CBIOMES 2025 (June 17th, 2025)
Biogeography from Metabarcoding Data (GRUMP)
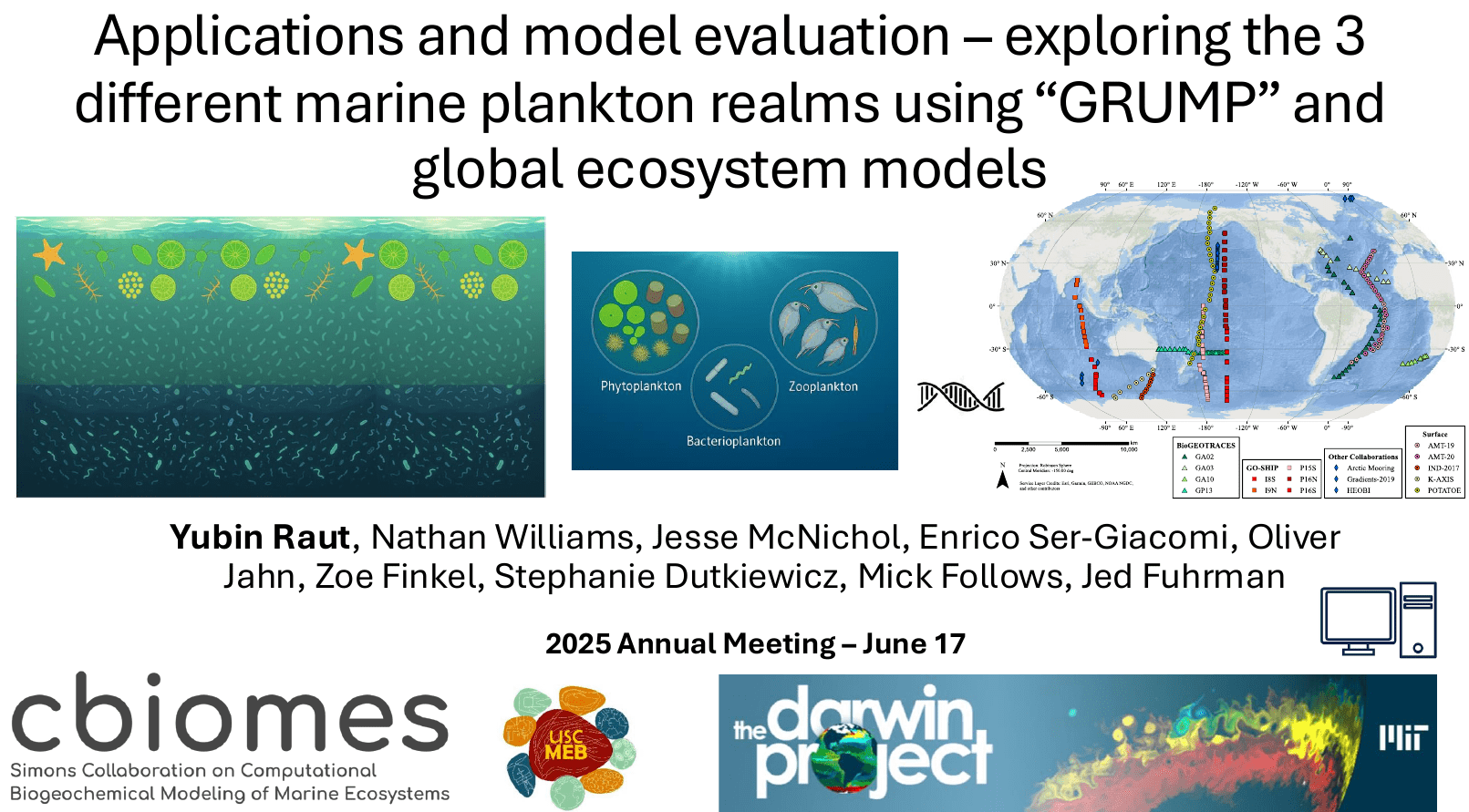
GRUMP began in 2018 with a request from Jed

Jesse, we need molecular data to compare with DARWIN, go find it

OK!
GRUMP began in 2018 with a request from Jed

OK then, let's generate the data ourselves...
[1-2 months later] ...ummm there isn't any*...
*That is easy to intercompare with DARWIN (not FAIR, size fractions, other methodological issues)

Jesse McNichol, Nathan Williams, Yubin Raut, Craig Carlson, Elisa Halewood, Kendra Turk-Kubo, Jonathan Zehr, Andrew Rees, Glen Tarran, Mary Gradoville, Matthias Wietz, Christina Bienhold, Katja Metfies, Sinhué Torres-Valdés, Thomas Mock, Sarah Lena Eggers, Wade Jeffrey, Joseph Moss, Paul Berube, Steven Biller, Levente Bodrossy, Jodie Van De Kamp, Mark Brown, Swan Sow, E. Virginia Armbrust, Jed Fuhrman










AMT
GRUMP is built on connections and collaborations




Yes, that means you!
- Ecosystem modellers
- Physical oceanographers
- Satellite ocean colour experts
- Math / stats nerds
- Fellow "gene jockeys"
- Other ocean enthusiasts
Lots of time for questions during / after talks
Basic questions welcomed!
GRUMP is "live" and we want people to use it!

