Institute for Comparative Genomics Guest Lecture
Dr. Jesse McNichol (Oct 31st, 2025)
'omics-Driven Marine Microbial Ecology: Tracking Food Web Structure, Climate Change Impacts, and Ecosystem Resilience
- Where I come from as a scientist and what motivates me
- Microbiome madness, and my view on the way out
- The "quiver" of methods I use
- The kind of questions that can be asked
- A microbial map of the ocean with 3-domain metabarcoding
- Global models of "heteroprokaryotes"
- Subtle patterns driving phytoplankton dynamics in open-ocean ecosystems
- My current research in Nova Scotia in the Bras d'Or Lakes
Talk outline

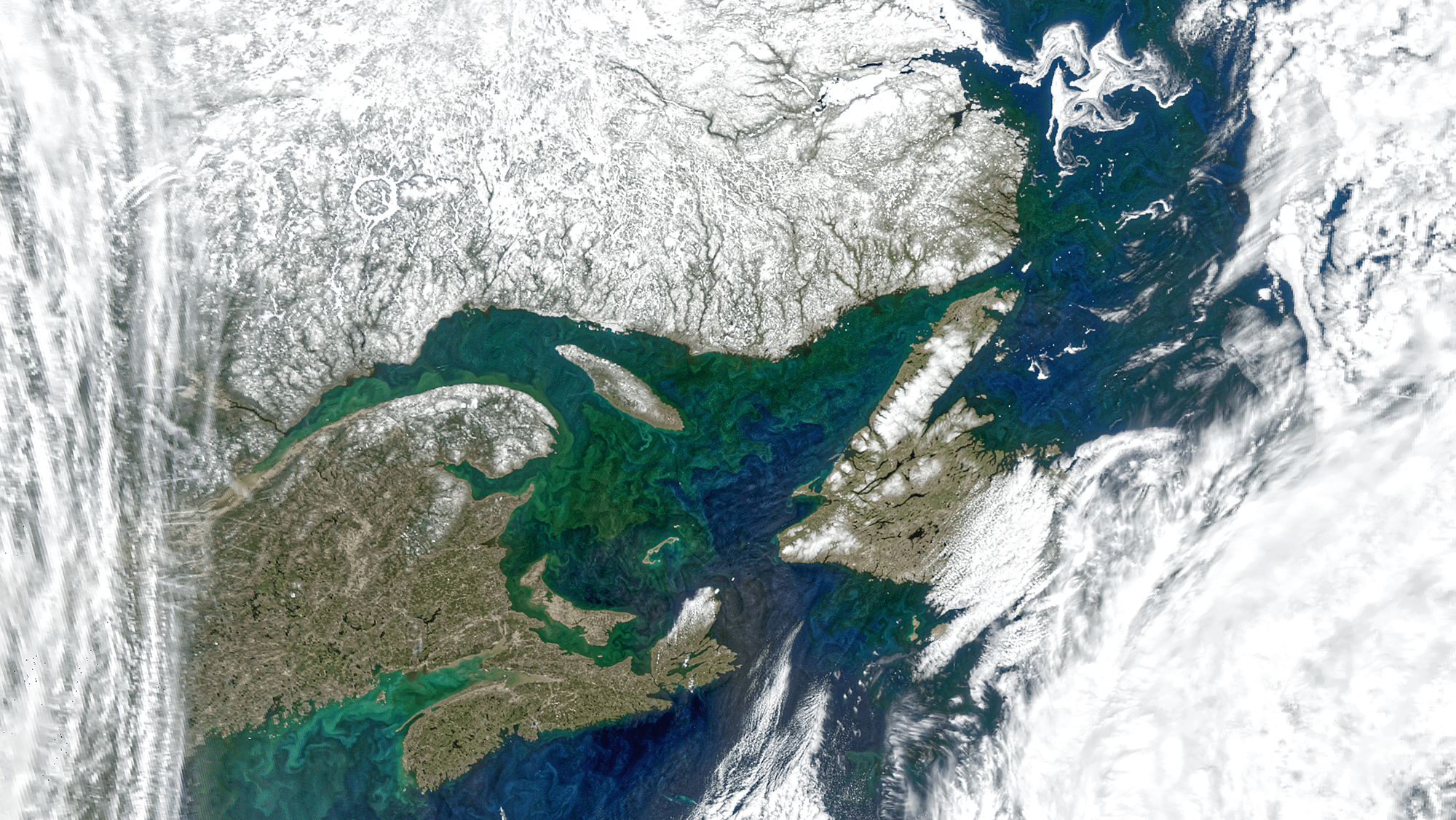
If you could step back to the edge of the solar system...

...and look at the light passing through our atmosphere...

...what would you see?
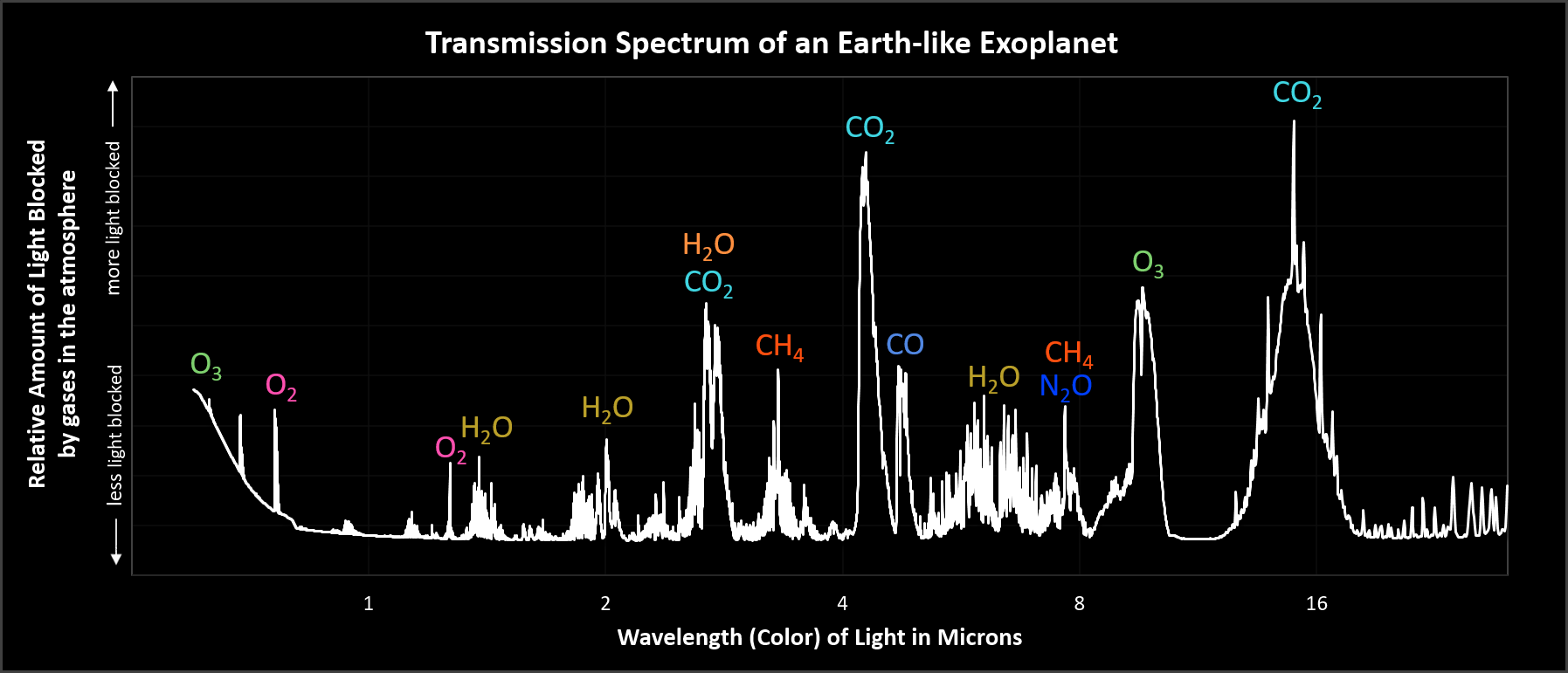
Chemical signatures of a living planet!
- Primary production
- Nutrient cycling
- GHG production / consumption
- Influence geology, climate, chemistry, and biology of Earth
Microbial life

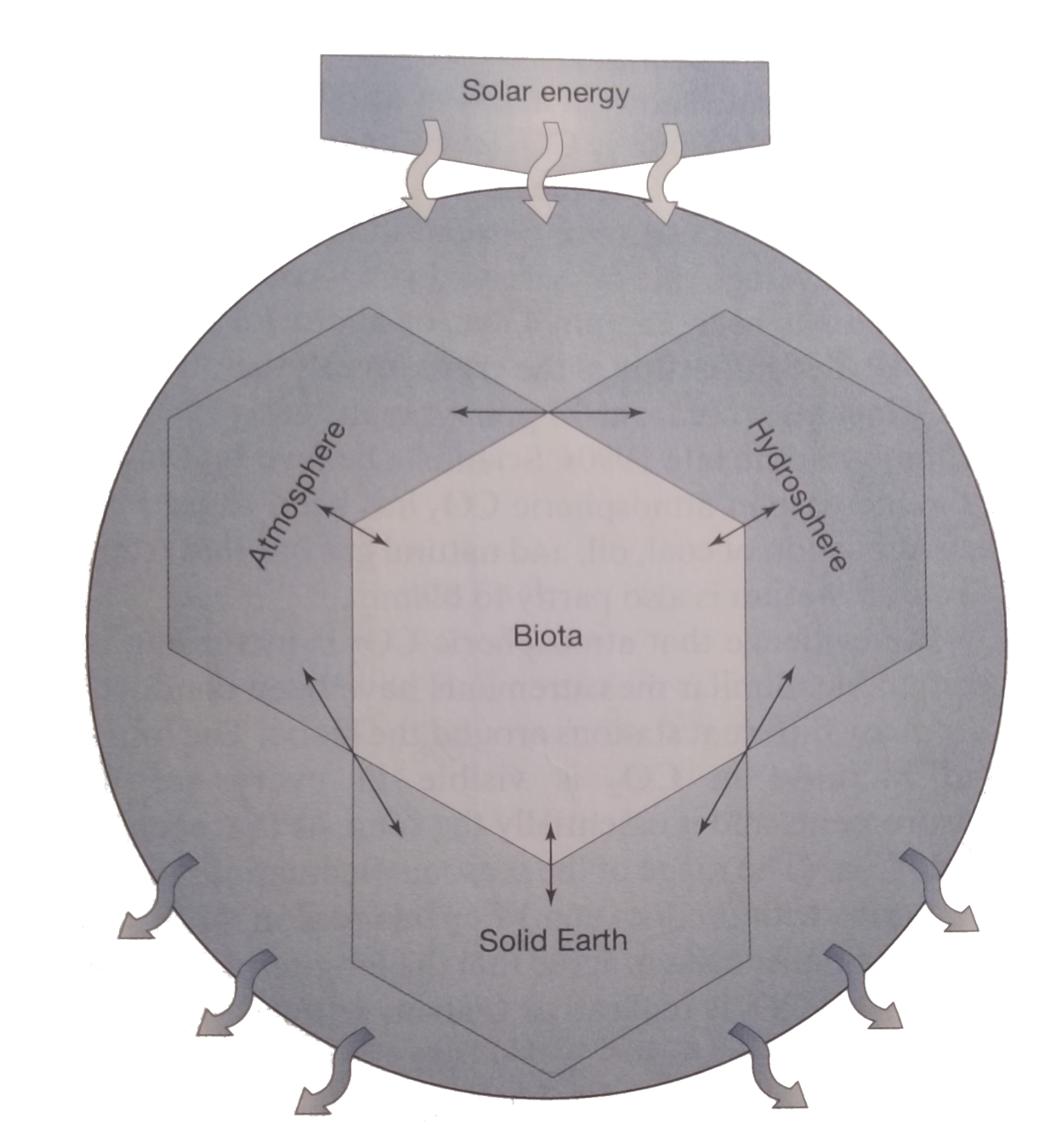
(Micro)
Kump, Kasting, and Crane, The Earth System
Microbial life keeps the big wheel turning...

Microbial life



(Micro)
Kump, Kasting, and Crane, The Earth System

My undergrad mentor Dr. Zoe Finkel introduced me to the "Earth System" and microbes' central role in it
Why do I study microbiology?
My academic background before StFX

BSc, Biology 2003-2008






PhD Biological Oceanography, 2011-2016

2009-2011 NRC (Algal Biofuels, Halifax)


Postdocs CUHK, USC, 2017-2023



My academic background before StFX

BSc, Biology 2003-2008






PhD Biological Oceanography, 2011-2016

2009-2011 NRC (Algal Biofuels, Halifax)


Postdocs CUHK, USC, 2017-2023




Earth Systems Science
Analytical Chemistry
Analytical Chemistry
Ecological Niches
Biogeochemistry
Microbial Ecology
NGS / 'omics
Bioinformatics
My academic background before StFX

BSc, Biology 2003-2008






PhD Biological Oceanography, 2011-2016

2009-2011 NRC (Algal Biofuels, Halifax)


Postdocs CUHK, USC, 2017-2023




Earth Systems Science
Analytical Chemistry
Analytical Chemistry
Ecological Niches
Biogeochemistry
Microbial Ecology
NGS / 'omics
Bioinformatics
Today's topic: My thoughts on how to bring all these together
My research is motivated by the elephant in the room...

