Ecosystem-centered microbial
ecology & biogeochemistry
Biology Department, StFX (Apr 4th, 2022)
Jesse McNichol (he/him) : PhD, Biological Oceanography
Postdoctoral Scholar, University of Southern California




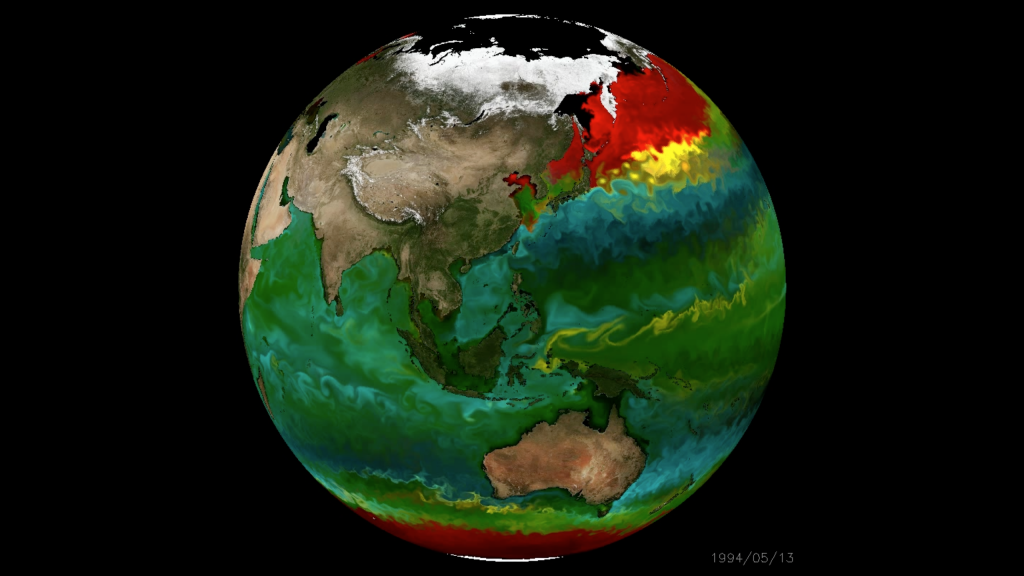
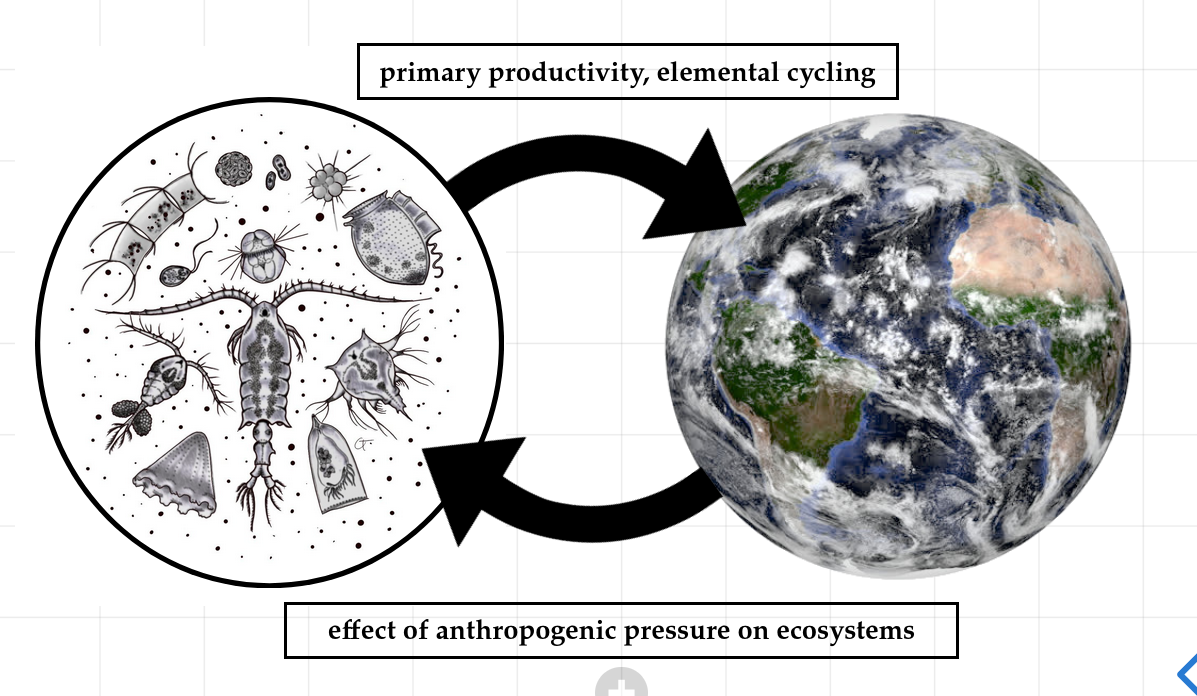
How do microbial communities* influence the Earth System**,
and vice-versa***?