TO USE GRUMP DATA





-
Jesse & Nathan - GRUMP: Promise and potential (25 min + 5)
-
Yubin - Applications and model evaluation (15 min + 5)
-
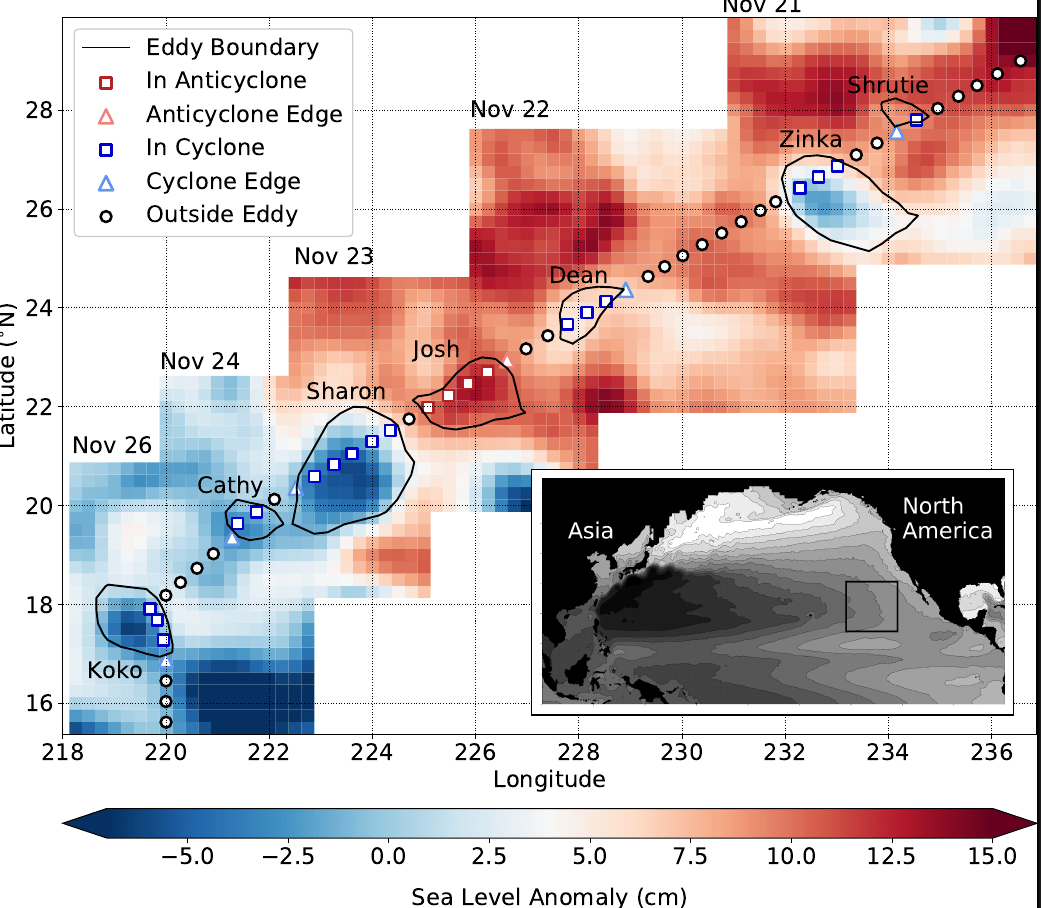
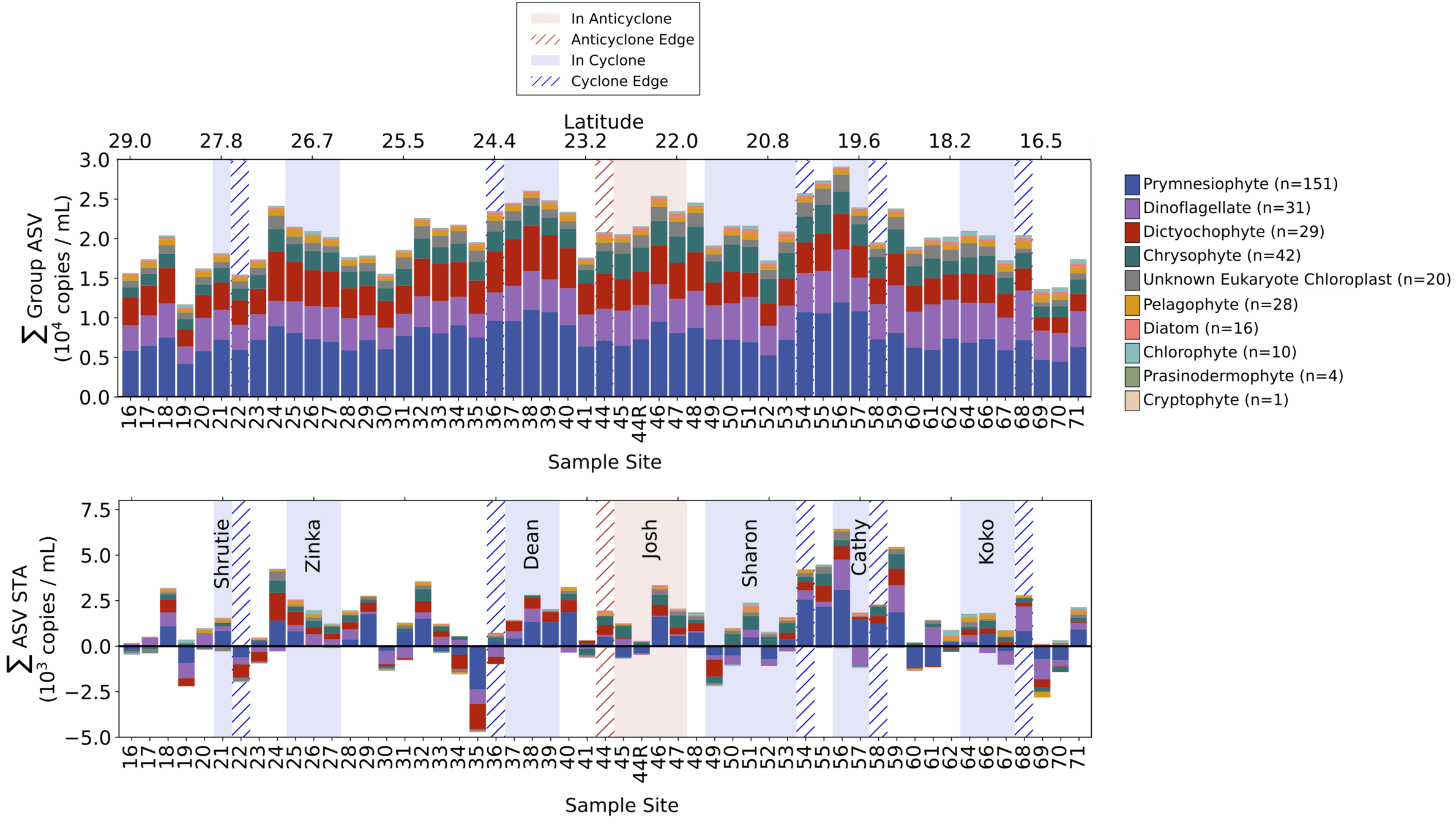
Lexi - Revealing mesoscale organization of communities with metabarcoding on Gradients 4 transect (15 min + 5)
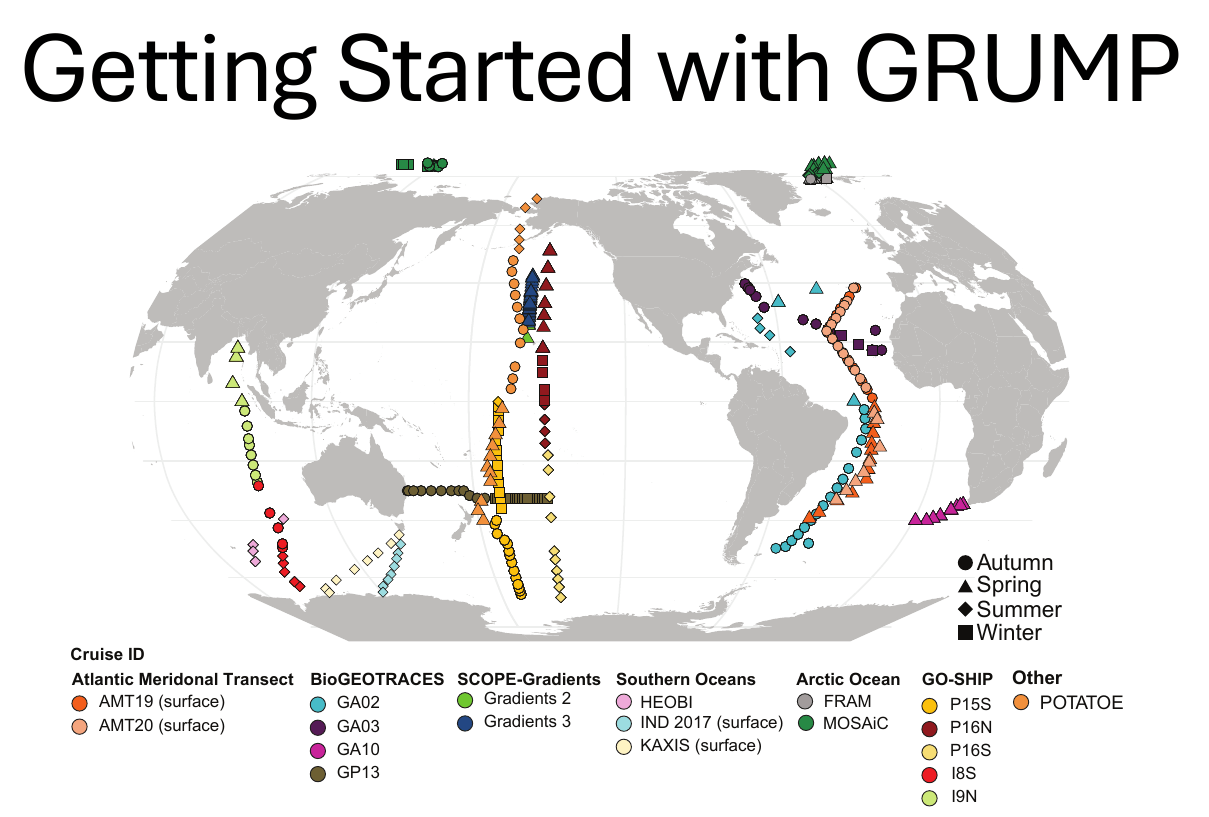
- Nathan - Getting started with GRUMP (setup for post-break hands-on opportunity; 10 min)
- Coffee Break (10:30)
-
Optional “Hands on with GRUMP” breakout led by Nathan (11:00 - 12:30)
Session lineup
CBIOMES 2025: Biogeography from Metabarcodes with GRUMP
Jesse McNichol and Nathan Williams (June 17th, 2025)
GRUMP: Promise and Potential
or... "What is this thing called GRUMP???"

- Methods refresher
- What makes GRUMP unique?
- GRUMP 2.0, now with internal standards!
- Open questions / The future of GRUMP
Talk outline
1. Methods refresher
Metabarcodes are short strings of DNA that identify taxa

Ecosystem
Ecosystem
(PCR)

Metabarcoding
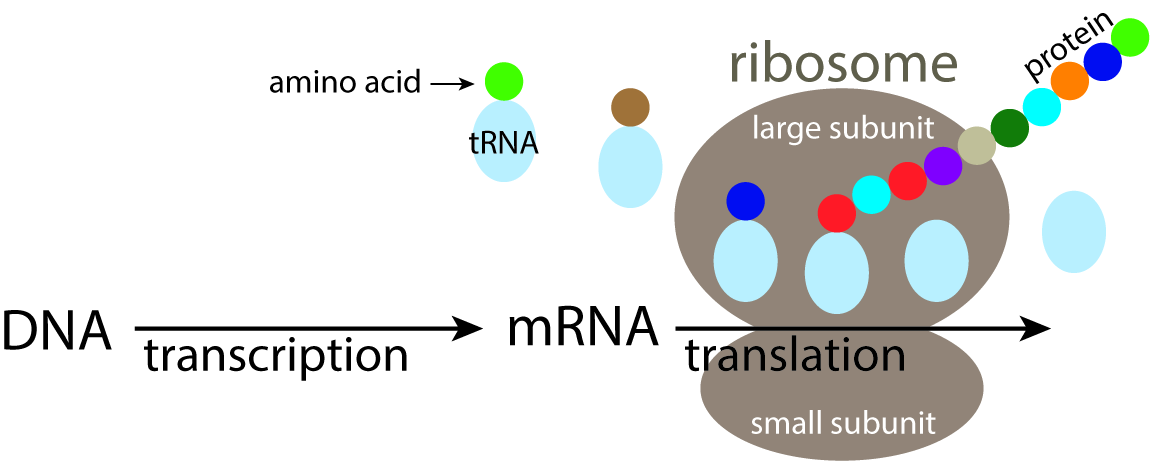
(rRNA)
(shotgun sequencing)

Metagenomics
Metabarcodes are short strings of DNA that identify taxa

Ecosystem
Ecosystem
(PCR)

Metabarcoding

(rRNA)
Metabarcoding is an ecosystem "census"

- Who is there and how abundant are they?
- How much diversity is within each taxon?
- How do these patterns change across space and time?