Microbial life



(Micro)
Kump, Kasting, and Crane, The Earth System
What effect will anthropogenic pressures such as climate change and coastal eutrophication have on microbial "ecosystem services"?
How to Escape the "Madness of the Microbiome"
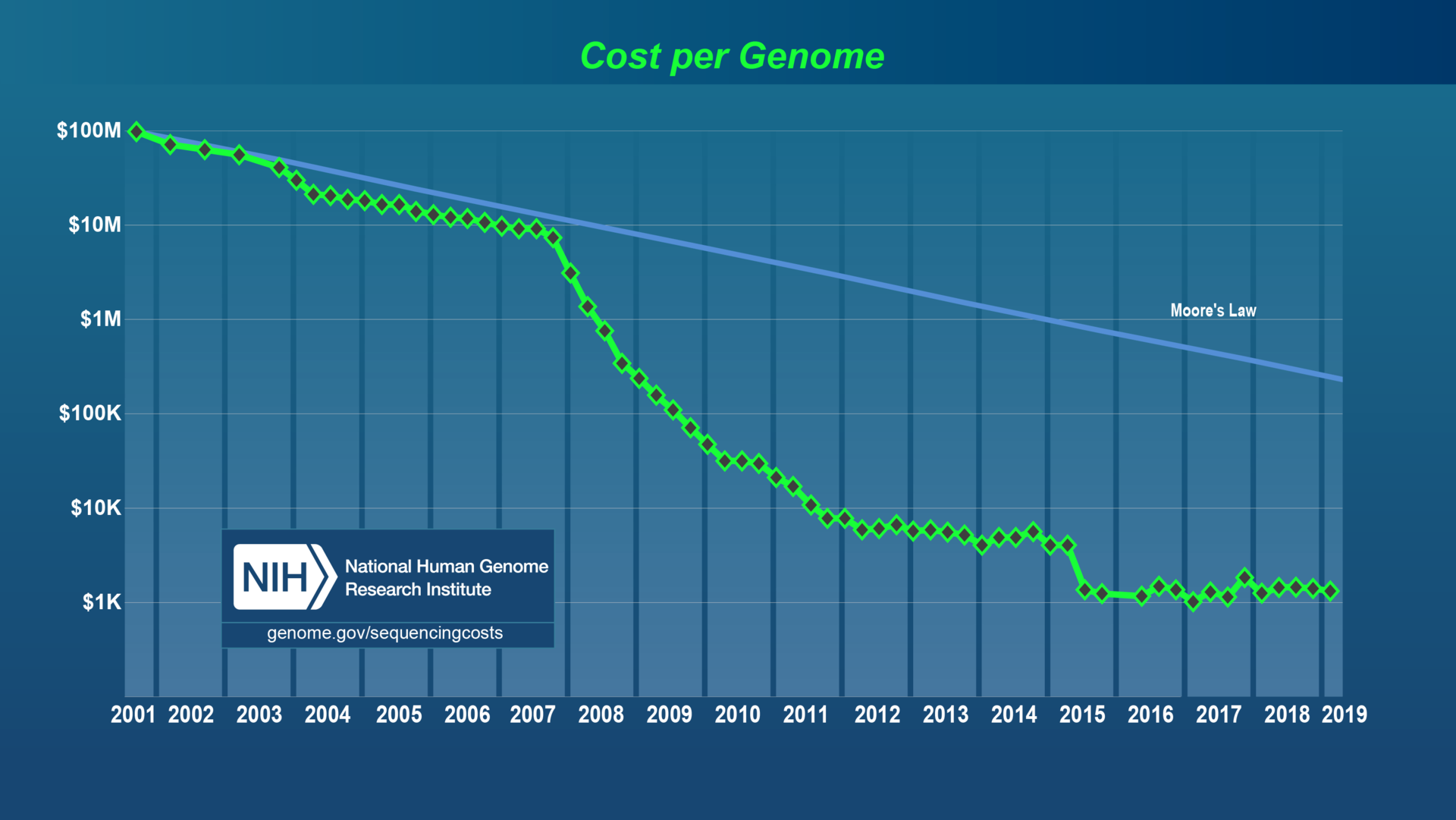
"Next Generation Sequencing" (NGS)
The era of cheap DNA sequencing
Early sequencing

An avalanche of data!
"Gee whiz!" era
"Arguing about methods" era
The era of arguing about methods

The era of arguing about methods

This gets tiring after a while (endless debates...)
- Intercalibration using standard reference material / mock communities so we know which methods really are best


My opinion: This will only end with intercalibration
But until this gets figured out, we should "move on with life"

"Gee whiz!" era
"Arguing about methods" era
"What does it all mean?" era
Let's enter the era of "what does it all mean?"

Currently = many microbial papers focus on describing community composition (who is there)
The era of "what does it all mean?"
Currently = many microbial papers focus on describing community composition (who is there)
The era of "what does it all mean?"


My view: Composition data = a map & compass!
The era of "what does it all mean?"




Schramm (2003) / Amann et al., 1995
"Full cycle rRNA analysis"

Sebastián & Gasol (2019), 10.1098/rstb.2019.0083
Single-cell activity assays
Classical techniques for context (microscopy, chemical assays, etc.)
The era of "what does it all mean?"
My view: Composition data = a map & compass!

- Generating sequencing data: easy
- Developing a good question: hard

A ‘good’ hypothesis has a number of desirable properties. It should be bold, risky and meaningful, addressing an important issue and not stating the obvious."
The era of "what does it all mean?"
Methods → Meaning
Ecosystem maps from eDNA

Ecosystem DNA
A Map


Metabarcoding
Vincent and Vardi, 2023
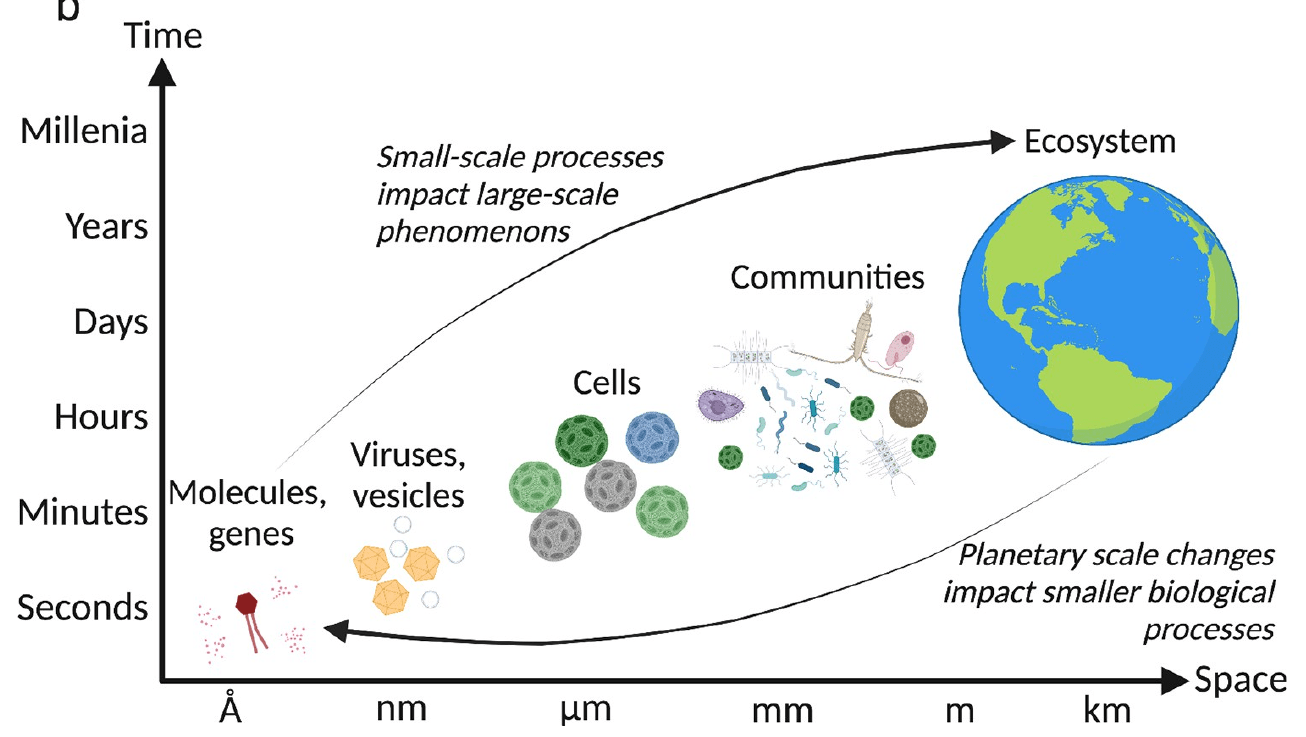
Ecosystem maps from eDNA → tackling big questions
Image credit: Chavez et al., 2021
Ecosystem maps with "eDNA assays" ( = amplicon sequencing = metabarcoding)
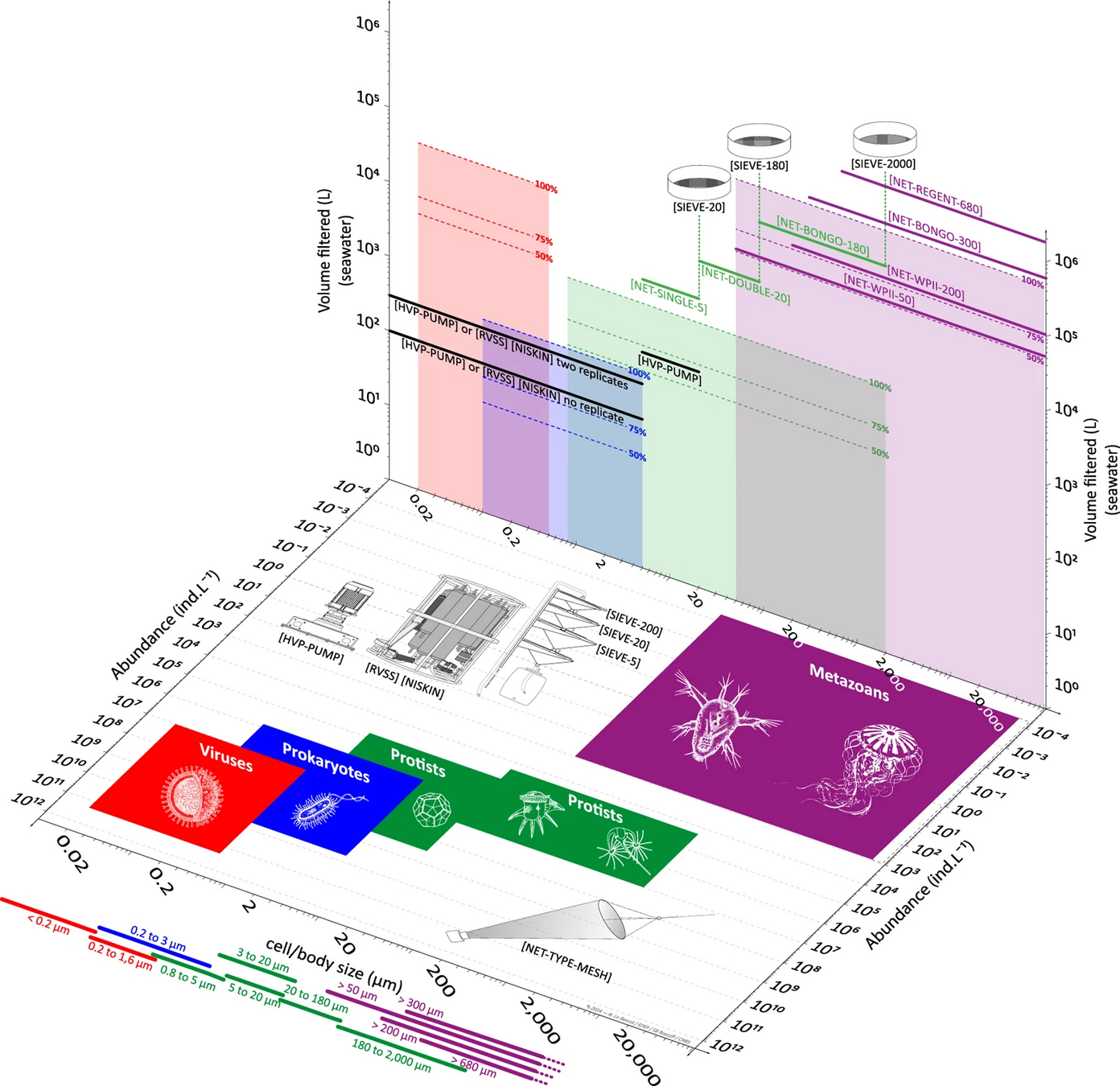
I don't size fractionate my samples (> 0.2 μm) because...

Microbe art: @claudia_traboni
Many size fractions complicate interpretation of metabarcoding data
Ecosystem maps from eDNA should be comprehensive
- Comprehensive ("Parada" primers target all rRNA from cellular life, including zooplankton & organelles)




Microbe art: @claudia_traboni
Ecosystem maps from eDNA should be comprehensive
plastid
16S rRNA
mito 16S
rRNA
nuclear 18S rRNA
A eukaryotic phytoplankter
Bacterium
16S rRNA
Archaeon 16S
rRNA
Bacteria and Archaea (prokaryotes)
mito 16S
rRNA
nuclear 18S rRNA
A eukaryotic protist or zooplankton cell
Ecosystem maps from eDNA should be comprehensive

p16S
e16S
18S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
Microbe art: @claudia_traboni
Sample x: dominated by prokaryotes (e.g. Sargasso Sea)

Ecosystem maps from eDNA should be comprehensive

Microbe art: @claudia_traboni
18S
p16S
e16S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
Sample y: dominated by eukaryotes (e.g. Southern Ocean)

Ecosystem maps from eDNA should be comprehensive
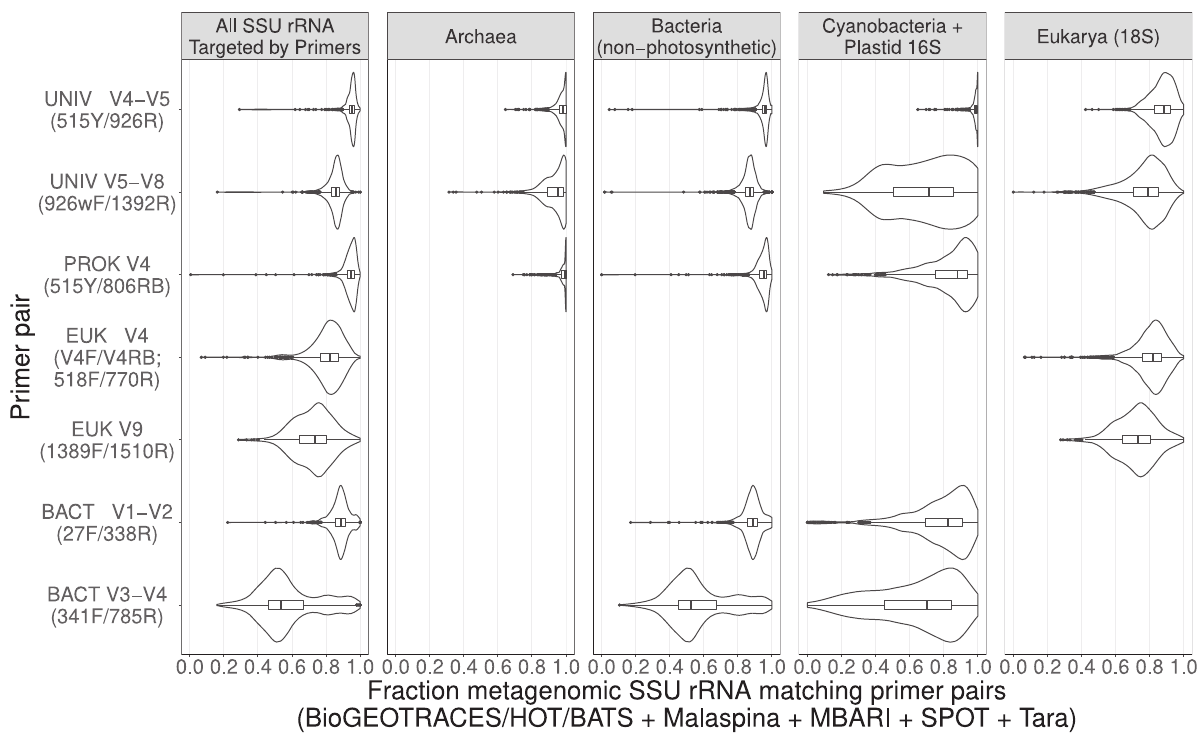