*whole community methods

**primary productivity, elemental cycling

***effect of anthropogenic pressure on ecosystems
Microbe art: @claudia_traboni
My core research question
Microbiology
Environmental science
Bioinformatics
Evolution, population genetics
Ecosystem modelling, global change biology
Microbial ecology, biogeochemistry
3 disciplinary "pillars"
Microbiology
Bioinformatics
How do microbial communities influence the Earth System, and vice-versa?

Pillars support core research question
Environmental science
Long term vision: A lab that links microbial diversity & biogeochemical function in the context of global ecosystem change with modern 'omics-enabled techniques
'omics enables my research questions
-
Which microbes catalyze specific bio(geo)chemical processes?
- What proteins / pathways are responsible?
- What are the rates / efficiency of these processes?
- What is the structure & function of the whole ecosystem, and how is it changing?
- How does speciation / evolution affect these processes?
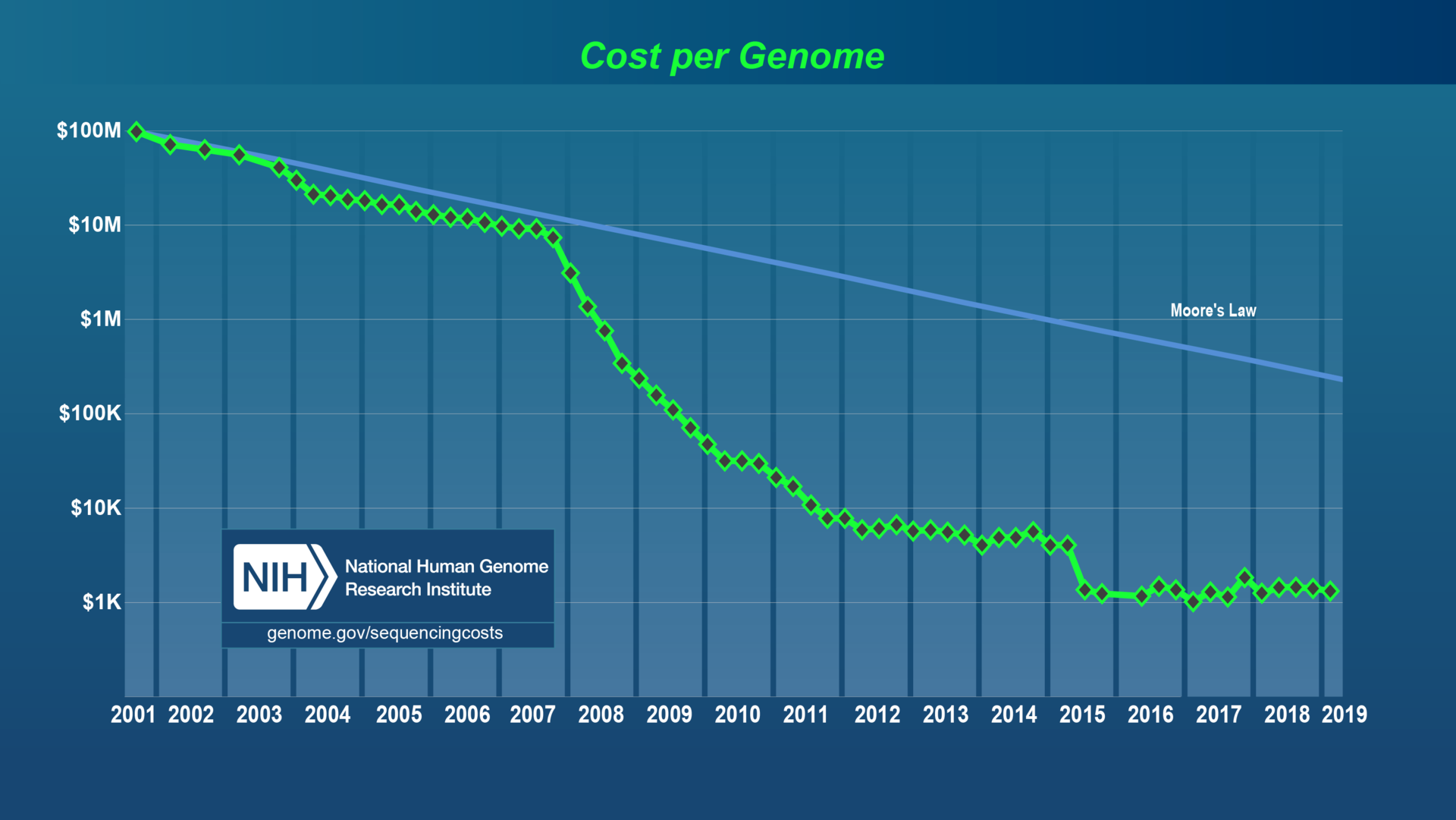
DNA sequencing is now low-cost...


...allowing us to sequence environmental "microbiomes"
Outline: My research experience & StFX

3. Current work




2. HK/Taiwan work




1. PhD work
Outline: A 5-year plan
|
|
Years 1-2 |
|---|---|
|
|
Years 3-4 |
|
|
Year 5 onwards |


Long term vision: A lab that links microbial diversity & biogeochemical function in the context of global ecosystem change with modern 'omics-enabled techniques
Hydrothermal vent work & StFX (Part I)




1. PhD work
Ecology & environmental science
Microbiology
Bioinformatics
Research question
- How do chemical / physical factors affect microbial growth and carbon fixation efficiency?