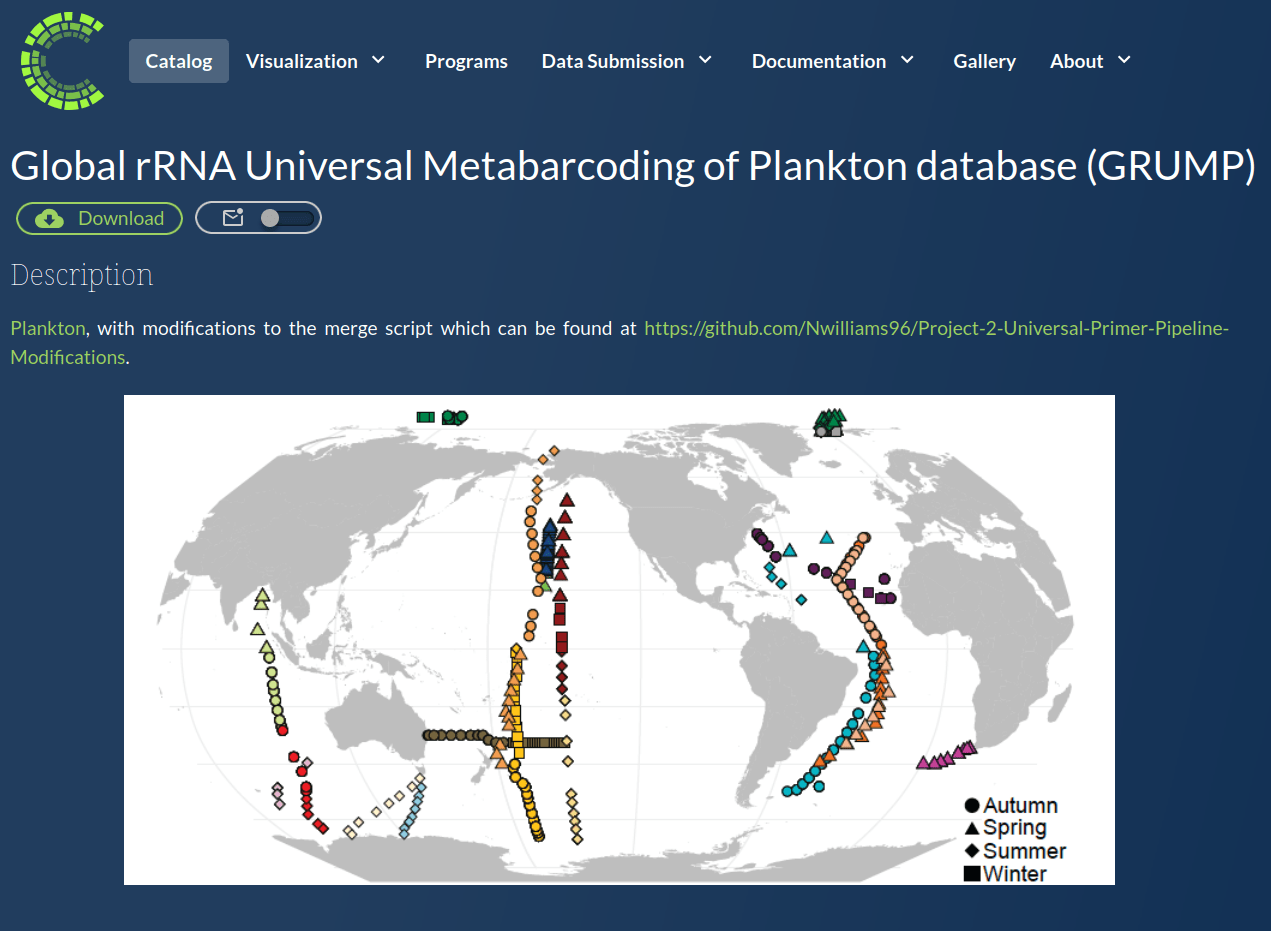
GRUMP = a metabarcoding census on global samples
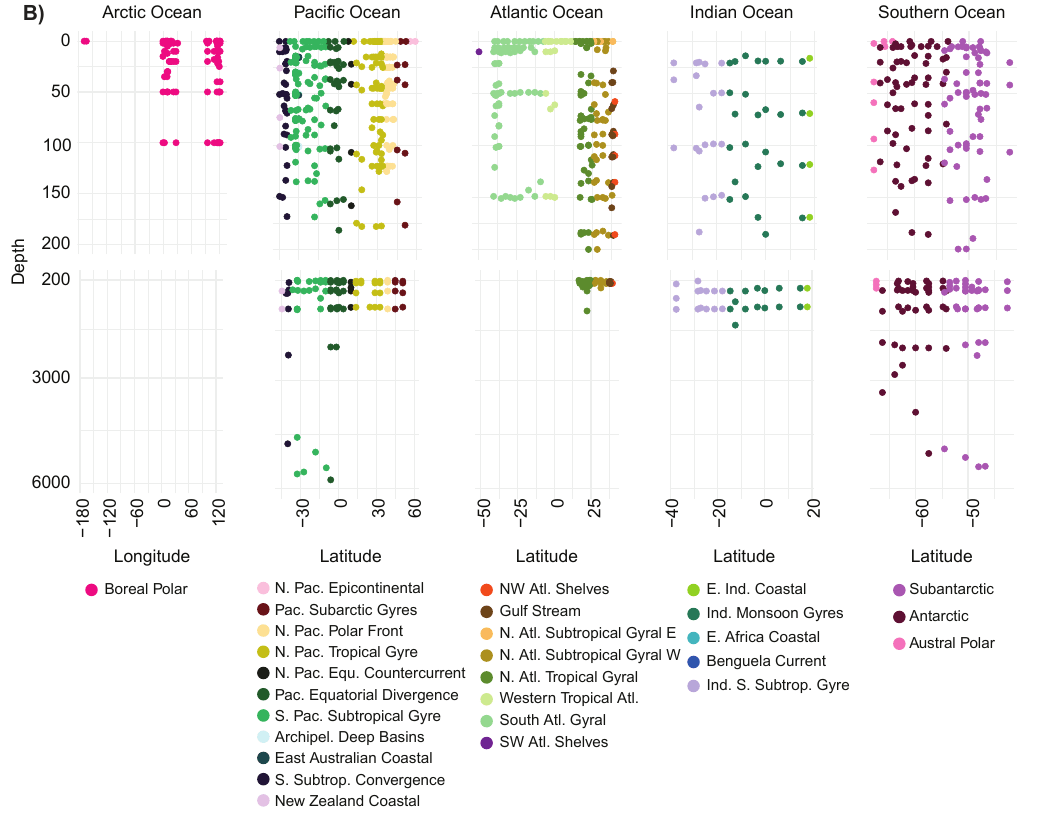
GRUMP = a metabarcoding census on global samples... across depth!
GRUMP = a metabarcoding census on global samples... in the context of global sampling campaigns
- Nathan has assembled a core set of covariates common to all cruises in GRUMP (T, S, Oxygen, Nutrients, Chlorophyll)
- Also, Longhurst Provinces, Ocean Basin, Depth Categories, Predicted Euphotic Depth
- Also... some of the campaigns also have their own unique data products you may be interested in (CMAP colocolalization)
Jesse McNichol, Nathan Williams, Yubin Raut, Craig Carlson, Elisa Halewood, Kendra Turk-Kubo, Jonathan Zehr, Andrew Rees, Glen Tarran, Mary Gradoville, Matthias Wietz, Christina Bienhold, Katja Metfies, Sinhué Torres-Valdés, Thomas Mock, Sarah Lena Eggers, Wade Jeffrey, Joseph Moss, Paul Berube, Steven Biller, Levente Bodrossy, Jodie Van De Kamp, Mark Brown, Swan Sow, E. Virginia Armbrust, Jed Fuhrman










AMT





2. What Makes GRUMP Unique?
- Comprehensive ("Parada" primers target all rRNA from cellular life, including zooplankton & organelles)
- Sensitive with deep sequencing (~200,000 reads per sample)
- Specific with "denoising" algorithms (→ "ASVs")




Microbe art: @claudia_traboni
GRUMP "unique selling points"
GRUMP: primers target all rRNA

SSU rRNA (we target the gene that encodes this)
plastid
16S rRNA
mito 16S
rRNA
nuclear 18S rRNA
A eukaryotic phytoplankter
GRUMP: primers target all rRNA
Bacterium
16S rRNA
Archaeon 16S
rRNA
Bacteria and Archaea (prokaryotes)
mito 16S
rRNA
nuclear 18S rRNA
A eukaryotic protist or zooplankton cell