Ecosystem maps from eDNA should be comprehensive - validation with metagenomes
Image credit: Chavez et al., 2021
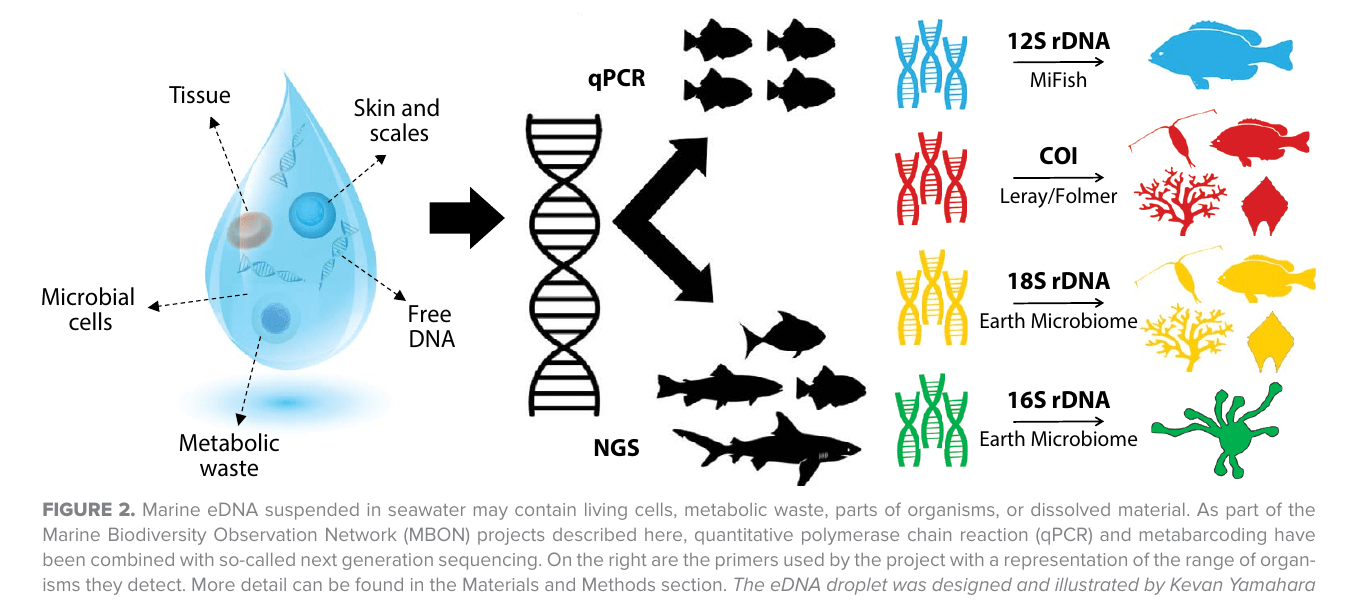
Ecosystem maps from "3 domain metabarcoding"- a mix of "eDNA" and "amplicon sequencing"

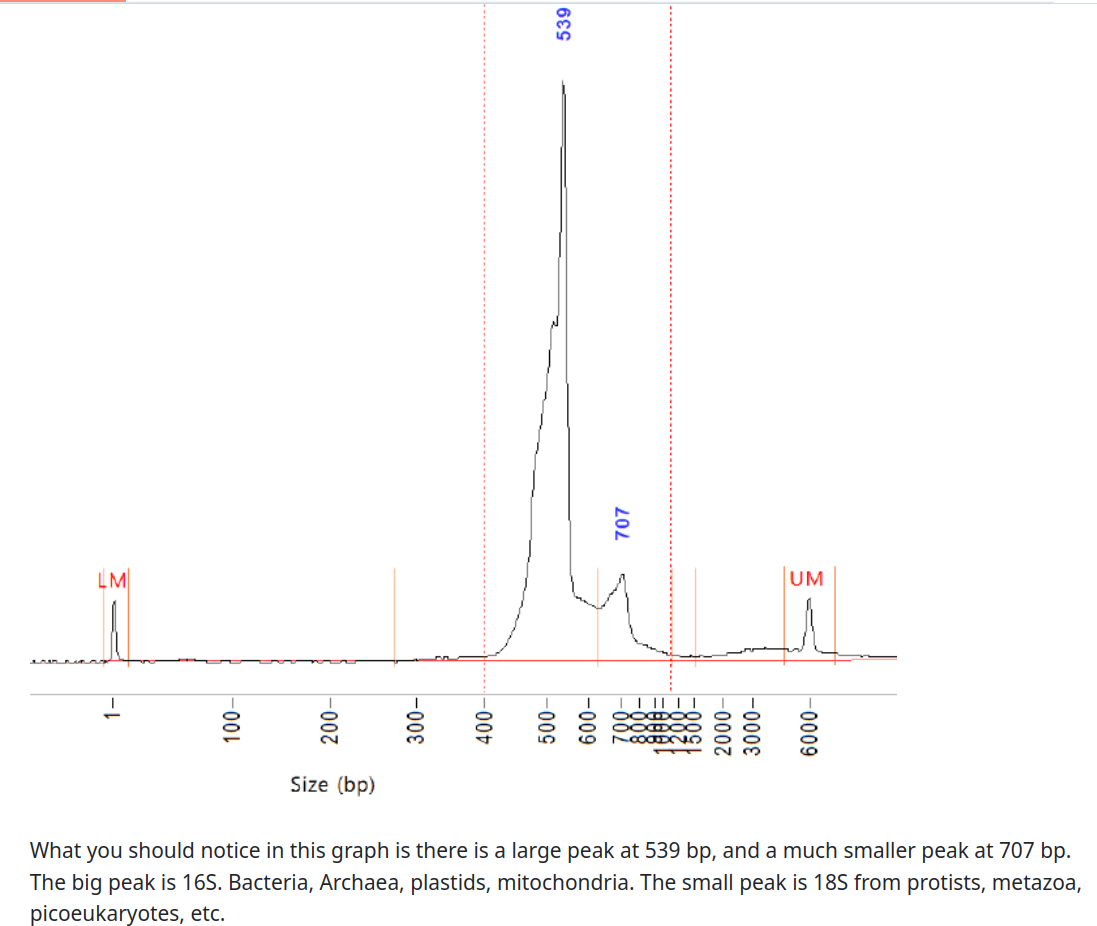
Ecosystem maps from "3 domain metabarcoding" - bioinformatic methods
Ecosystem maps can be cheaply generated from a "quiver" of DIY methods that I like to use
- Kit-free, non-toxic DNA extraction with linear acrylamide as co-precipitant, allows for recovery of very small amounts of DNA for PCR (~10 mL seawater*)
- Cheap, simple protocol for removal of PCR inhibitors with linear acrylamide (for difficult samples, Zymo 1-step is cheap & works)
- Protocols for decontamination of reusable plasticware for molecular protocols (reduce waste, guarantee quality)
- 1-step PCR reaction for library prep in lab (advantages = speed, lower cost, flexibility)
*Other methods for as little as 1 µL exist
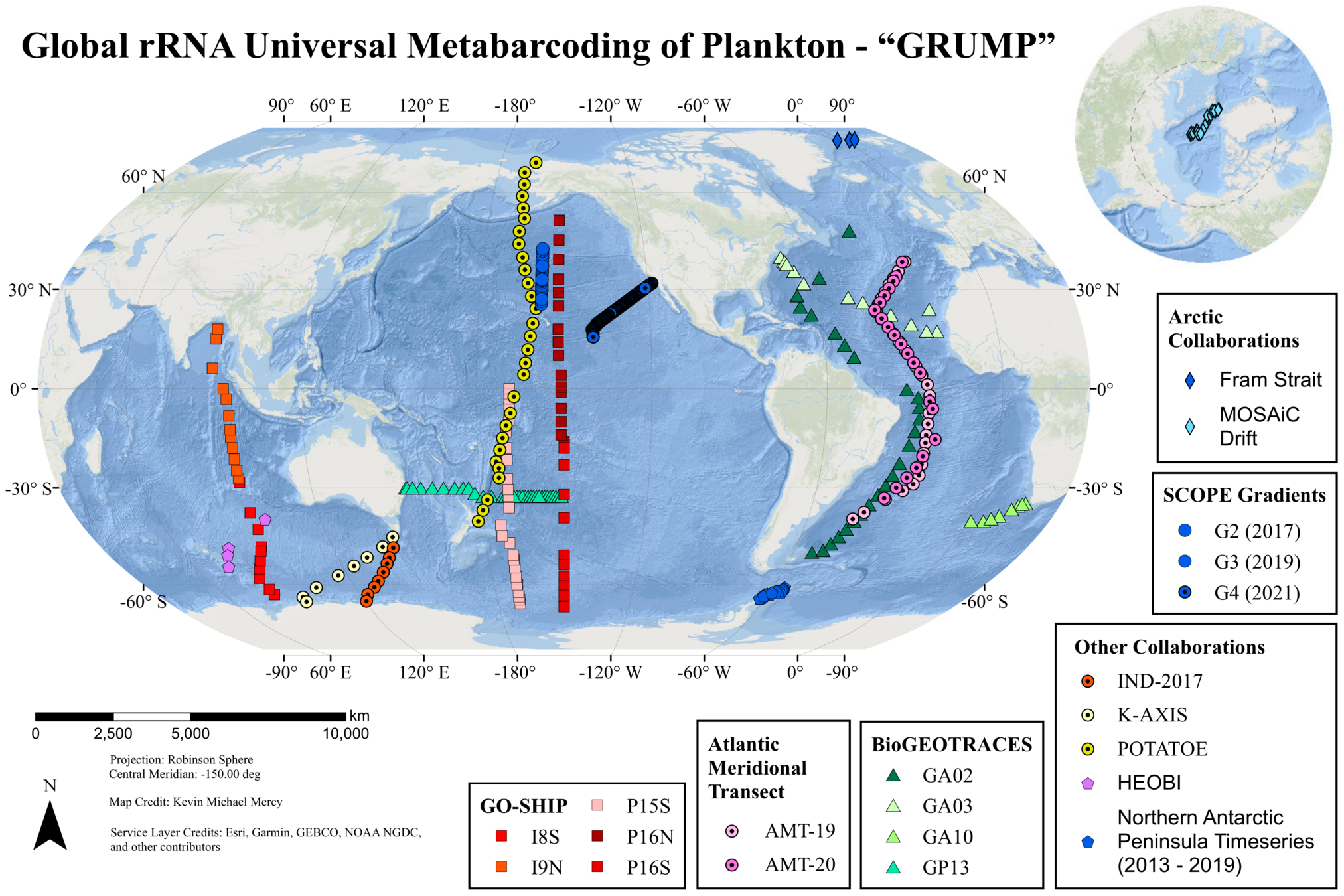
Application of 3DMB → GRUMP
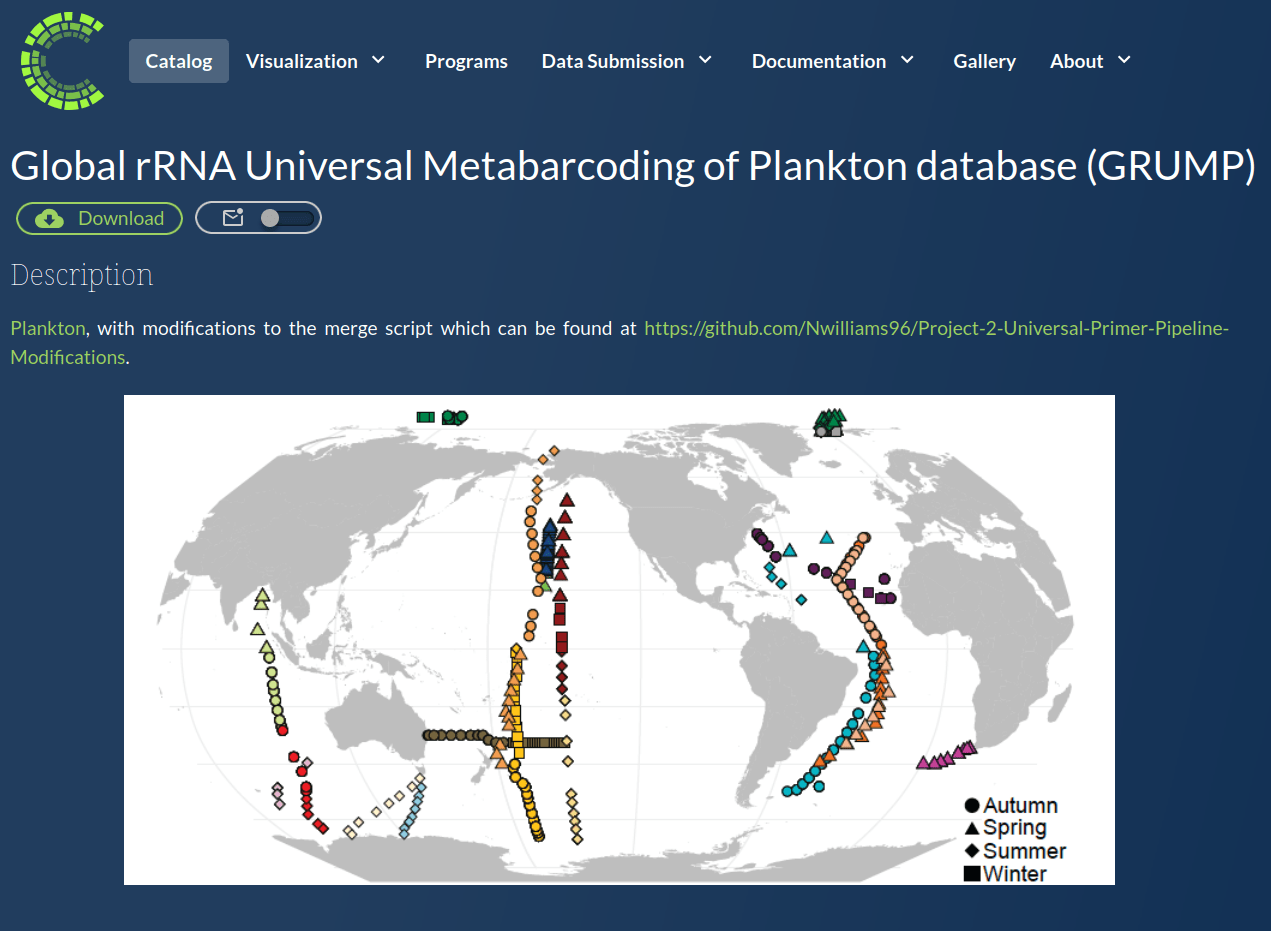

