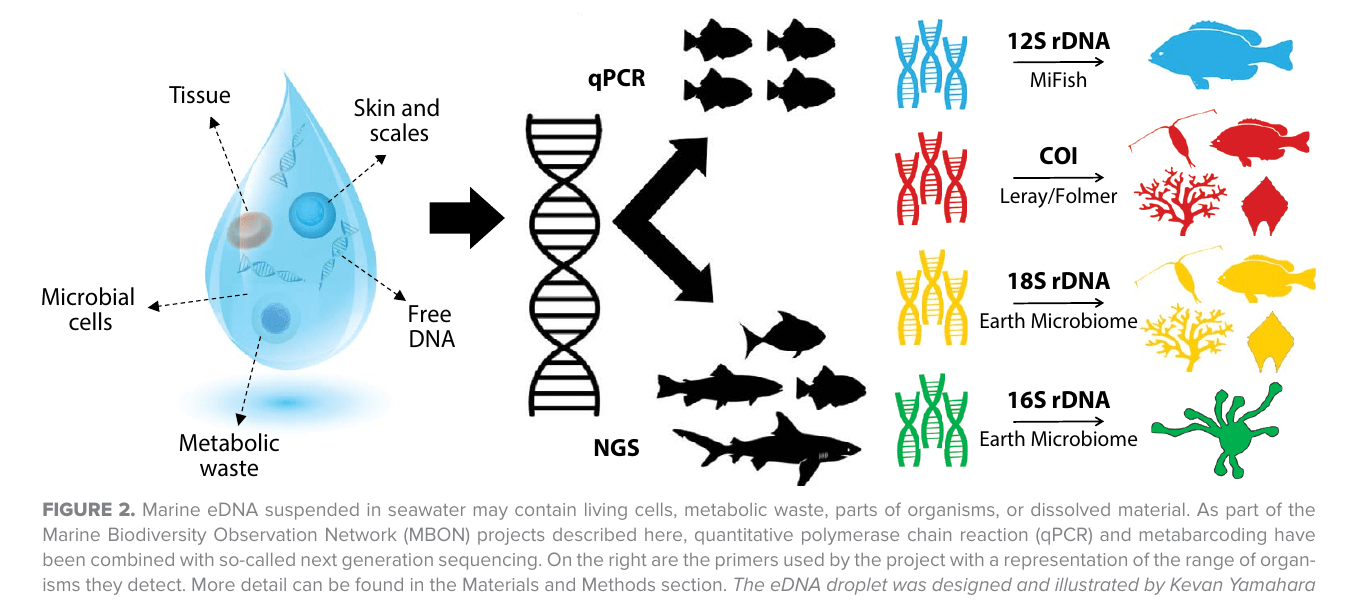
Cheap DNA sequencing → "eDNA assays" ( = amplicon sequencing = metabarcoding)

"Gee whiz!" era
"Arguing about methods" era
The era of arguing about methods
The era of arguing about methods

This gets tiring after a while (endless debates...)
- Intercalibration using standard reference material / mock communities so we know which methods really are best


My opinion: Arguing about methods will only end with community intercalibration efforts
But until this gets figured out, we should "move on with life"

"Gee whiz!" era
"Arguing about methods" era
"What does it all mean?" era
Let's enter the era of "what does it all mean?"
Let's move from complex descriptive data to ecological understanding
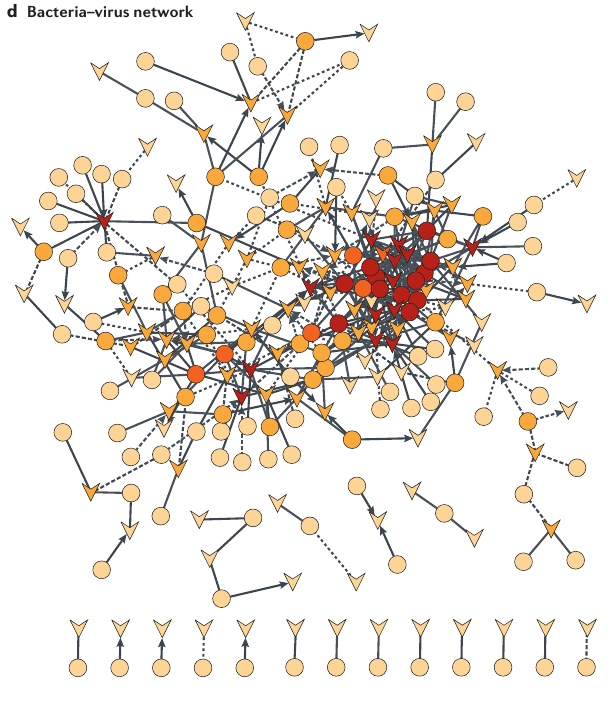
How do we go from complex data to ecological understanding?
Vincent and Vardi, 2023