GRUMP: primers target all rRNA
GRUMP: DNA from samples > 0.2 µm
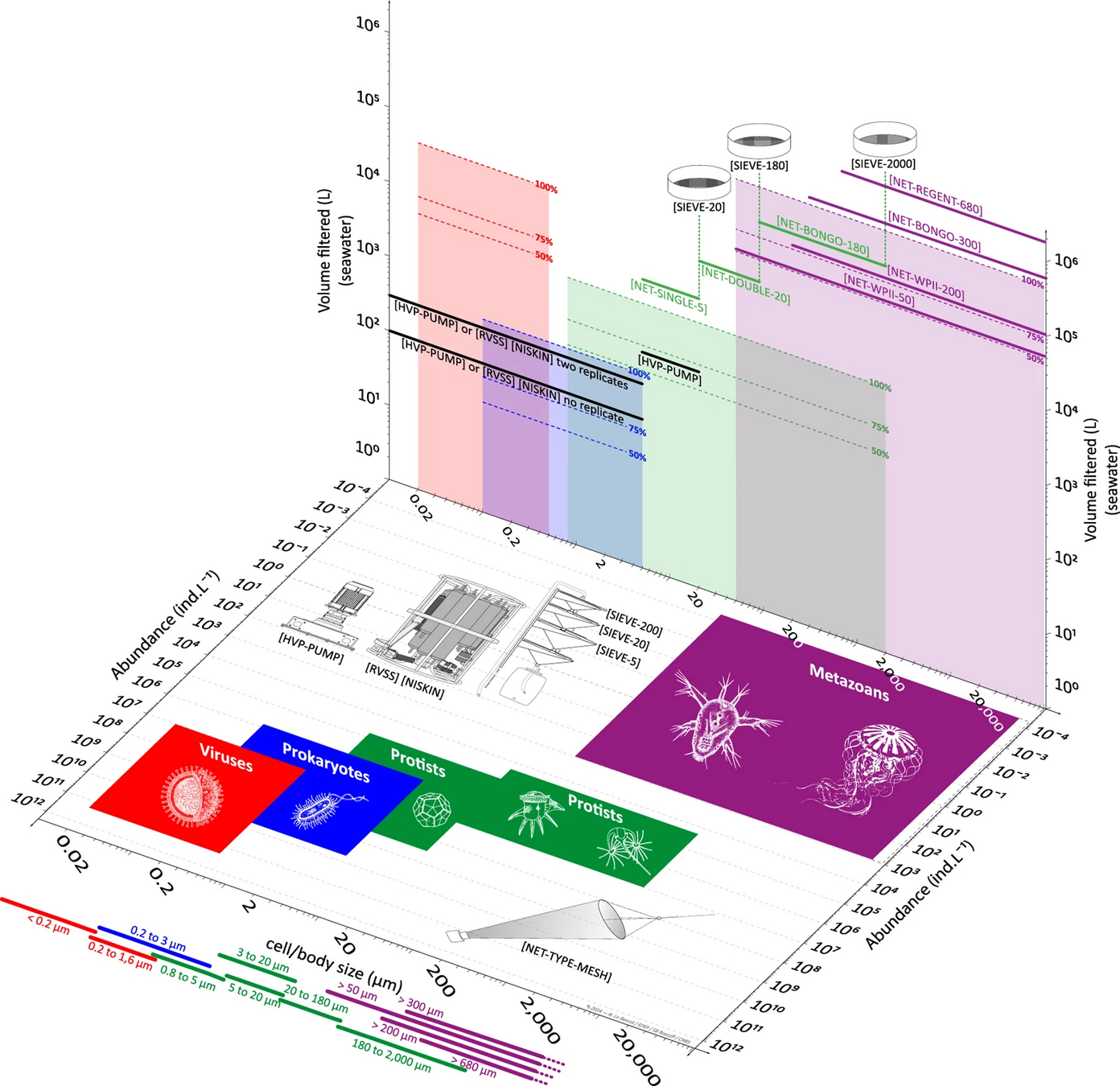
GRUMP: No size fractionation

Microbe art: @claudia_traboni
TARA: Many size fractions (complicates interpretation)

p16S
e16S
18S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
Microbe art: @claudia_traboni
Sample x: dominated by prokaryotes (e.g. Sargasso Sea)
GRUMP: whole community perspective


Microbe art: @claudia_traboni
18S
p16S
e16S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
Sample y: dominated by eukaryotes (e.g. Southern Ocean)
GRUMP: whole community perspective

Craig Carlson, Elisa Halewood, UCSB
18S / (16S + 18S)

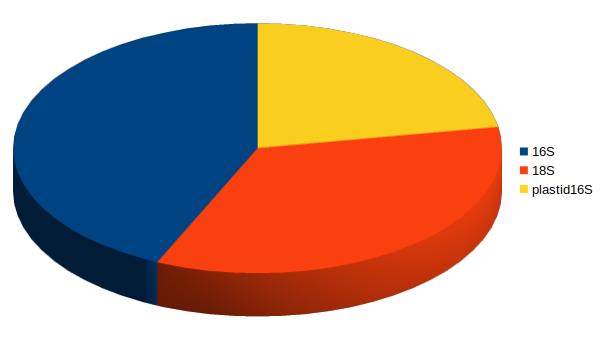
16S
plastid 16S
18S

Jan-Feb 2005
Feb-Mar 2006
With deep sequencing, good coverage for all 3 domains
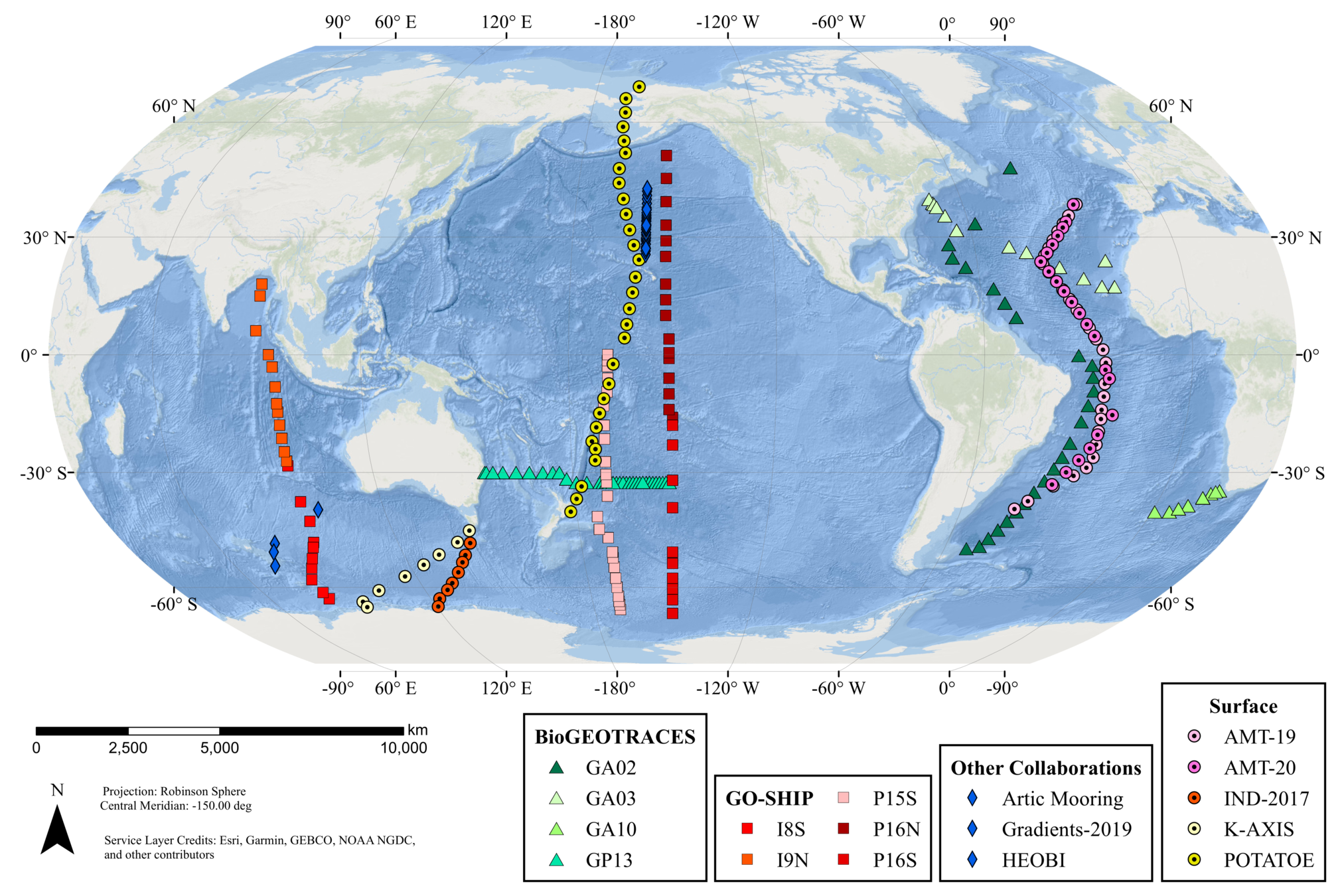
GRUMP: whole community perspective
Kendra Turk-Kubo, Rosie Gradoville, Jon Zehr, UCSC

AMT
Glen Tarran, Andy Rees, PML

Trichodesmium (AMT 20)
nifH gene copies / L
16S ASV relative abundance
GRUMP: specific taxa abundances also possible
With 200k sequences per sample, practical detection limit = 1 copy nifH / mL
nifH gene copies / L (x 106)
16S ASV relative abundance