GRUMP = a global ecosystem map

GRUMP: No size fractionation

Microbe art: @claudia_traboni
TARA: Many size fractions (complicates interpretation)
GRUMP = a global ecosystem map... of whole seawater
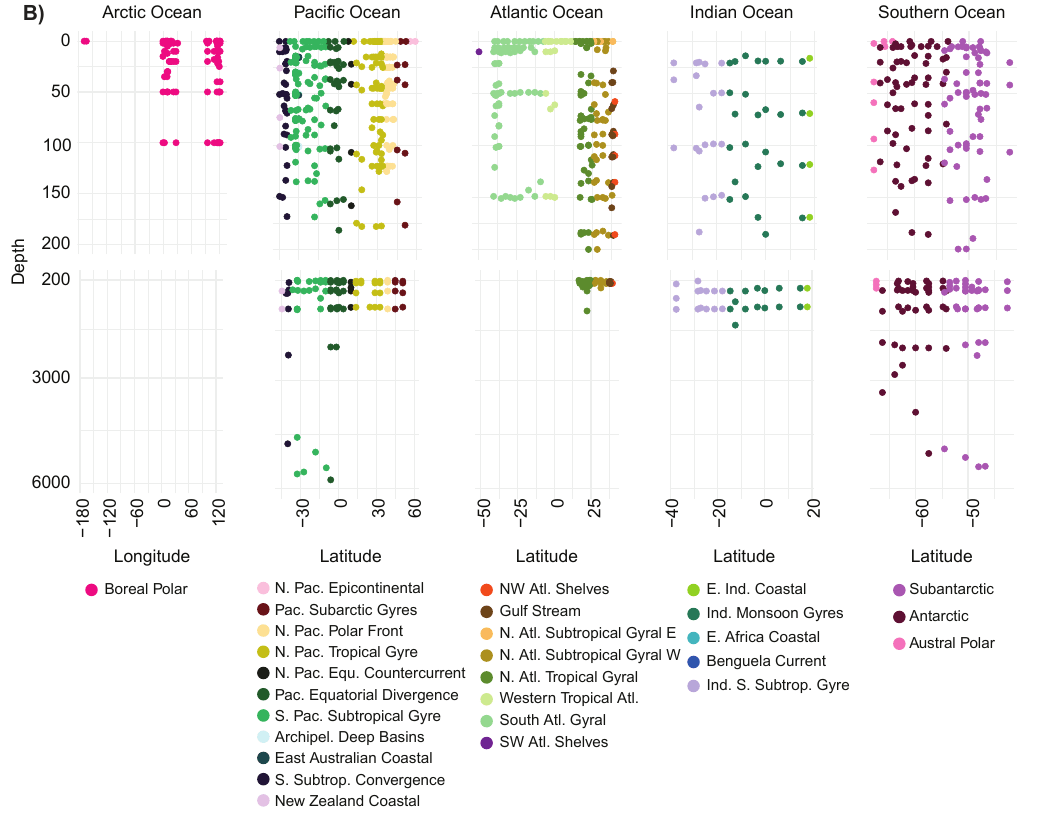
GRUMP = a global ecosystem map... across depth!
- Comprehensive ("Parada" primers target all rRNA from cellular life, including zooplankton & organelles)
- Sensitive with deep sequencing (~200,000 reads per sample)
- Specific with "denoising" algorithms (→ "ASVs")




Microbe art: @claudia_traboni

GRUMP = a global ecosystem map with many uses
With 200k sequences per sample, practical detection limit = 1 copy nifH / mL
nifH gene copies / L (x 106)
16S ASV relative abundance

UCYN-A (AMT 20)
Kendra Turk-Kubo, Rosie Gradoville, Jon Zehr, UCSC

AMT
Glen Tarran, Andy Rees, PML
GRUMP = a global ecosystem map... with good detection limits
Jesse McNichol, Nathan Williams, Yubin Raut, Craig Carlson, Elisa Halewood, Kendra Turk-Kubo, Jonathan Zehr, Andrew Rees, Glen Tarran, Mary Gradoville, Matthias Wietz, Christina Bienhold, Katja Metfies, Sinhué Torres-Valdés, Thomas Mock, Sarah Lena Eggers, Wade Jeffrey, Joseph Moss, Paul Berube, Steven Biller, Levente Bodrossy, Jodie Van De Kamp, Mark Brown, Swan Sow, E. Virginia Armbrust, Jed Fuhrman










AMT



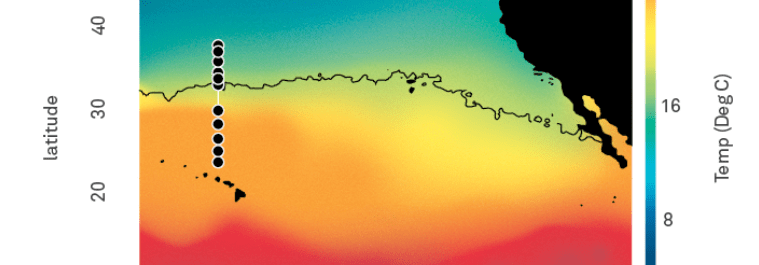

GRUMP = a global ecosystem map built on collaborations
- Nathan has assembled a core set of covariates common to all cruises in GRUMP (T, S, Oxygen, Nutrients, Chlorophyll)
- Also, Longhurst Provinces, Ocean Basin, Depth Categories, Predicted Euphotic Depth
- Also... some of the campaigns also have their own unique data products you may be interested in
Jesse McNichol, Nathan Williams, Yubin Raut, Craig Carlson, Elisa Halewood, Kendra Turk-Kubo, Jonathan Zehr, Andrew Rees, Glen Tarran, Mary Gradoville, Matthias Wietz, Christina Bienhold, Katja Metfies, Sinhué Torres-Valdés, Thomas Mock, Sarah Lena Eggers, Wade Jeffrey, Joseph Moss, Paul Berube, Steven Biller, Levente Bodrossy, Jodie Van De Kamp, Mark Brown, Swan Sow, E. Virginia Armbrust, Jed Fuhrman










AMT





GRUMP = a global ecosystem map with high-quality metadata


Ecologically-relevant annotations aggregate complex ASV data into sensible groupings (makes plotting, intercomparison easier)
- Bacterioplankton
-
Phytoplankton
- Prochlorococcus (ecotype)
McNichol, Williams, et al., 2025 (in revision)
GRUMP = a global ecosystem map with ecologically-relevant annotations
Vast majority of Prochlorococcus ASVs consistent with genome phylogeny and associated with a particular ecotype
*Berube et al. eLife 2019;8:e41043.



ASV hits to clade
GRUMP = a global ecosystem map with ecologically-relevant annotations
Yes, that means you!
- Ecosystem modellers
- Physical oceanographers
- Satellite ocean colour experts
- Math / stats nerds
- Fellow "gene jockeys"
- Other ocean enthusiasts
GRUMP is "live" and we want people to use it!

TO USE GRUMP DATA





What else can we do with these maps?
- Develop global models of heterotrophic prokaryotes!
A story about ecosystem maps + meta'omics + models
OK! I'm generating some data...


Let's collaborate! I'm building a model...
Emily Zakem, Carnegie
A story about ecosystem maps + meta'omics + models



Hey guys, can I join? I'm a stats / metagenome whiz
JL Weissman, Stony Brook
Emily Zakem, Carnegie
A story about ecosystem maps + meta'omics + models


Emily Zakem, Carnegie

JL Weissman, Stony Brook
Hey guys, I'm good at comparing models and data!

Yubin Raut, MIT

A story about ecosystem maps + meta'omics + models

- Street map = GRUMP data
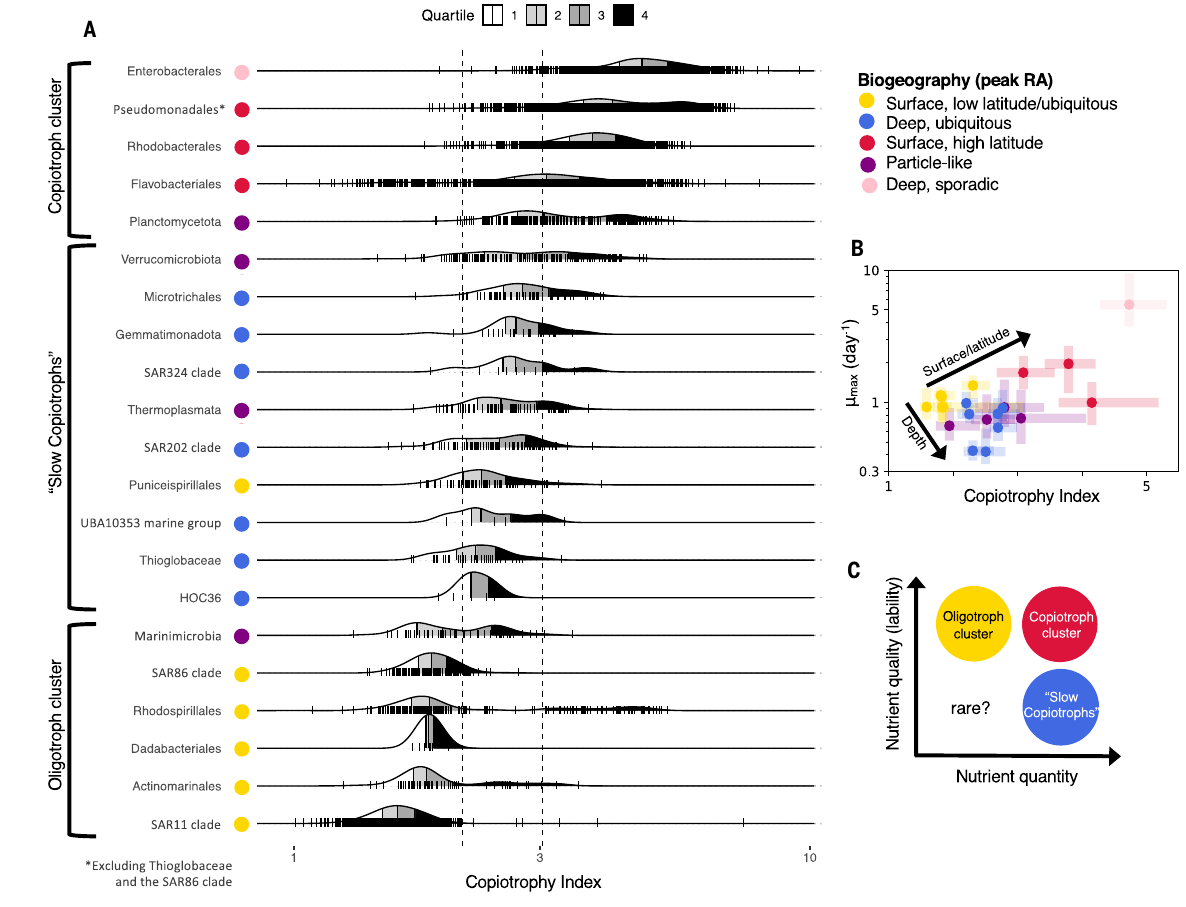
- Detail in between = MAG/SAG/genome data, derived from BLASTing @ 95% ID vs GTDB
- Genome size, growth rate, CAZymes → "index of copiotrophy"
- Compared with a biogeochemical model (DARWIN + heteroproks)
A story about ecosystem maps + meta'omics + models
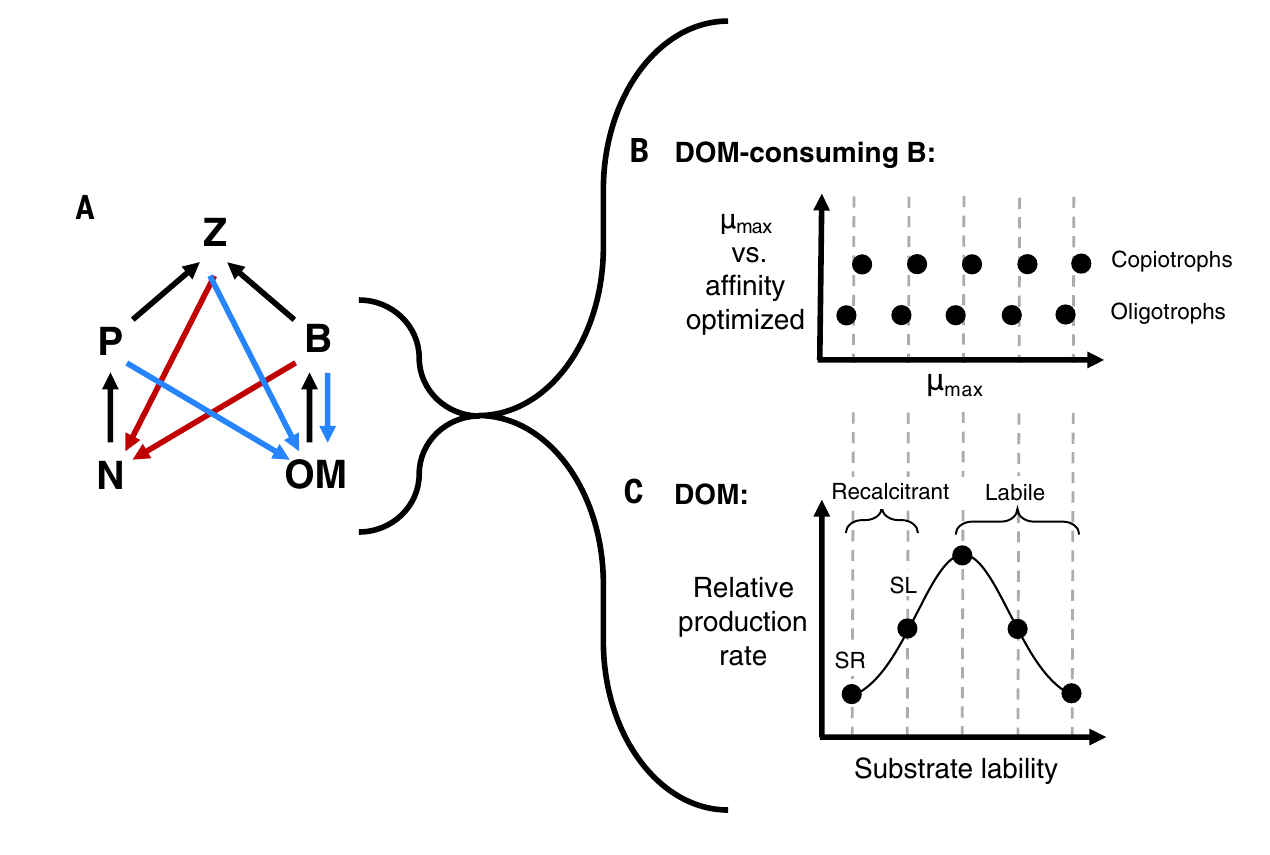