Part IV: Thoughts on doing good coastal microbiome work
Easy to get overwhelmed! Remainder of presentation = my thoughts on how to navigate this complexity

Complex ecosystems + complex methods / data

- Part I: "Carl Sagan" intro
- Part II: Coastal Complexity
- Part III: Microbiome madness
- Part IV: How not to go mad: CAFCA
- Contextualized
- Affordable / Accessible
- FAIR
- Comprehensive
- Archived
- My thoughts on the future of coastal microbial ecology
- Questions / discussion
CAFCA
- Part IV: How not to go mad: CAFCA
- Contextualized
- Affordable / Accessible
- FAIR
- Comprehensive
- Archived
Bedford basin


Saanich inlet

SPOT (Los Angeles)


Minas basin
Contextualized: metadata-rich time-series

Long-term increase in Primary Productivity


Contextualized: metadata-rich time-series
Contextualized: collaborate!
Microbial ecology
Marine chemists
Physical oceanographers
Ecosystem modellers
Organismal biologists
Contextualized: focus on "bold hypotheses" that build knowledge
- Generating sequencing data: easy
- Developing a good question: hard

A ‘good’ hypothesis has a number of desirable properties. It should be bold, risky and meaningful, addressing an important issue and not stating the obvious."

Accessible: Use open-source workflows
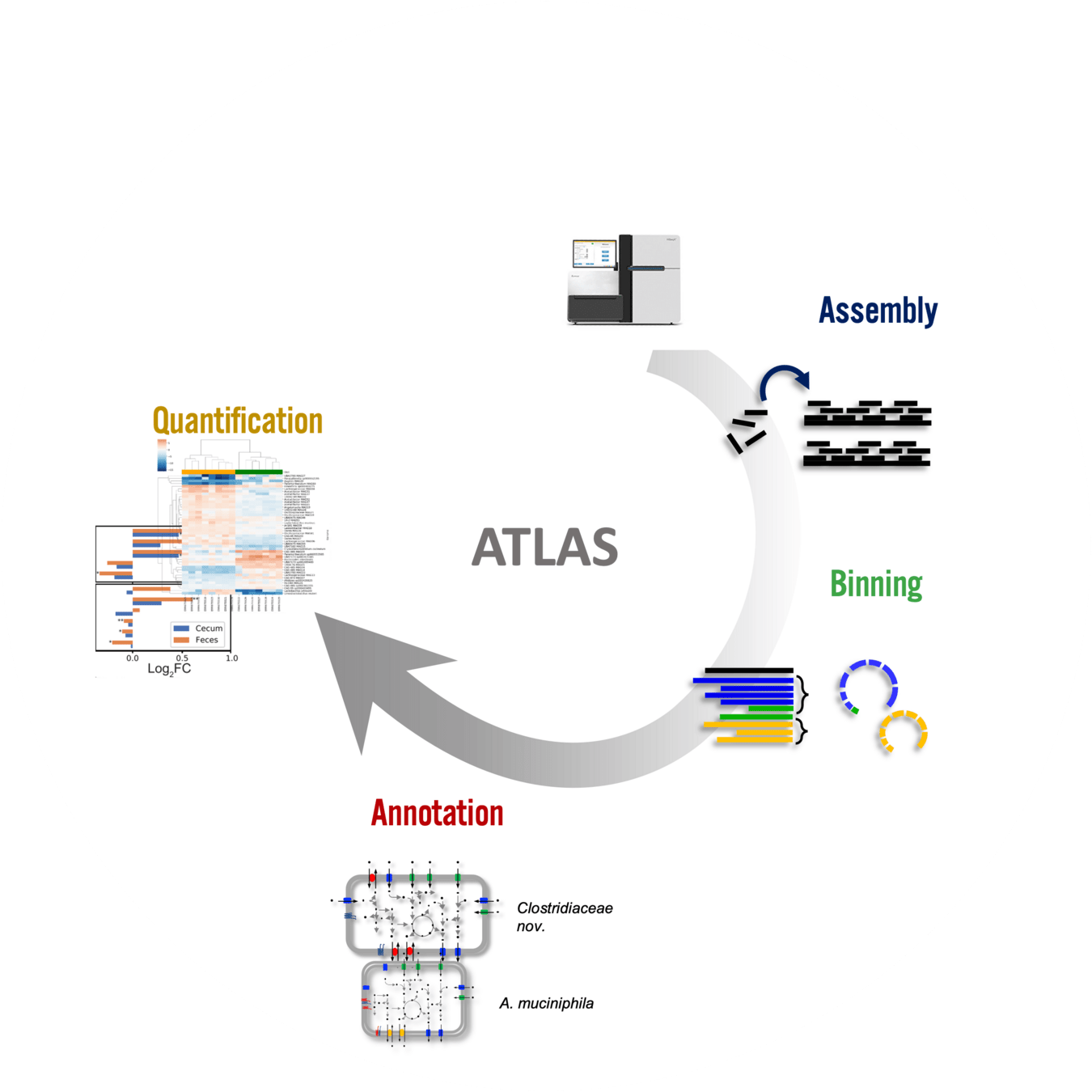


Accessible: Use open-source workflows
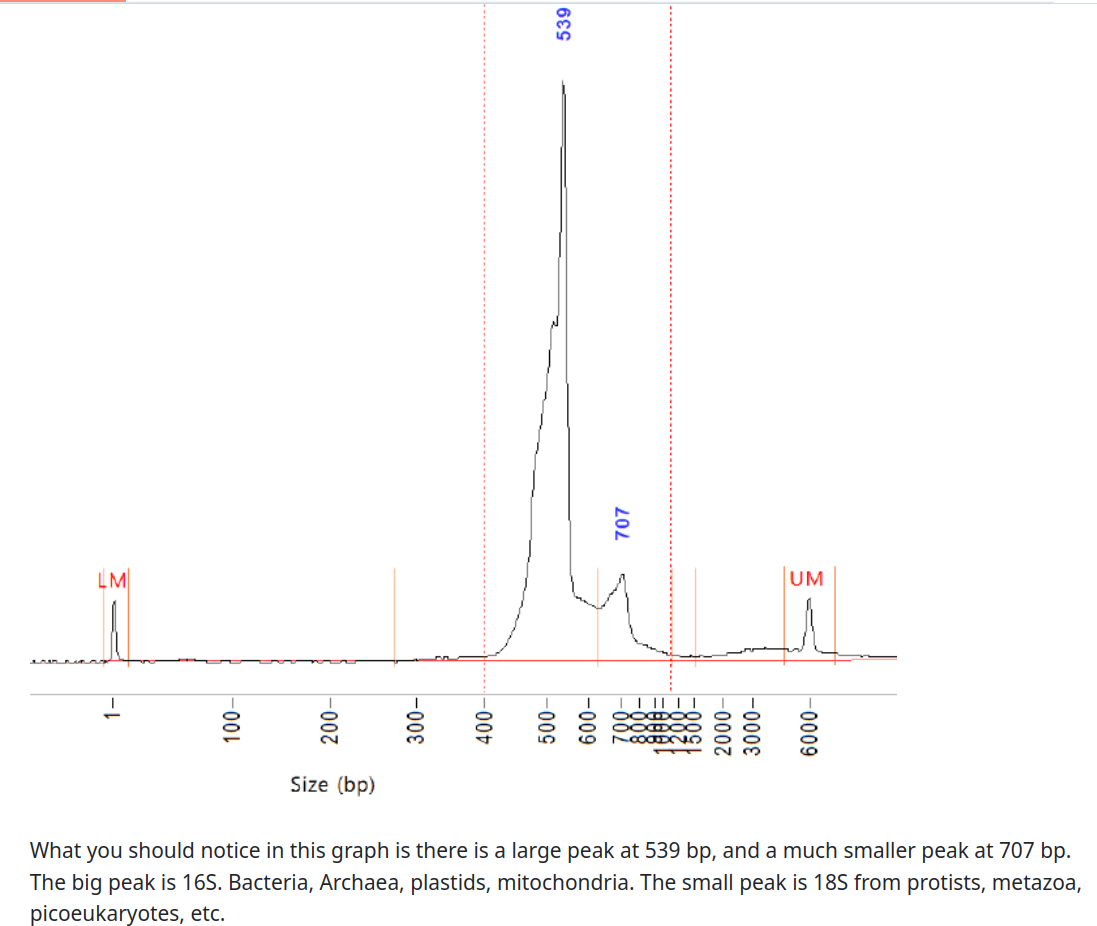
Accessible: Use expert-curated databases