UCYN-A (AMT 20)
Kendra Turk-Kubo, Rosie Gradoville, Jon Zehr, UCSC

AMT
Glen Tarran, Andy Rees, PML
GRUMP: specific taxa abundances also possible


Ecologically-relevant annotations aggregate complex ASV data into sensible groupings (makes plotting, intercomparison easier)
- Bacterioplankton
-
Phytoplankton
- Prochlorococcus (ecotype)
GRUMP: expert-curated annotations of ASVs
3. GRUMP 2.0, now with internal standards!
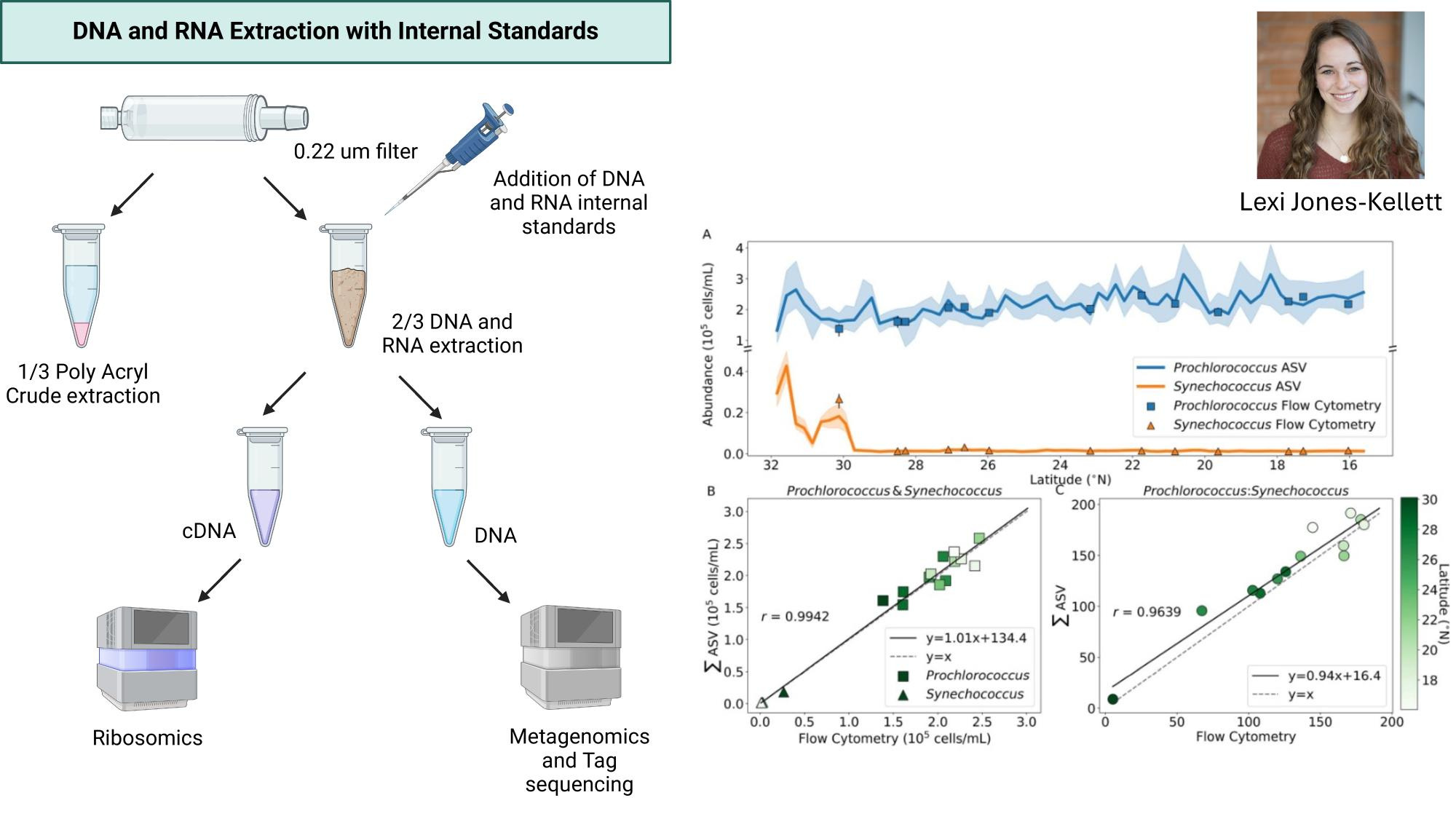
GRUMP 2.0: moving towards absolute abundances
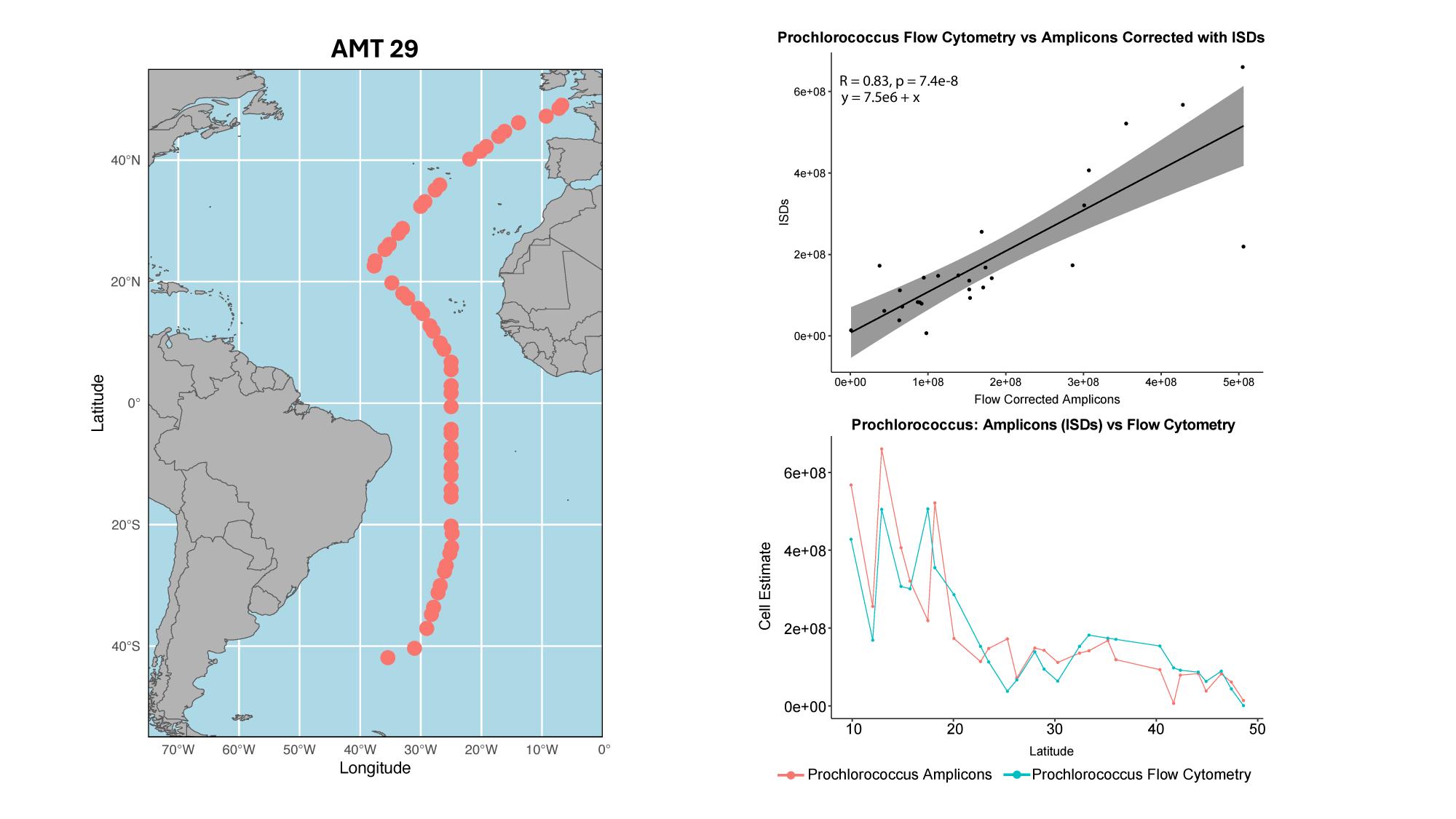
GRUMP 2.0: moving towards absolute abundances
Williams, unpublished
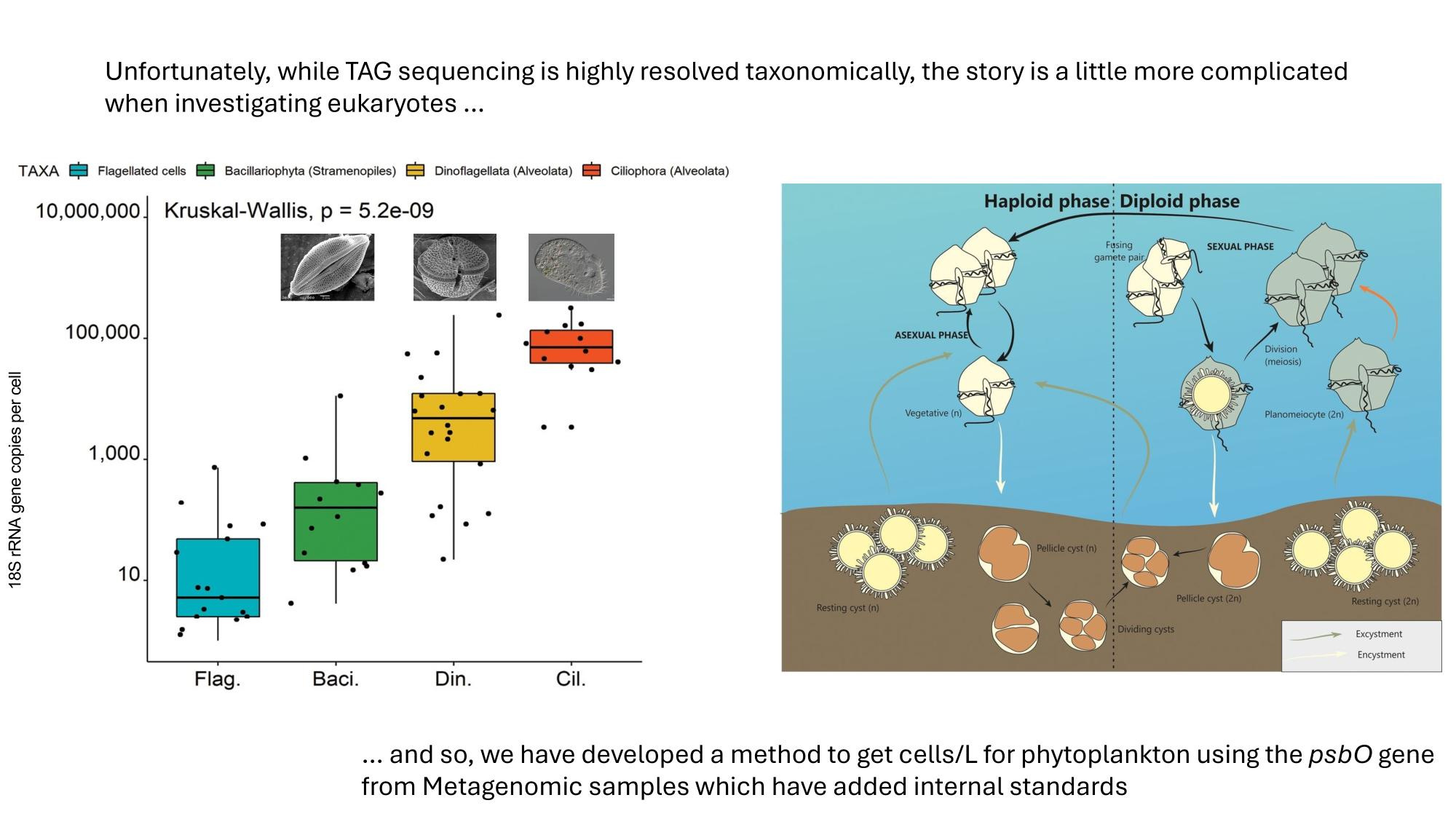
GRUMP 2.0: moving towards absolute abundances
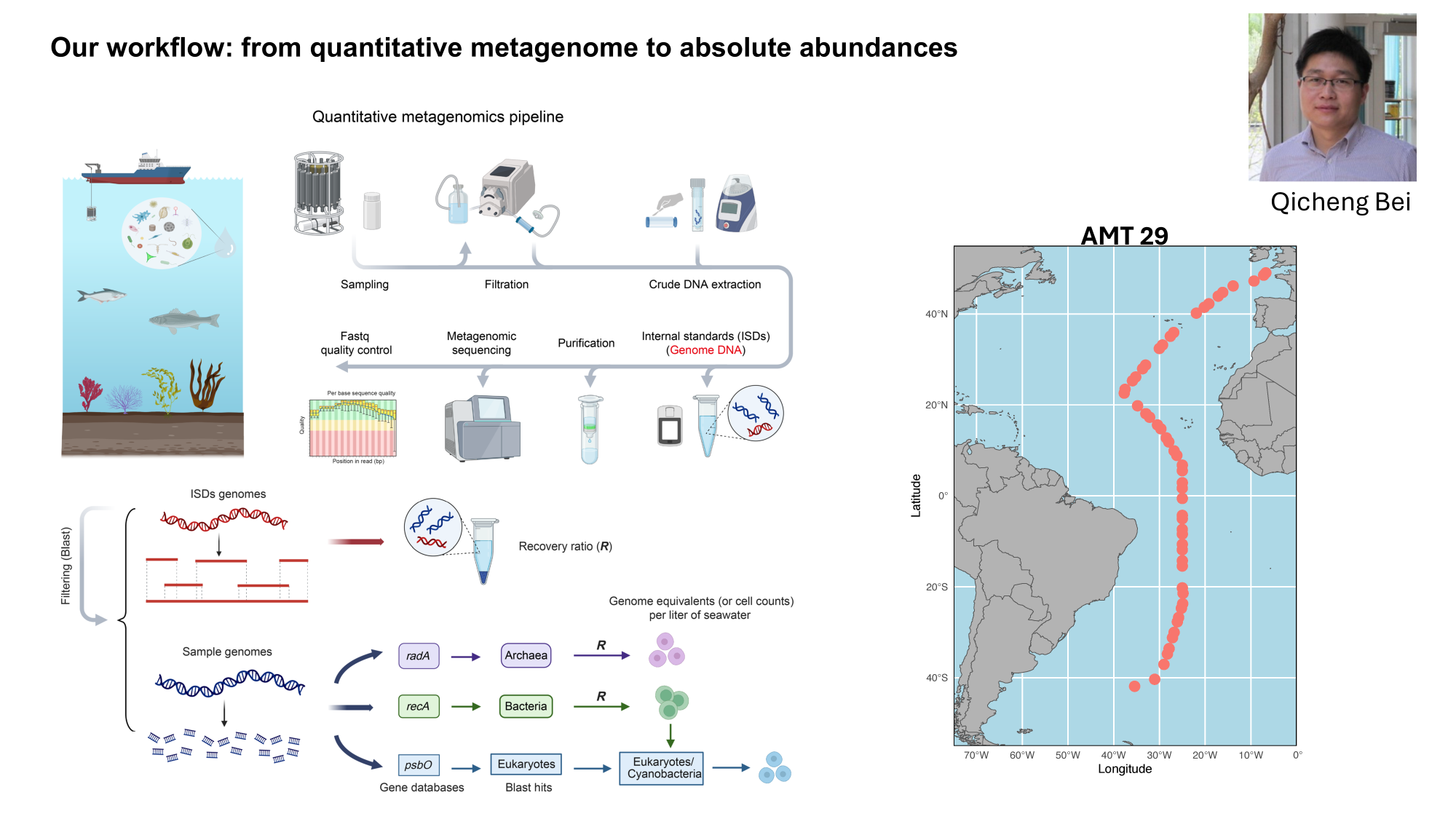
GRUMP 2.0: moving towards absolute abundances
Bei, 2025 (in review)