Emily's model:
DARWIN
+ "heteroprokaryotes"
+ DOM
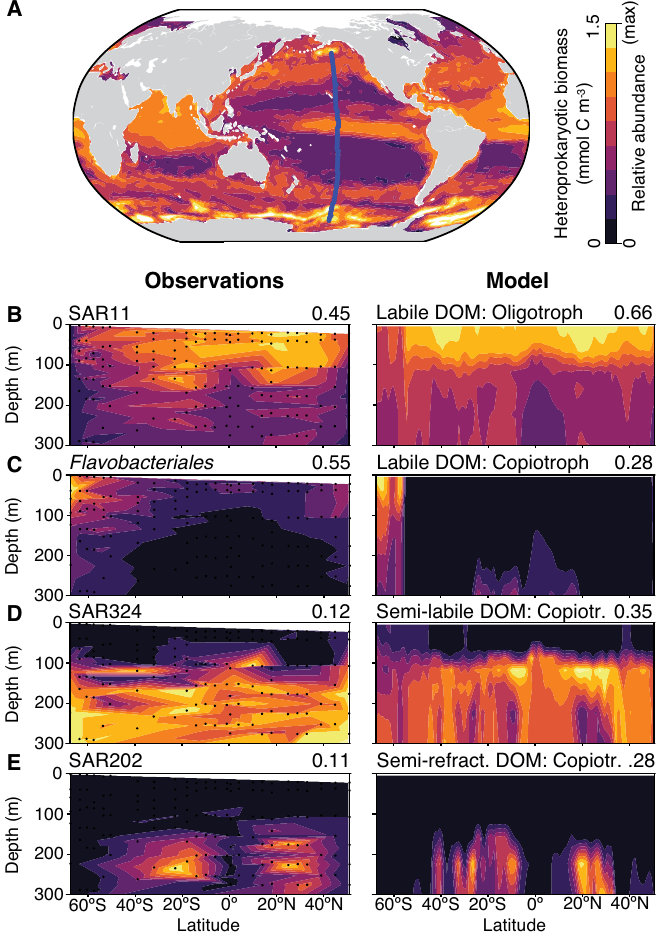
- Emily's model showed "emergent complexity" and had high qualitative correspondence to meta'omics data
- Analysis revealed insight into new metabolic classification in deep ocean → "slowiotrophs"
- Predictions of growth rates, could be tested experimentally
A story about ecosystem maps + meta'omics + models
A story about ecosystem maps + meta'omics + models





What else can we do with these maps?
- Make them quantitative to find subtle biological patterns
- Near perfect match between high-quality flow cytometry data and our eDNA quantification method with internal standards