Accessible: Use expert-curated datasets
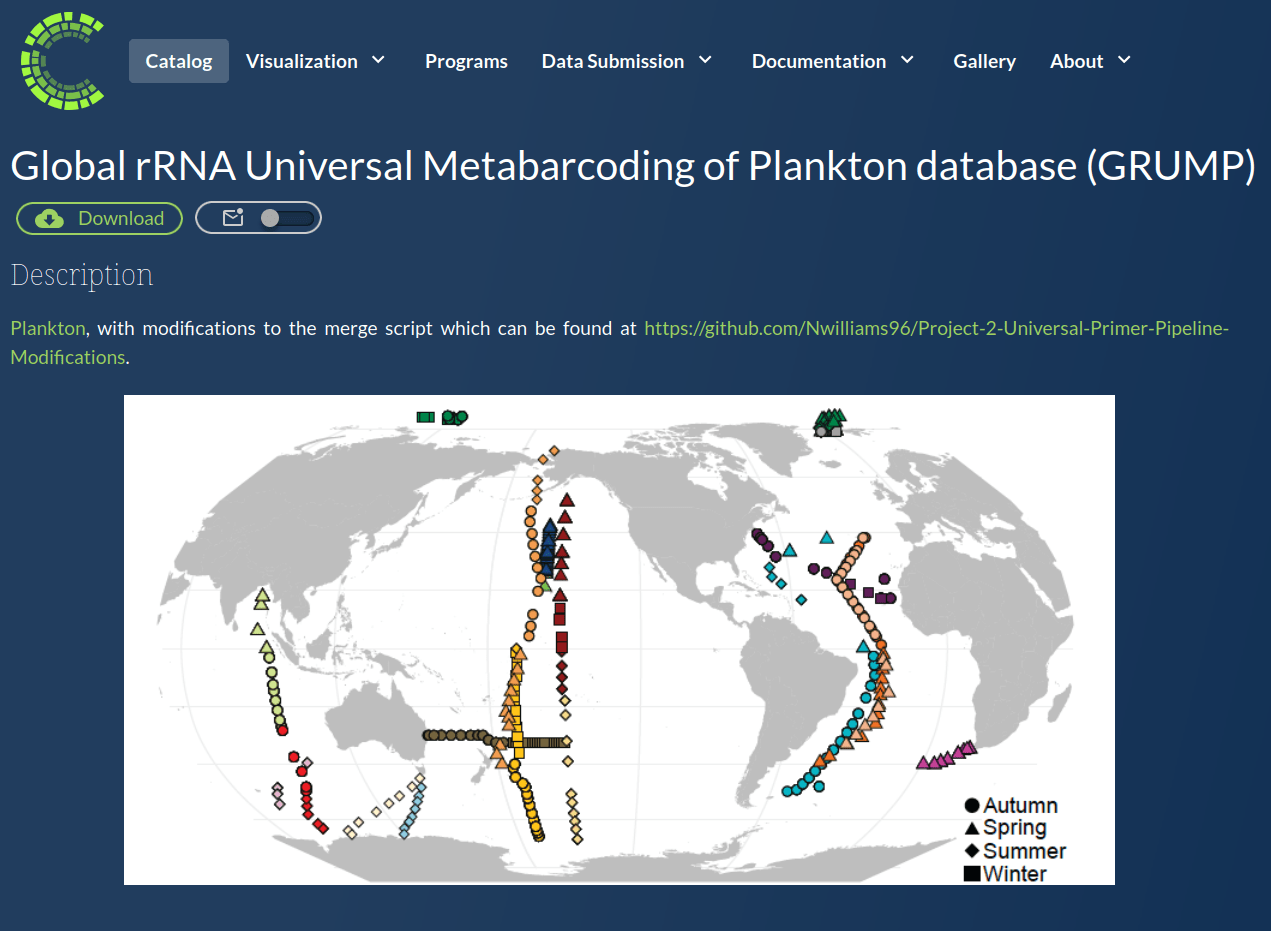

Ecologically-relevant annotations aggregate complex ASV data into sensible groupings (makes plotting, intercomparison easier)
- Bacterioplankton
-
Phytoplankton
- Prochlorococcus (ecotype)
McNichol, Williams, et al., 2025 (in revision)
Affordable: My "quiver" of DIY methods
- Kit-free, non-toxic DNA extraction with linear acrylamide as co-precipitant, allows for recovery of very small amounts of DNA for PCR (~10 mL seawater*)
- Cheap, simple protocol for removal of PCR inhibitors with linear acrylamide