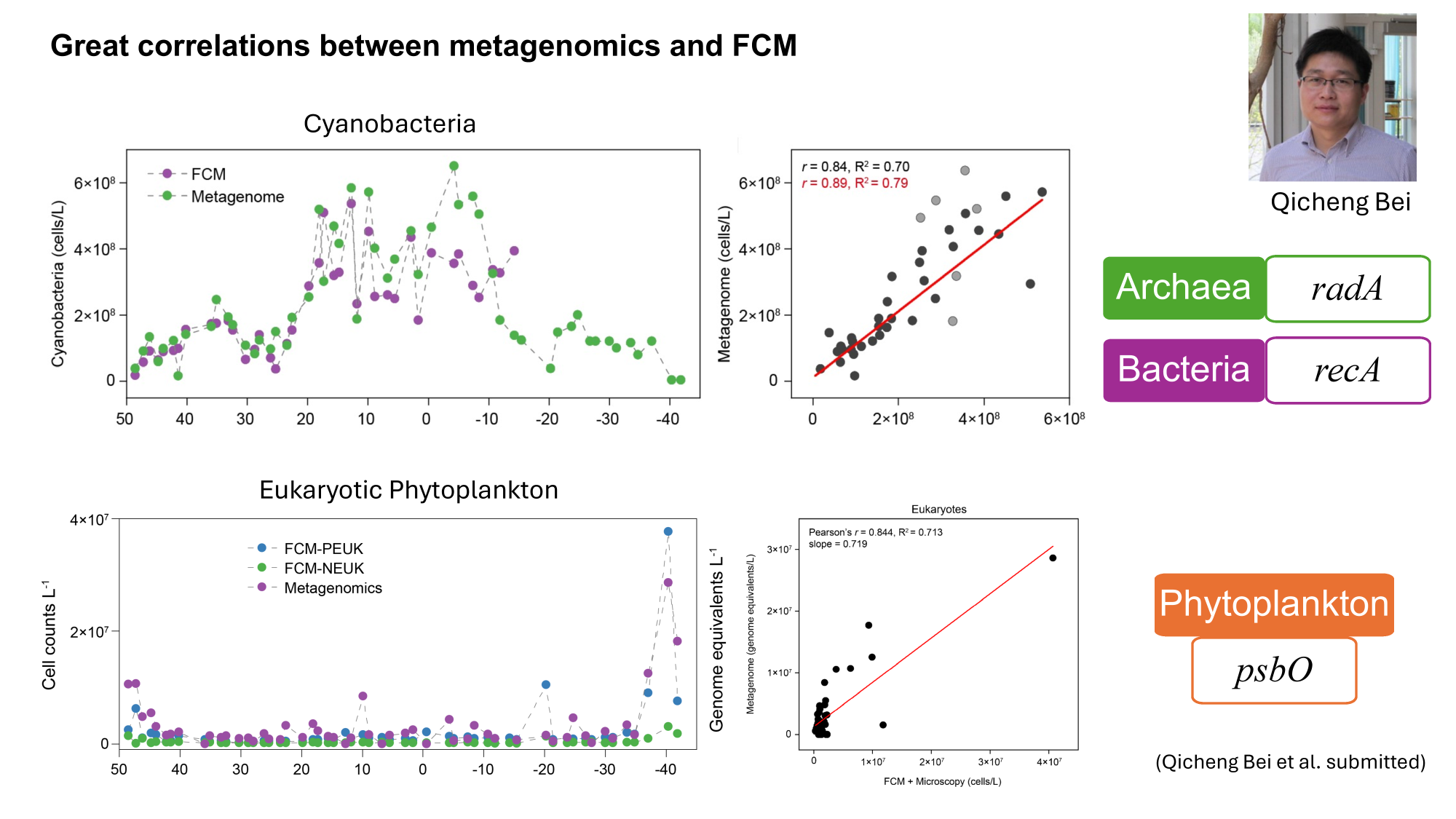
GRUMP 2.0: moving towards absolute abundances
Bei, 2025 (in review)
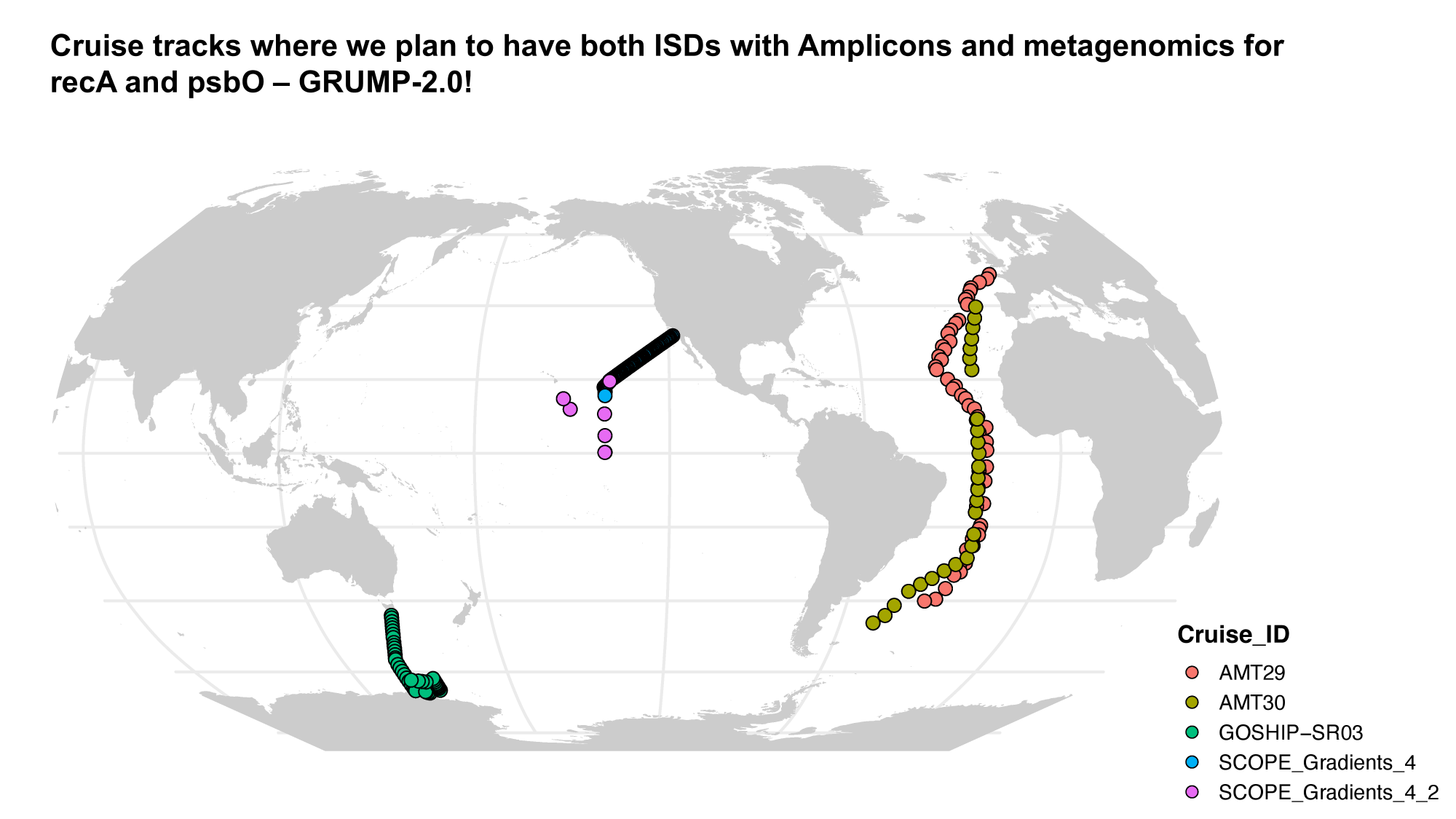
GRUMP 2.0: moving towards absolute abundances
Williams, unpublished
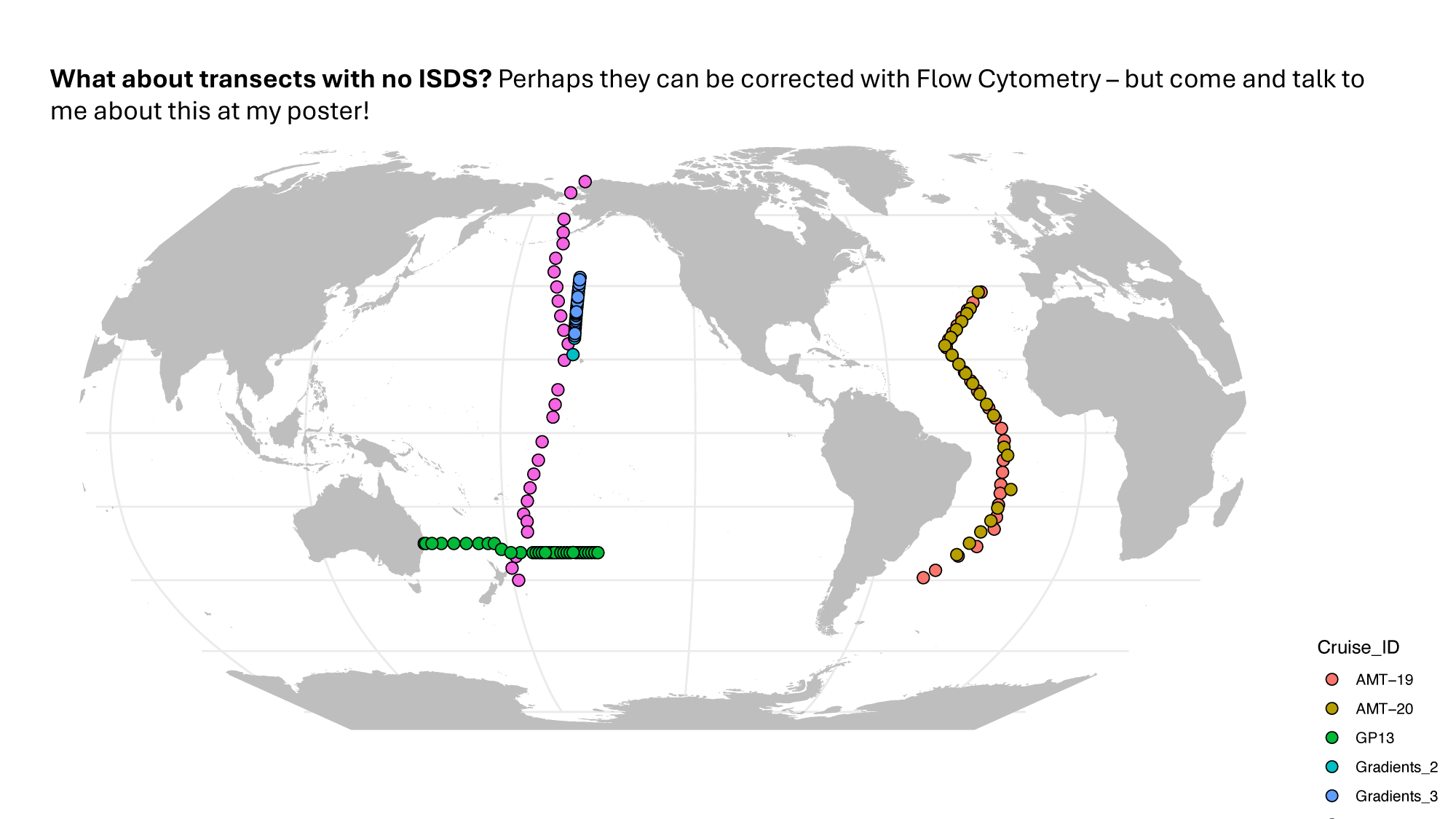
GRUMP 2.0: moving towards absolute abundances
Williams, unpublished
4. Open questions / The future of GRUMP
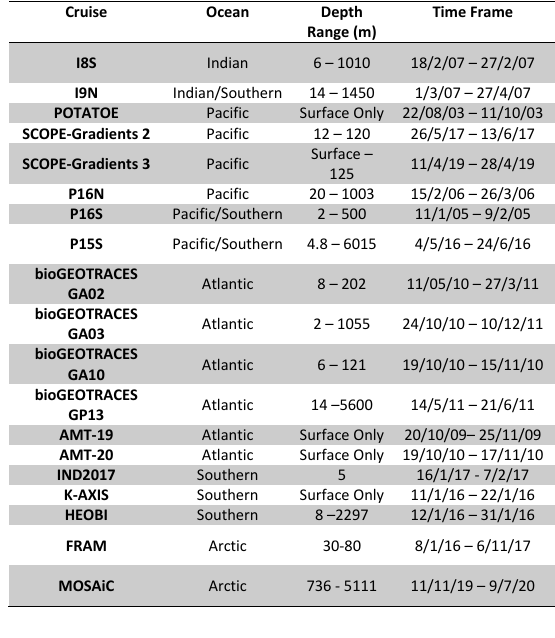
- Data as far back as 2003 (POTATOE)
- How to use GRUMP to infer ecosystem change (present or future)?
- If we need more data (most likely from older cruises), how can we get it? Should we be putting out an "APB" for more legacy samples sitting in -80°C freezers?
- Should we be in the "business" of generating new data? If so, who would do it?
Open question: how are marine communities changing?