- Workshop on internal standard methods with Dr. Julie Laroche, rescheduled date TBA
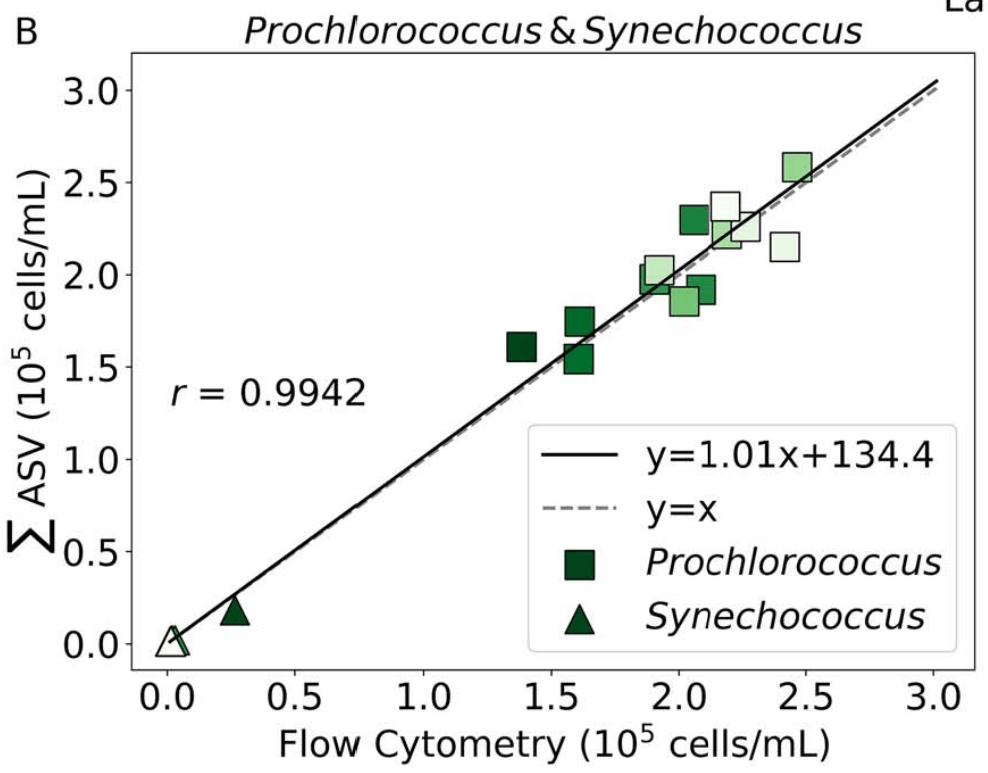
GRUMP 2.0 = a story about absolute units

GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)
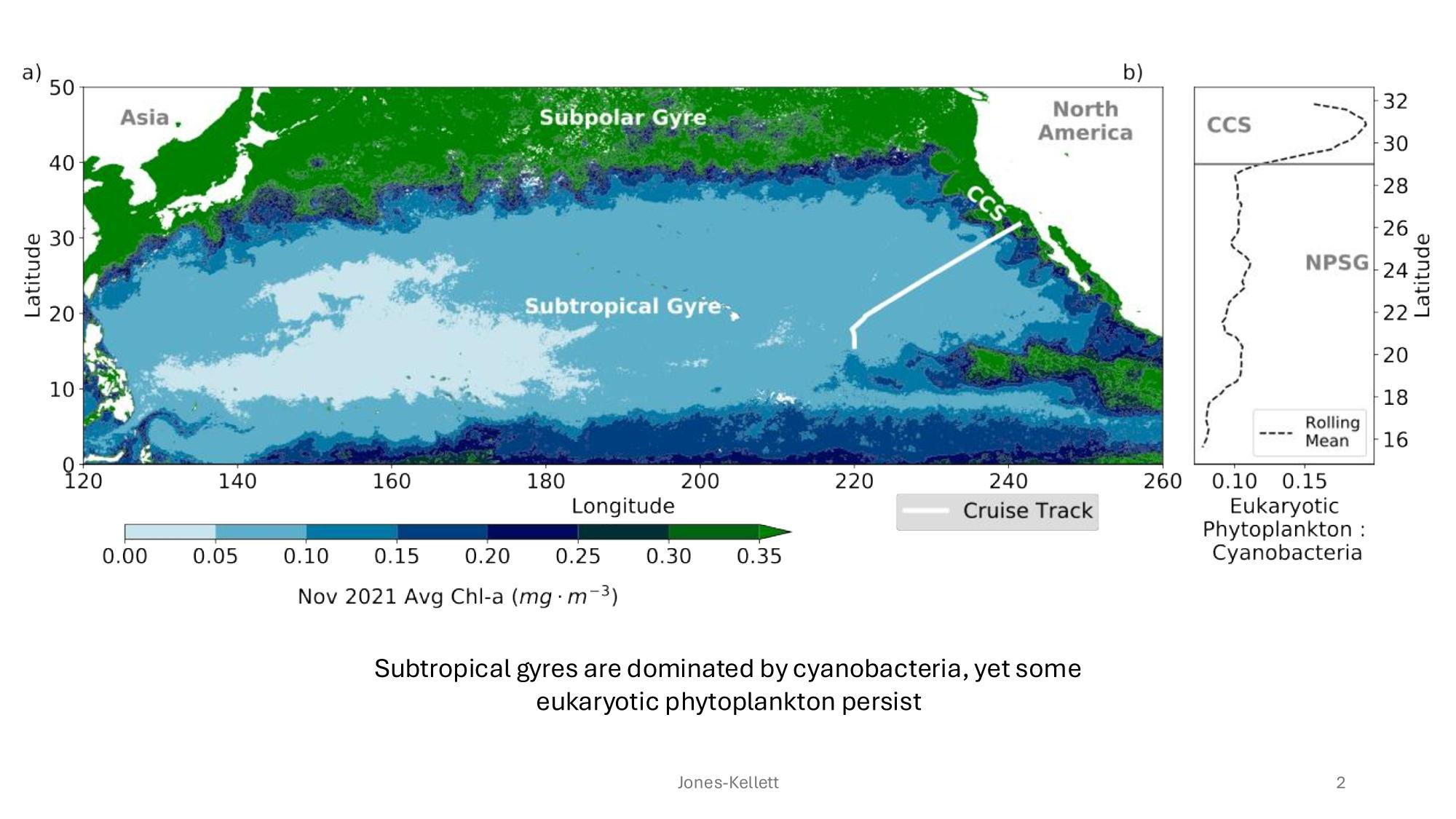
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)
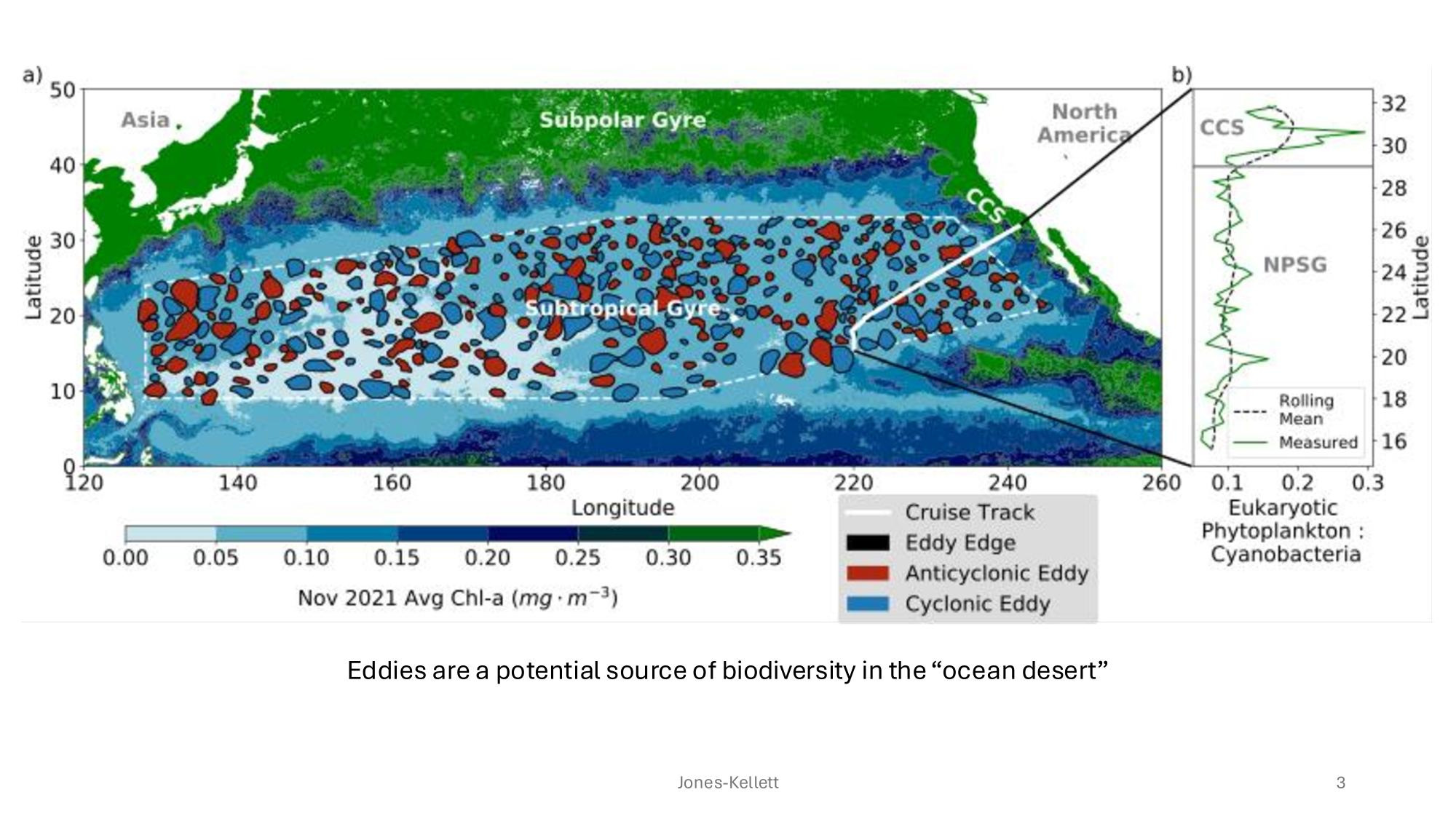
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)
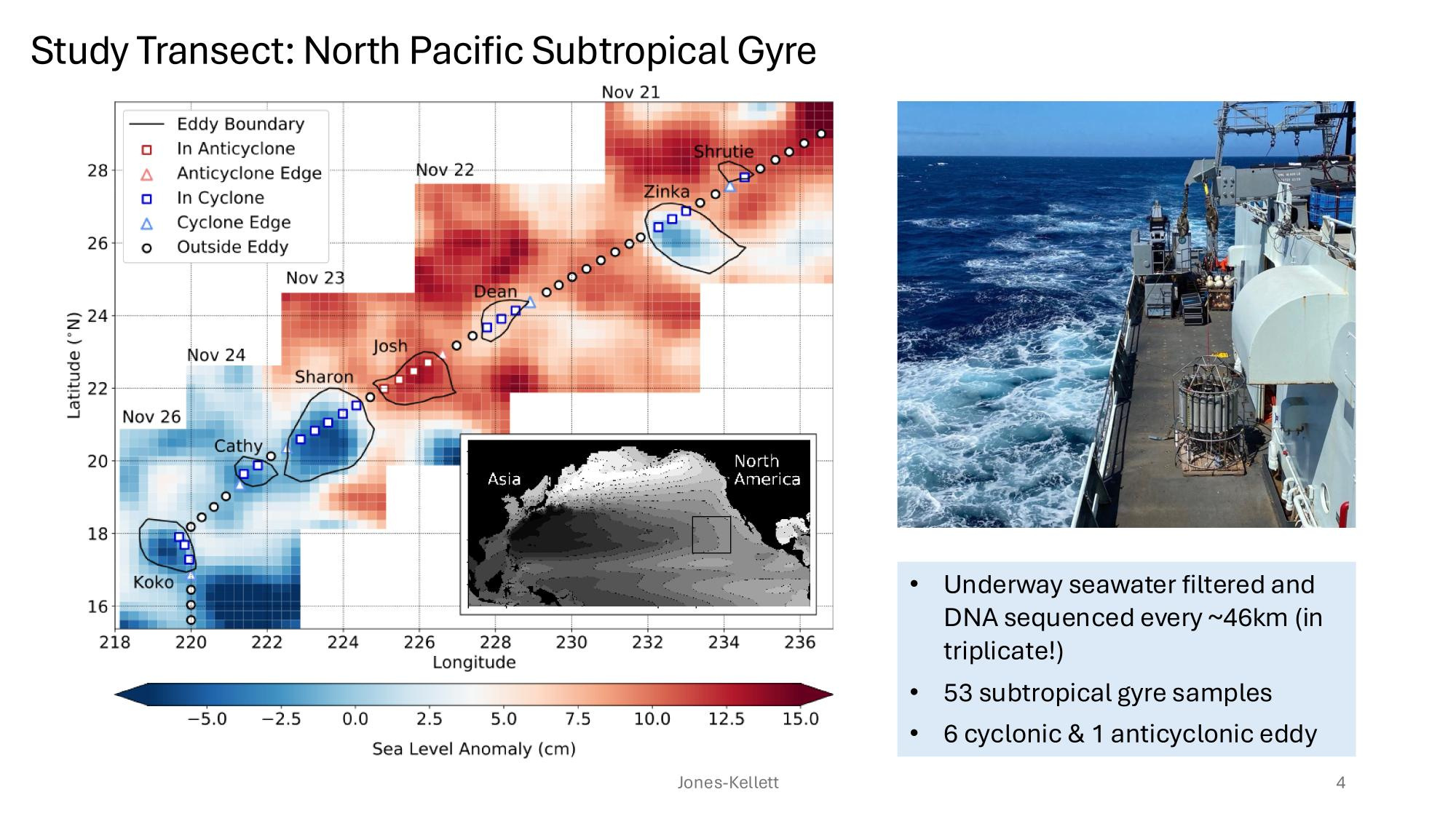
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)
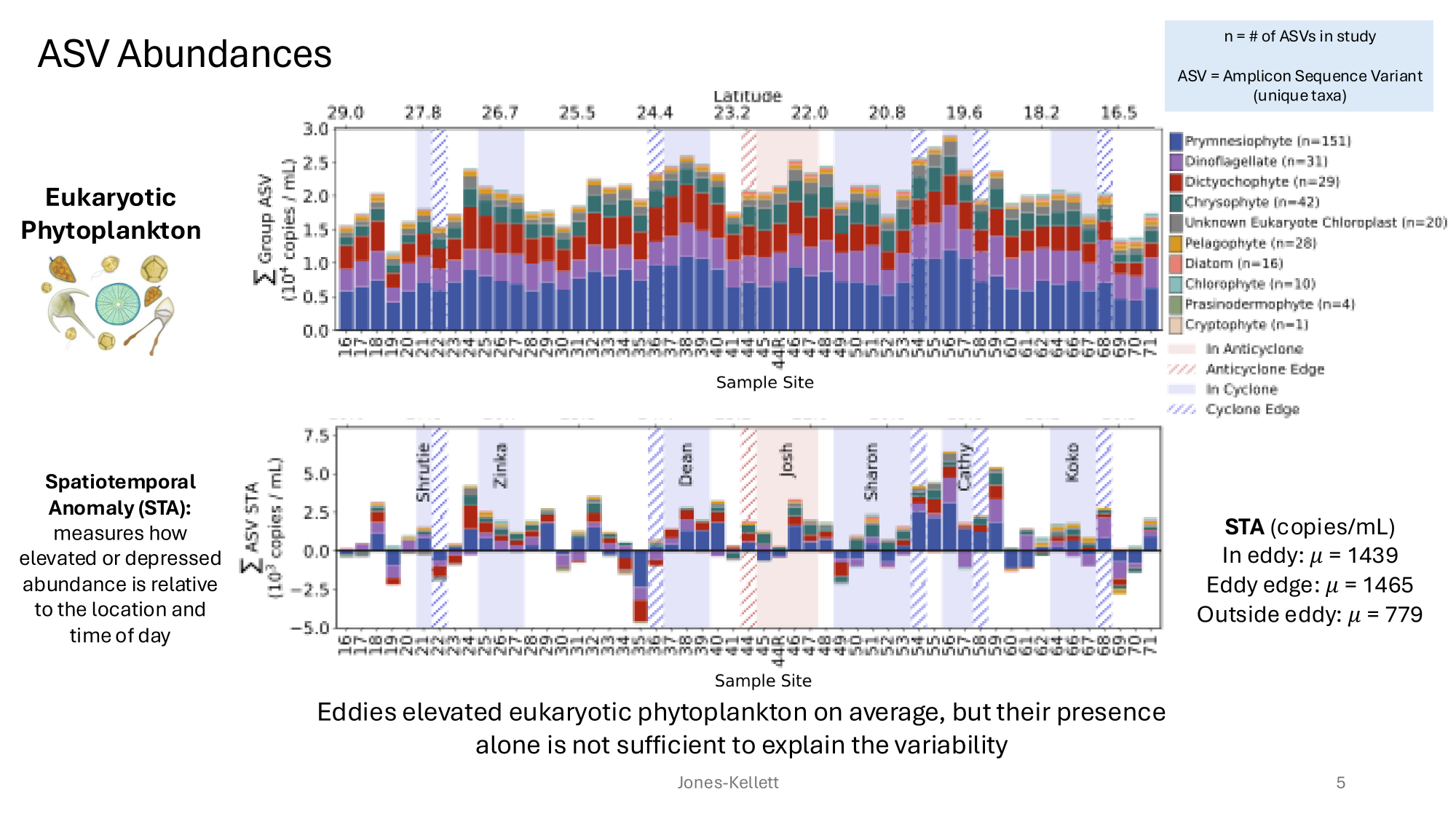
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)