- Protocols for decontamination of reusable plasticware for molecular protocols (reduce waste, guarantee quality)
- 1-step PCR reaction for library prep in lab (advantages = speed, lower cost, flexibility)
*Other methods for as little as 1 µL exist
Wilkinson et al., 2016
FAIR
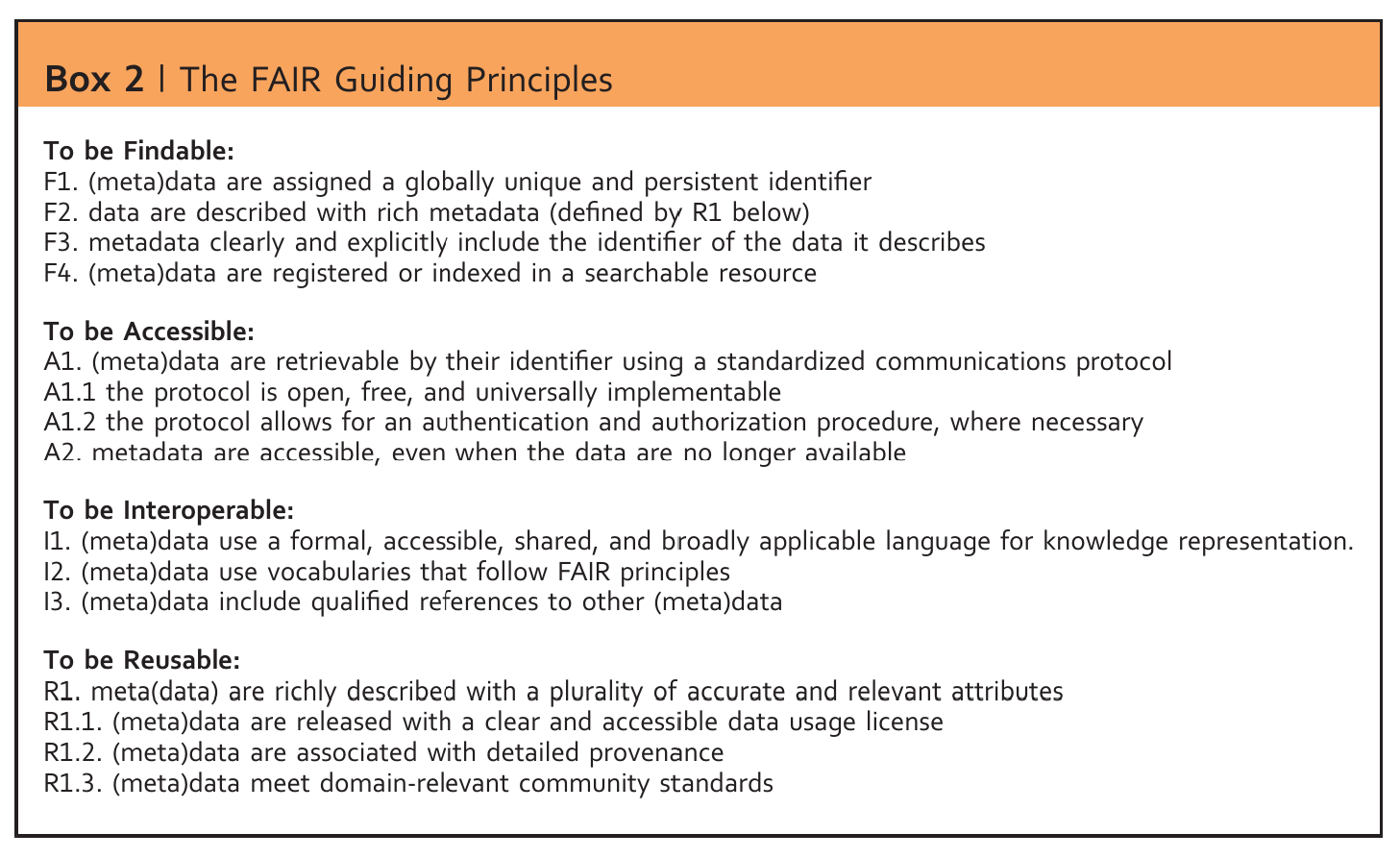
Wilkinson et al., 2016
FAIR
After the lapse of many years, possibly a century, the student of the future will have access to the original record of faunal conditions in California."
-Annie Montague Alexander, from "Why Ecology Needs Natural History"
FAIR data and Natural History