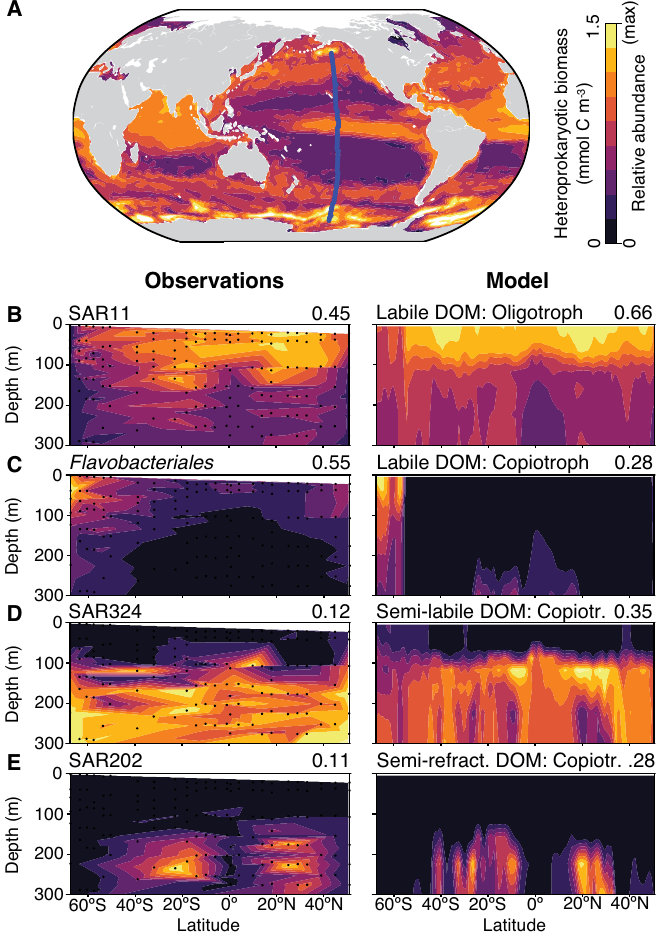
- For any given cruise, the consistency of broad-scale biogeographic patterns is always striking (at least to me)
- Are we being blinded by these abundant taxa and missing information hidden beneath (e.g. rare, sporadic taxa)?
- Could we discover real ecologically-relevant patterns that are not just stochastic noise?
Open question: what information lies beneath the broad and consistent patterns we see?
GRUMP future: Should we be doing more intercomparisons?
- How well do methods other than flow cytometry (e.g. imaging flow cytobot) align with 3-domain metabarcoding?