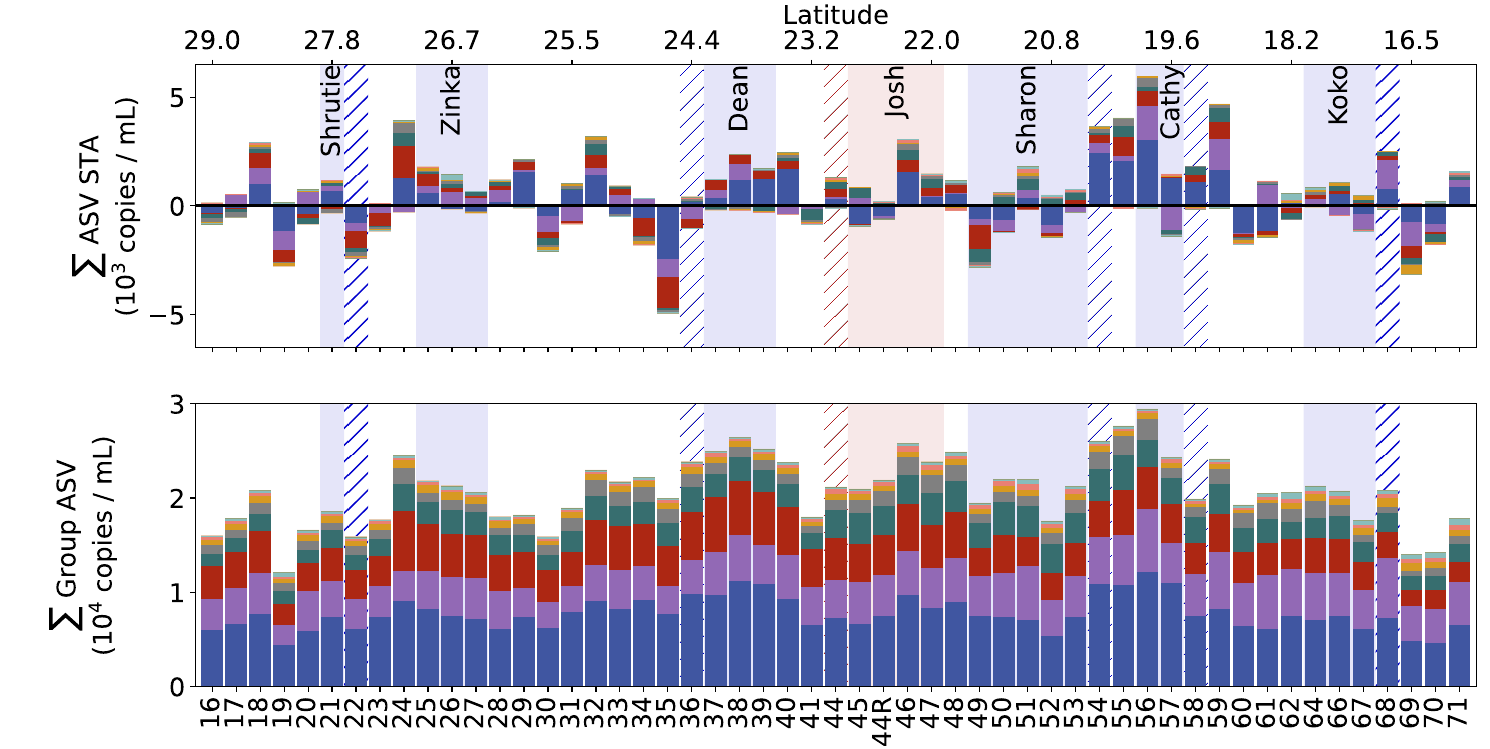






GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)
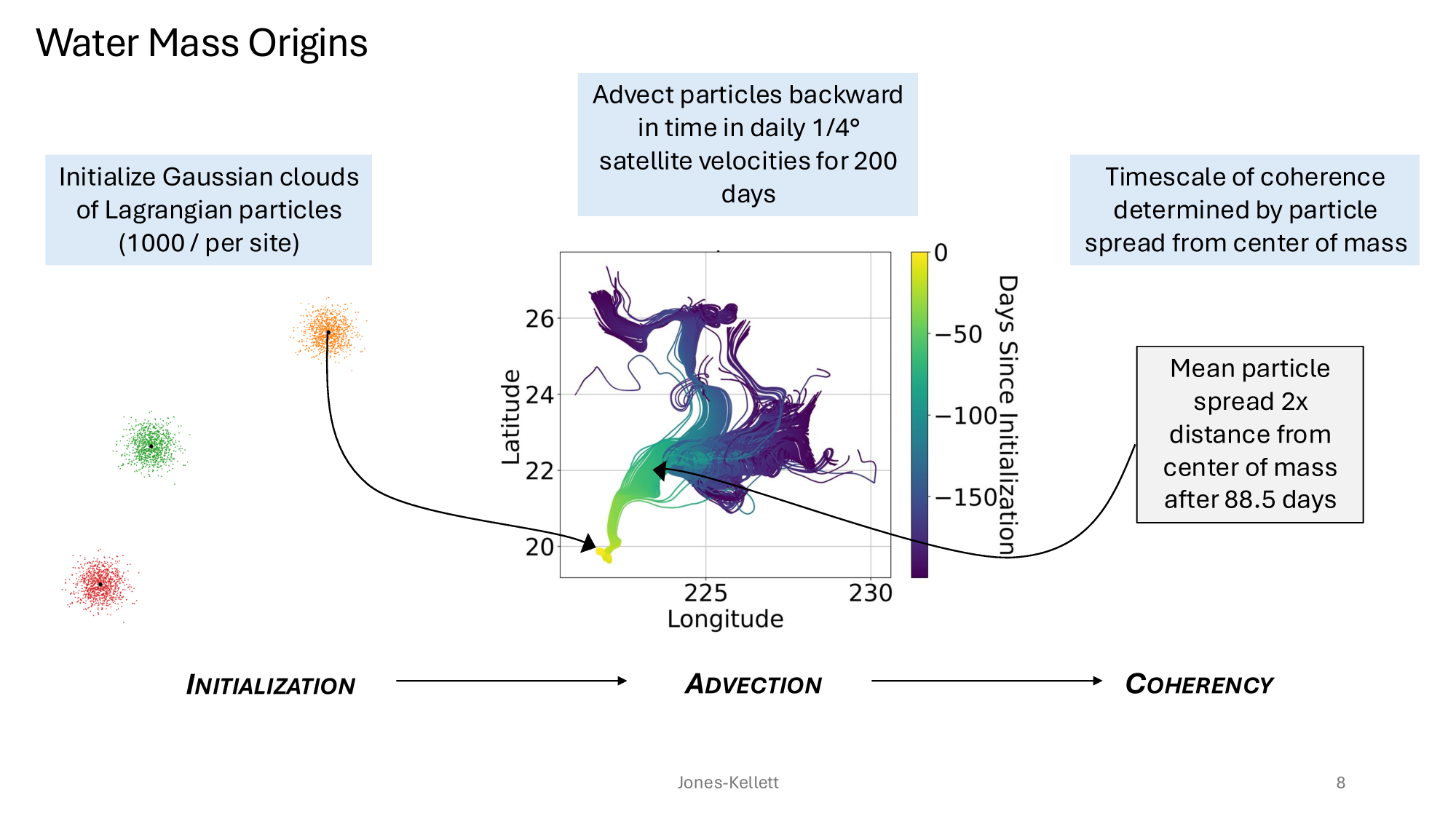
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)
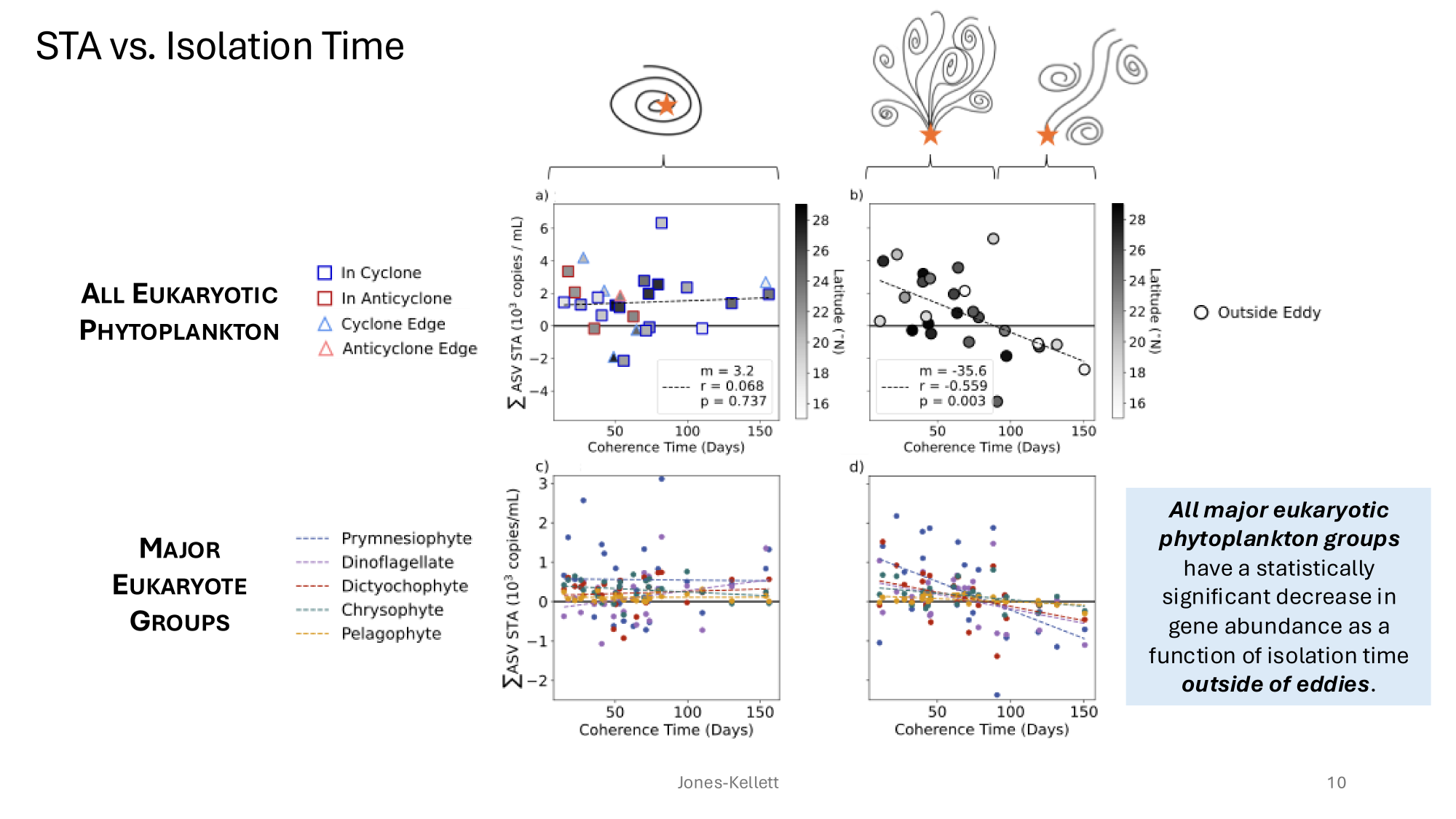
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)

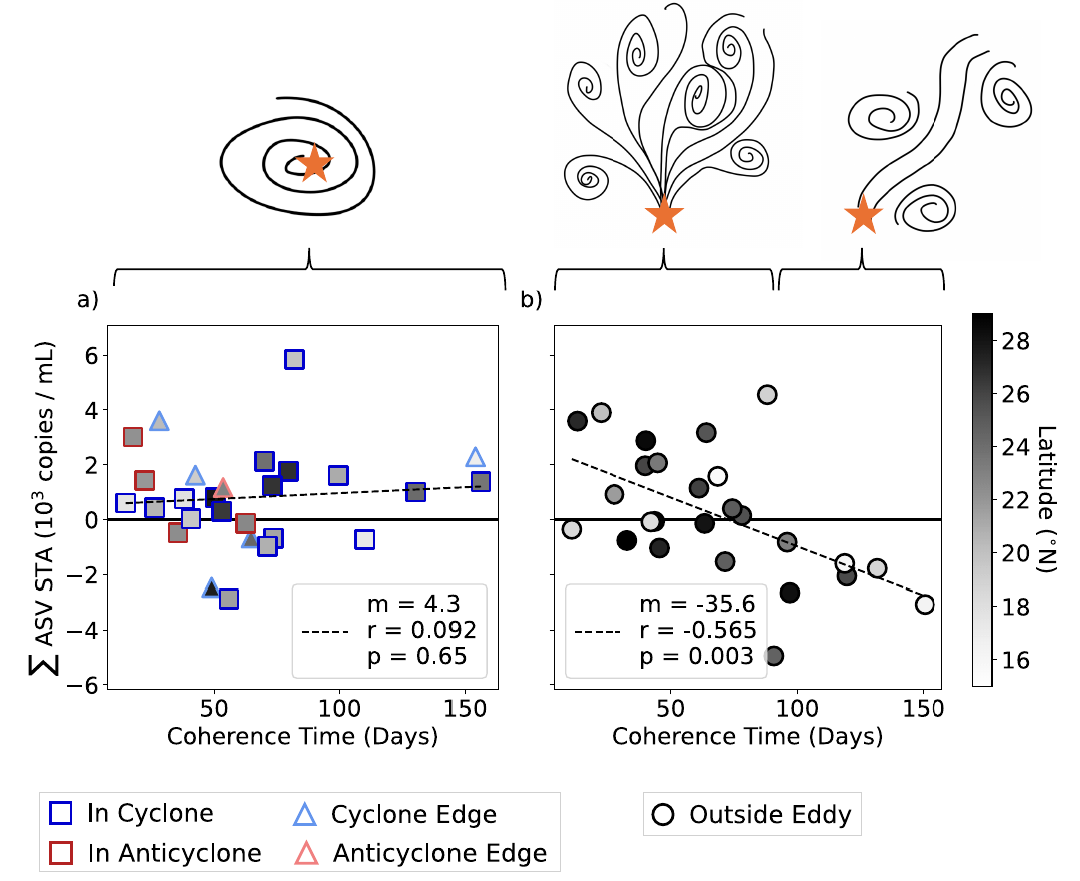




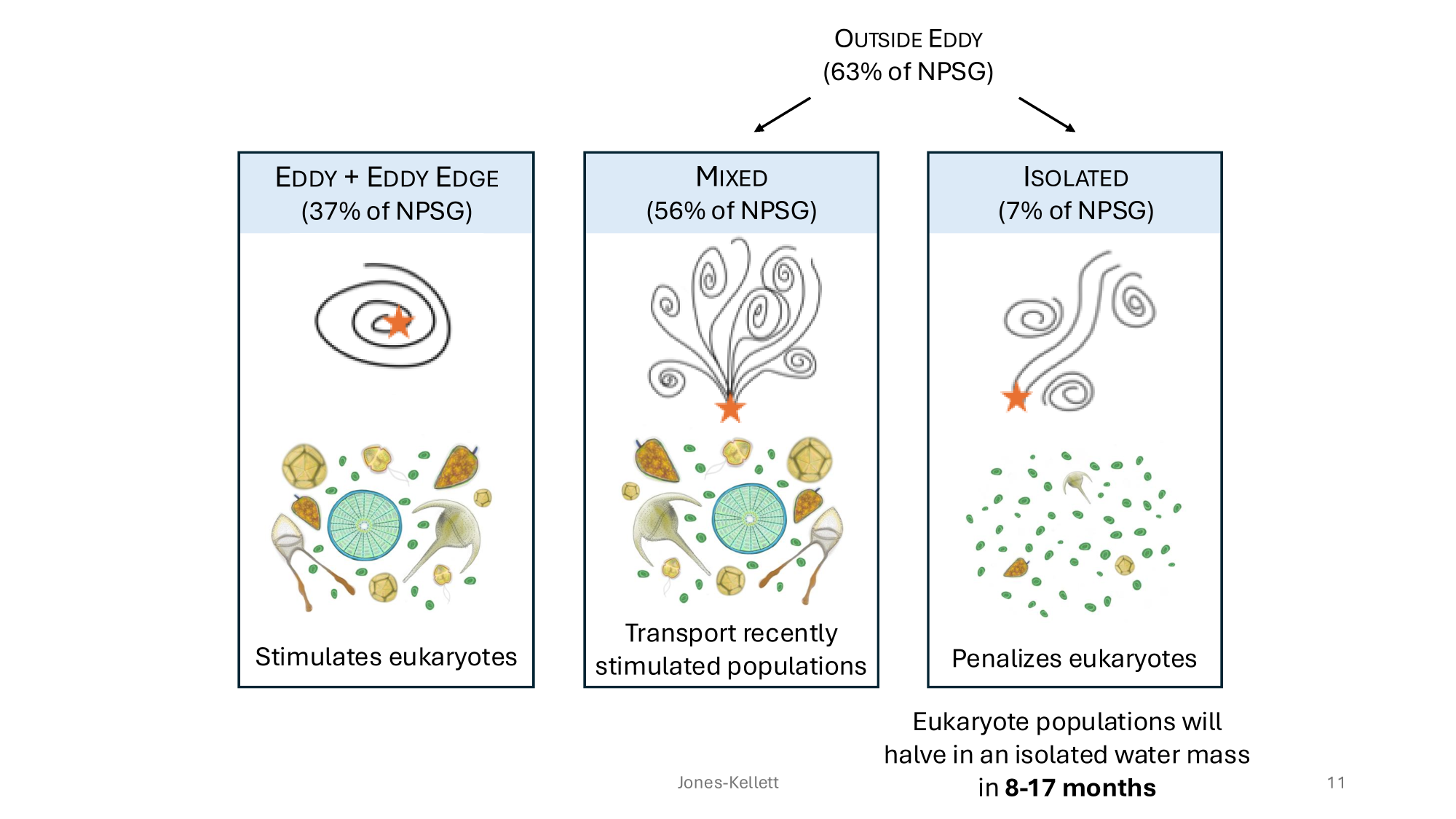
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)
Bringing it all together
- (What I'm doing in NS)

NASA PACE
Coastal systems are now my research focus
Bedford basin


Saanich inlet

SPOT (Los Angeles)


Minas basin
Coastal systems make great "model ecosystems"
A "model ecosystem" in Cape Breton
- Stratified basin with anoxic, sulfidic waters ~ 17 m (historically ~1000 μM H2S)
- Temperature and oxygen gradients very steep at redoxcline
- Beginning time-series (2024-present)
- With Katherine Rutherford (StFX) we are using metabarcoding, metagenomics to understand taxa present, dynamics, functional potential

Whycocomagh Bay (WB)
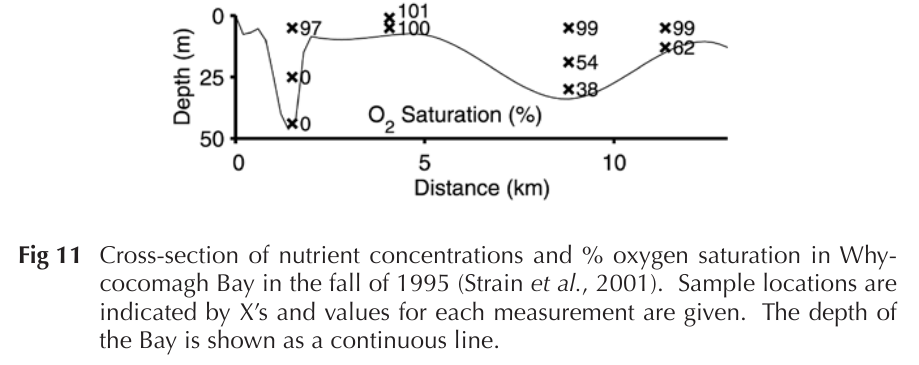
A "model ecosystem" in Cape Breton
A "model ecosystem" in Cape Breton


Me aboard Dr. Bruce Hatcher's research vessel Exocet in 2024
A "model ecosystem" to ask questions about the link between diversity and function


Microbes have a lot of genetic diversity, but we know little about the relationship between this diversity and function outside of a few model species (e.g. Prochlorococcus)
A "model ecosystem" to ask questions about the link between diversity and function
...it is unclear and controversial how multiple disturbances affect microbial community stability and what consequences this has for ecosystem functions."