FAIR Computation

Think of the joy you can bring...
Comprehensive: whole-community methods

Ecosystem
Ecosystem

Metagenomics

Metabarcoding
Metabarcoding

p16S
e16S
18S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
Microbe art: @claudia_traboni
Comprehensive: whole-community methods
Sample x: dominated by prokaryotes (e.g. Sargasso Sea)

Microbe art: @claudia_traboni
18S
p16S
e16S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
Comprehensive: whole-community methods
Sample y: dominated by eukaryotes (e.g. Southern Ocean)
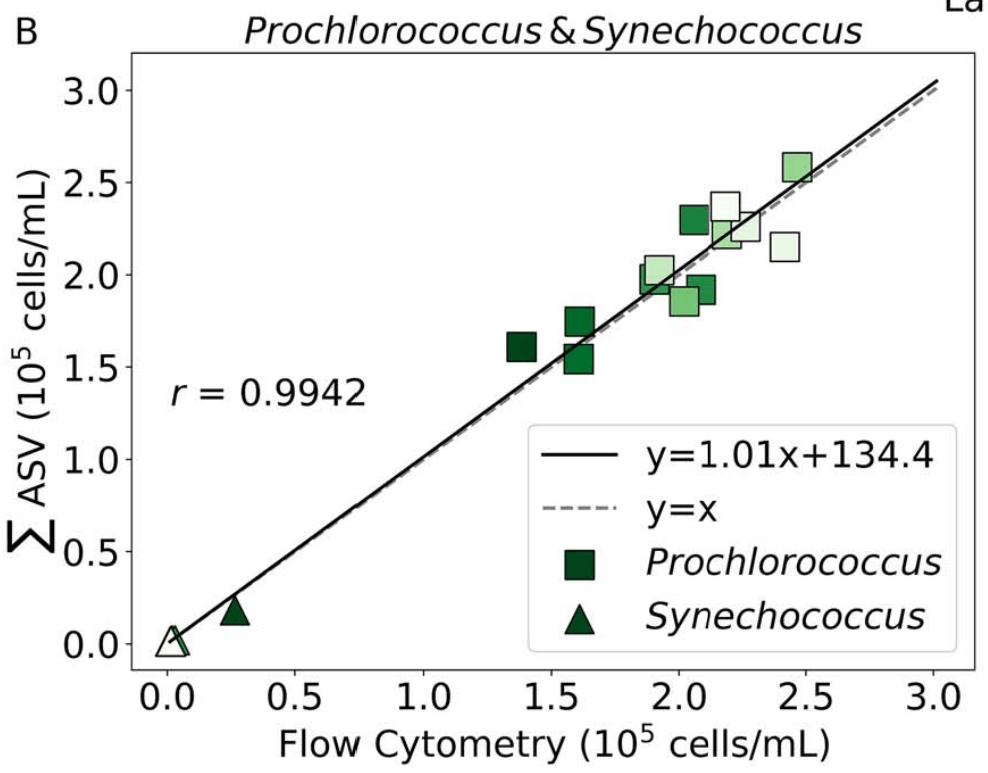
Comprehensive: absolute units
- Near perfect match between high-quality flow cytometry data and our eDNA quantification method with internal standards
- Workshop on internal standard methods with Dr. Julie Laroche (Sept 12-13th 2025, Halifax)
Archived
- DNA extracts from time-series studies may be incalculably valuable in future (new technologies, ecological restoration)
- Current methods of long-term preservation are energy intensive and/or not stable in very long term - should we be archiving DNA to create "microbial herbaria" as a long-term record?
Some final thoughts on the future of microbial ecology
- Accessible to non-experts, community organizations, affordable and sustainable
- Leading to collaborative, interdisciplinary research asking exciting new questions
- A record of changing ecosystems, and a possible warning for state changes