Kalmbach et al. (2017), arXiv:1703.07309v1

- DNA extracts from time-series studies may be incalculably valuable in future (new technologies, ecological restoration)
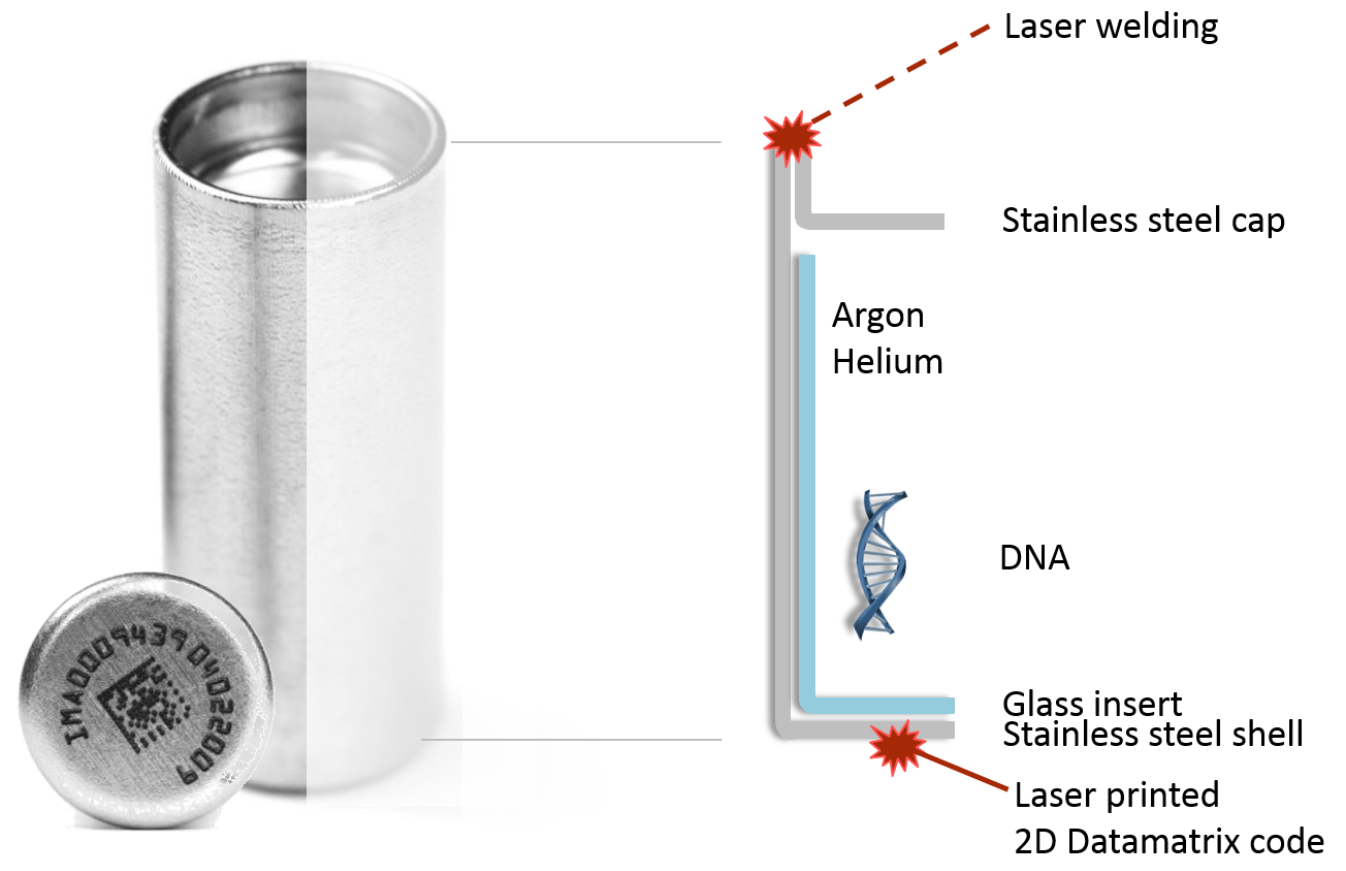
- Current methods of long-term preservation are energy intensive and/or not stable in very long term - should we be archiving DNA to create "microbial herbaria" as a long-term record?
GRUMP future: should we be archiving the DNA itself?

- GRUMP data used as a "scaffold" to recruit GTDB genome data to predict growth rates, index of copiotrophy
- Study provides context for experimental approaches that test new hypotheses about basic biology & microbe - climate interactions
GRUMP future: new research directions
Questions?


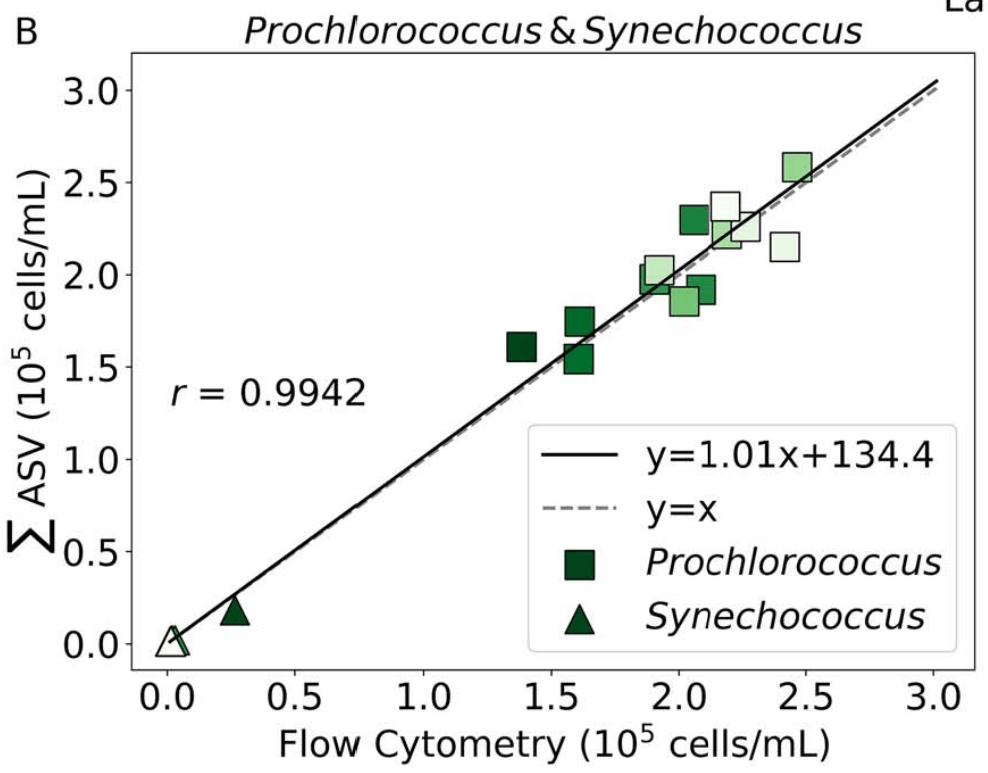
- Near perfect match between high-quality flow cytometry data and our eDNA quantification method with internal standards
- Lexi's talk will discuss this in more detail, Nathan generating new datasets with internal standards
GRUMP 2.0: moving towards absolute abundances