Ecosystem
Ecosystem

Metagenomics
The fuzzy link between diversity and function
Work thus far in literature:
- If metagenomes show different organisms have similar metabolic pathways → ecosystem resilience
- My view: maybe, but this is "potential", not measured resilience
- Ignores other aspects of microbial niches (T optimum, for e.g.)
- In this ecosystem, sulfur oxidation is an "ecosystem service"
- "Microbial firewall"
- Prediction: stable stratification means limited functional diversity
- If disturbance (e.g. water overturning) occurs → limited resilience of function
- Goal: experiments to generate empirical data to test this prediction

Whycocomagh Bay (WB)

How resilient are microbial ecosystem services to disturbance?
Vincent and Vardi, 2023
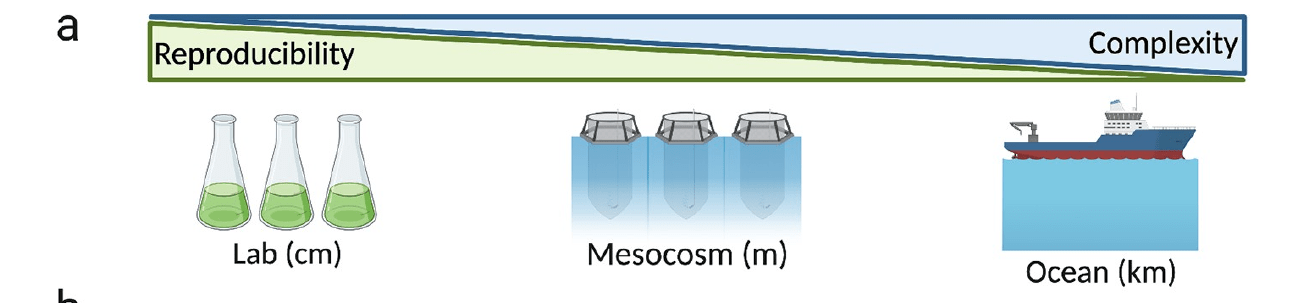
A "middle way" for understanding microdiversity
Pure cultures
Pros: Highly controlled
Cons: Selects for "lab rats", reduced diversity vs. environment
Environmental 'omics
Pros: Observe everything
Cons: Static measurement, limited functional information
Vincent and Vardi, 2023

A "middle way" for understanding microdiversity
Short-term incubation experiments of seawater
Pro: Generate empirical data for testing hypotheses
Con: The longer you incubate, the less the composition resembles the natural community. Need very short-term incubations, and single-cell activity measurements
A "middle way" for understanding microdiversity


In this paper, fluorescence in situ hybridization (FISH; targeted to rRNA) was used in combination with radiocarbon incubations to infer taxon-specific rates and response to different chemical conditions in short-term incubations
How we plan to do the work
[H2S]
[O2]
depth
EUX ~3°C
RXL ~10°C
SRF ~22°C
Field sampling
DNA
-3DMB
-MAGs
-FL-16S
Lab processing
OOI
(function,
microdiversity)
OOI-specific measurements:
-Growth rate
-Respiration
-Resilience
Field incubations
FISH probes
(broad group level,
microdiverse subcluster level)

Katherine Rutherford (StFX)

Bruce Hatcher (CBU)


- Street map = meta'omics data from Whycocomagh (which organisms are involved in sulfur cycling)
- Allows us to design FISH probes for particular organisms (including microdiversity within clade)
- FISH probes allows us to derive:
- Cell size, biovolume, morphological variability
- In situ growth rates
- Metabolic rates during incubations
How we plan to do the work
How we plan to do the work: in situ growth rates
FISH probes + FODC method → OOI growth rate (including microdiversity)
Could be compared with MAG estimates (gRodon)
How we plan to do the work: incubations
[H2S]
[O2]
depth
EUX ~3°C
RXL ~10°C
SRF ~22°C
Simulate upwelling to surface by subjecting RXL or SRF communities to increased [H2S], temp, [O2] (or combinations of all) by mixing at different ratios and incubating at controlled temperatures for ~6-12 h. Response (compared to control) measured by:
- Single-cell respiration rates
- Bulk H2S oxidation
- Bulk community respiration

in situ conditions (control)

"disturbance" conditions
How we plan to do the work: organism-specific respiration
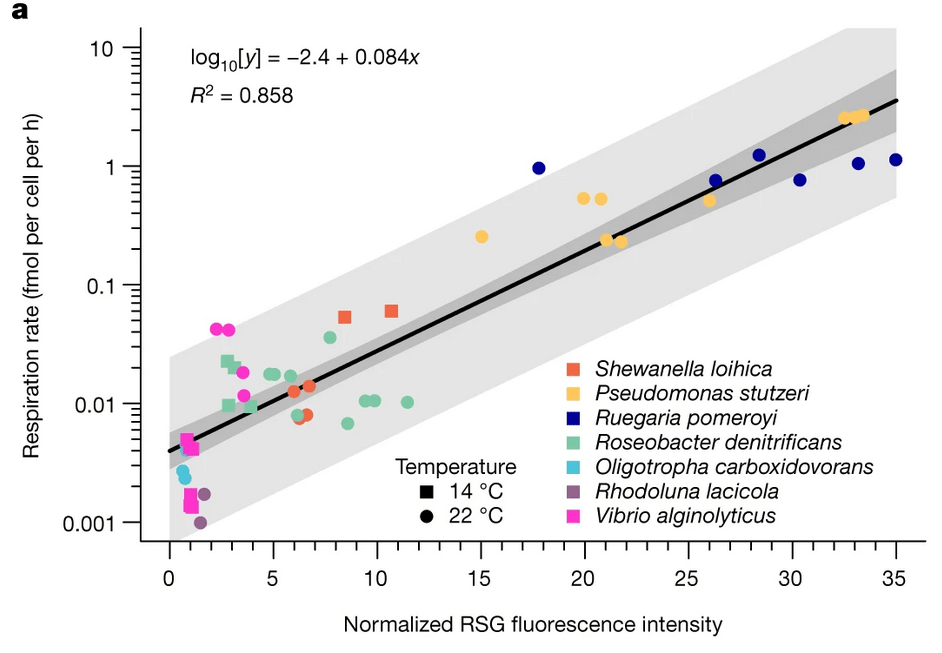
FISH probes + Redox Sensor Green → OOI respiration rate changes after short-term incubations

Microbial life



(Micro)
Kump, Kasting, and Crane, The Earth System
What effect will anthropogenic pressures such as climate change and coastal eutrophication have on microbial "ecosystem services"?
STAY TUNED over the next 2-3 years!
Questions?
Thanks to:
- Current and former CBIOMES collaborators (Fuhrman, Follows, Levine, Zakem labs)
- Dr. Bruce Hatcher (CBU)
- Katherine Rutherford and Jillian Davies (StFX)
- All of you for listening!


stfxmicroeco.ca | jmcnicho@stfx.ca
https://slides.com/jcmcnch/icg-dal-2025



Figure 1: Overview of sampling and data processing workflow. Oxygen and sulfide depth profiles shown in blue and orange, respectively. SRF=surface, RXL=redoxcline, EUX=euxinic. 3DMB=3-domain metabarcoding, MAGs = metagenome assembled genomes, FL-16S = full-length 16S. OOI = organism of interest. FISH = fluorescence in-situ hybridzation.
[H2S]
[O2]
depth
EUX ~3°C
RXL ~10°C
SRF ~22°C
DNA
-3DMB
-MAGs
-FL-16S
OOI
(function,
microdiversity)
(broad group level,
microdiverse subcluster level)
FISH probes
OOI-specific measurements:
-Growth rate
-Respiration
-Resilience
Field sampling
Lab processing
Field incubations

Vincent and Vardi, 2023
Coastal systems are flexible: experiment across scales, depending on question
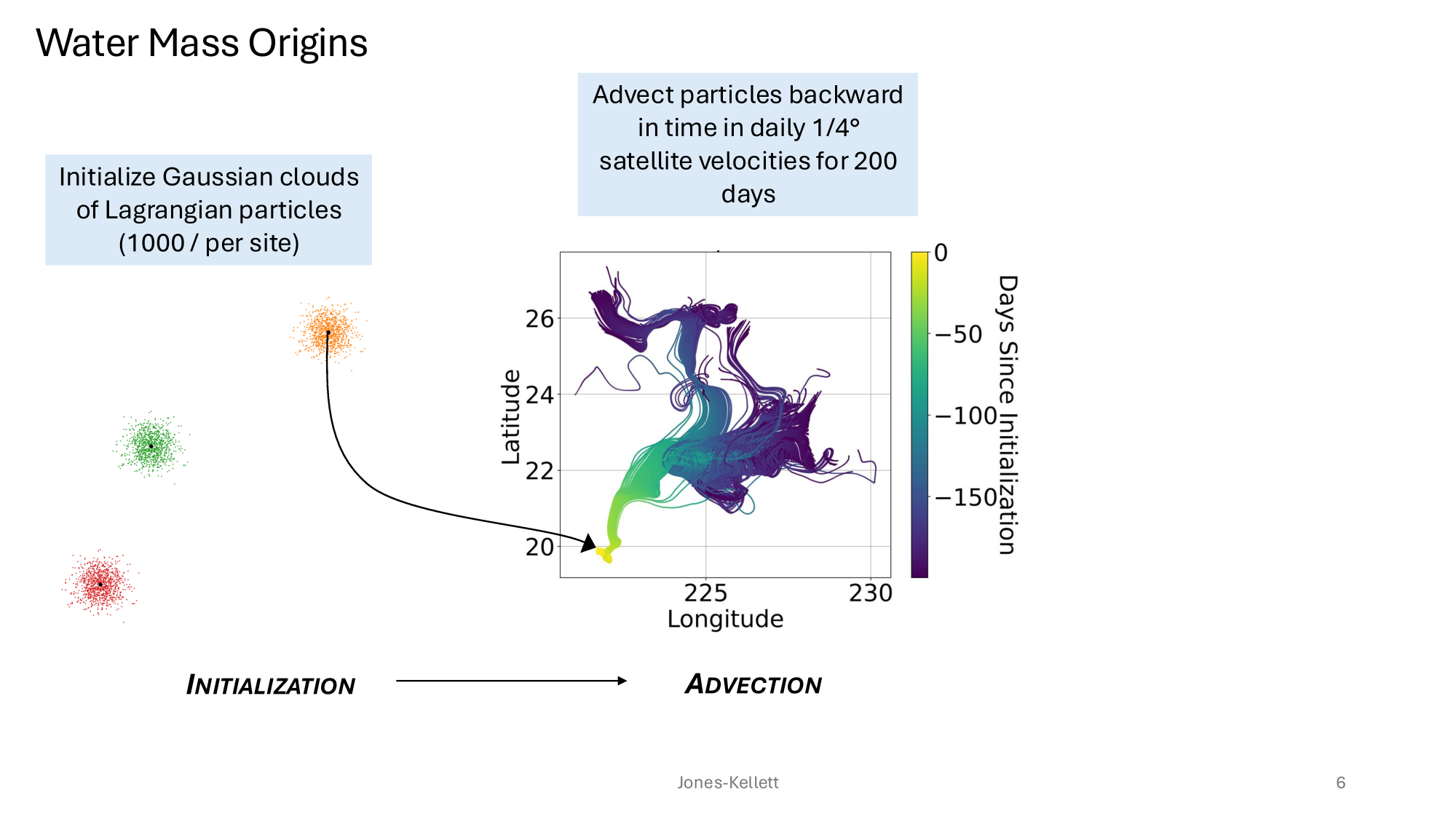
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)
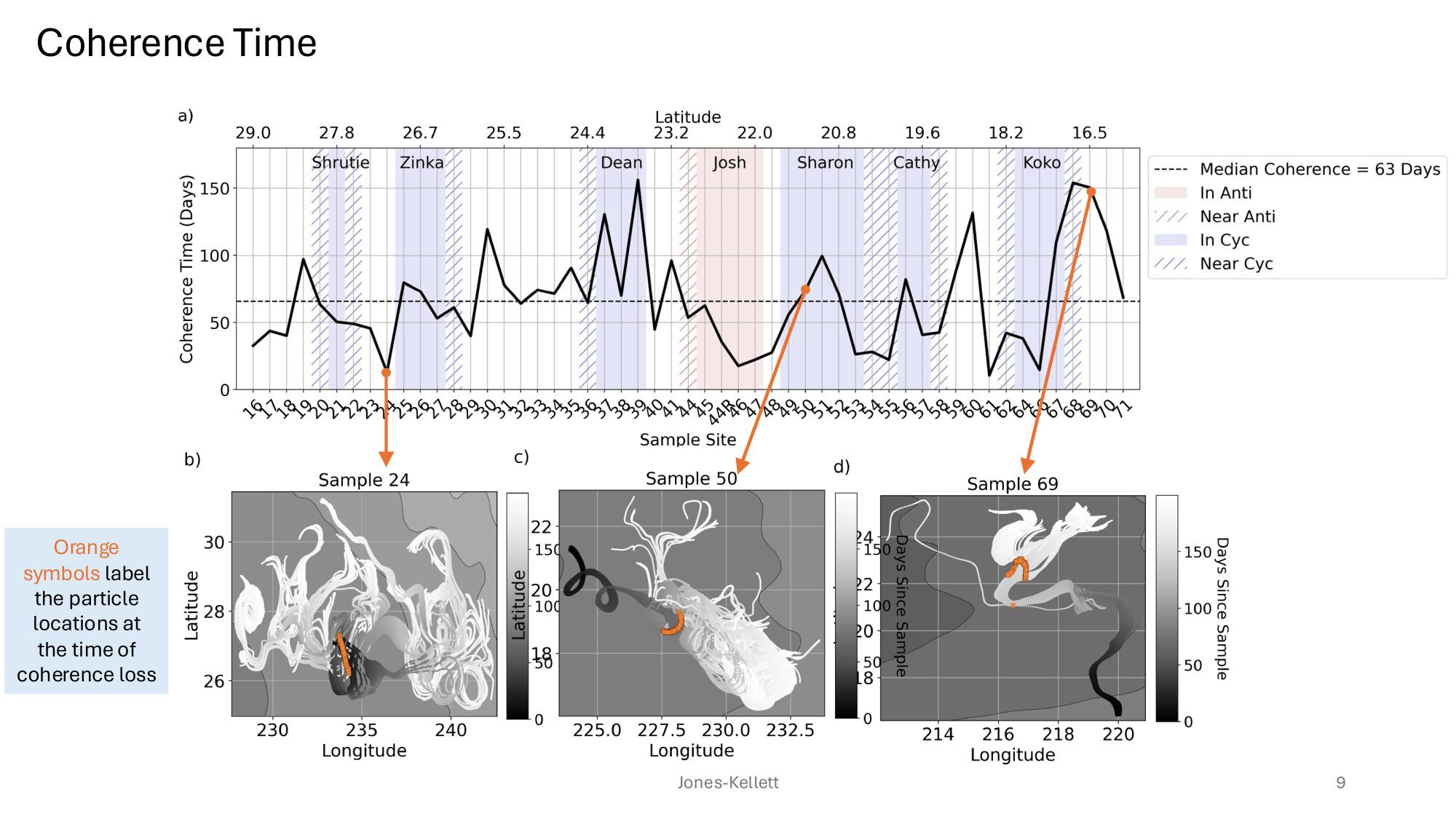
GRUMP 2.0 = a story about absolute units
Slide credits: Dr. Lexi Jones-Kellett (aejk@alum.mit.edu)