My vision for microbial ecology in the Anthropocene
The future


Community composition data : a map & compass!



Schramm (2003) / Amann et al., 1995
"Full cycle rRNA analysis"

Sebastián & Gasol (2019), 10.1098/rstb.2019.0083
Single-cell activity assays
Classical techniques for context (microscopy, chemical assays, etc.)
Currently = many microbial papers focus on describing community composition (who is there)



The future
The future
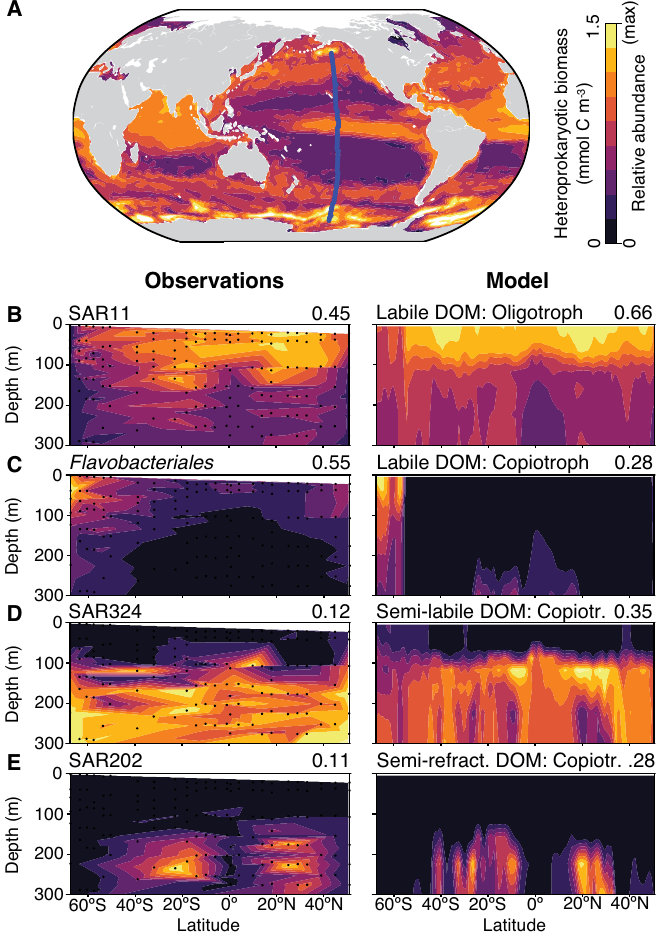
- Global eDNA data used as a "scaffold"
- GTDB genome data recruited to predict growth rates, niches
- Compared with models, showing high correspondence
- Observations → "bold hypotheses" about basic biology & microbe - climate interactions
The future: closing thought
At a time when forces are trying to distract and disrupt the scientific enterprise, doing the important work of finding and sharing the truth is now a great act of resistance."
-H. Holden Thorp, 2025 (link)
Questions?
Thanks to:
- Current and former CBIOMES collaborators (Fuhrman, Follows, Levine, Zakem labs)
- Dr. Bruce Hatcher (CBU)
- Katherine Rutherford and Jillian Davies (StFX)
- All of you for listening!


stfxmicroeco.ca | jmcnicho@stfx.ca
slides.com/jcmcnch/access-2025-plenary



Interested in aquatic microbial research at StFX?




Microbe art: @claudia_traboni
stfxmicroeco.ca | jmcnicho@stfx.ca
Contact me if you're interested in research experience