Deep-sea hydrothermal vents: Oases fueled by geochemical energy


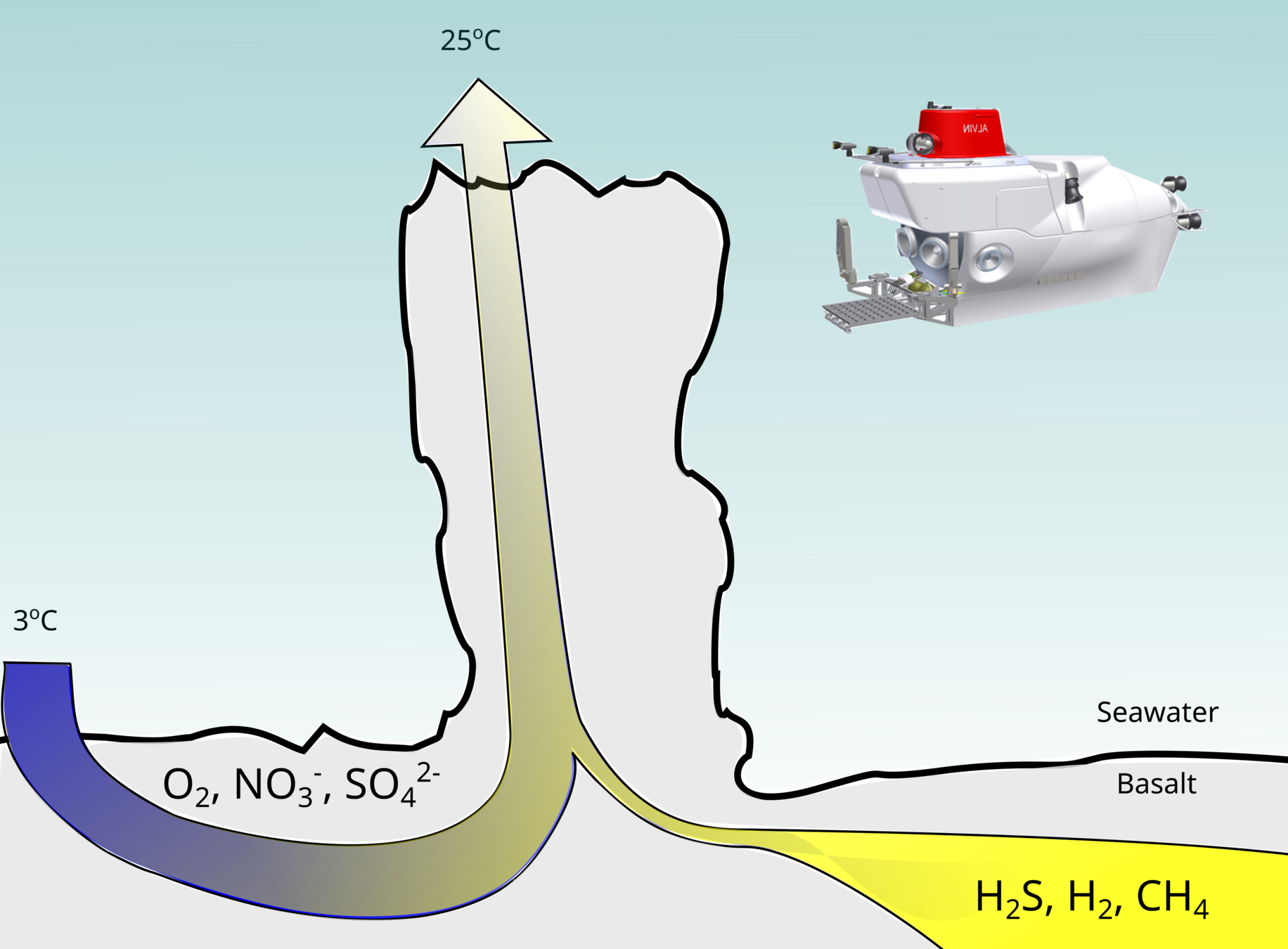
- Developed novel high-pressure incubations
- Quantified bulk and single-cell CO2 fixation
- Used DNA to measure community change

FISH
SIP



'omics data in an experimental, biogeochemical context
Process studies put 'omics in context


Example student project #1
- What is the genetic basis of differential oxygen tolerance in hydrothermal vent microbes?
Advantages for mentees:
- Clear experimental & environ-mental context, data available
- Unique publication
- Implications for evolution over geological time scales
O2 caused major shift in community over ~ 24 h

Use single-cell genomes to investigate genetic basis of this response







2. HK/Taiwan work
Ecology & environmental science
Microbiology
Bioinformatics
Hydrothermal vent work & StFX (Part II)
Research question and methods

1. Genomes from pure cultures
2. (Pan)genomic analysis with anvi'o

3. Diversity, evolution, biogeochemistry


- How does speciation affect biogeochemistry?

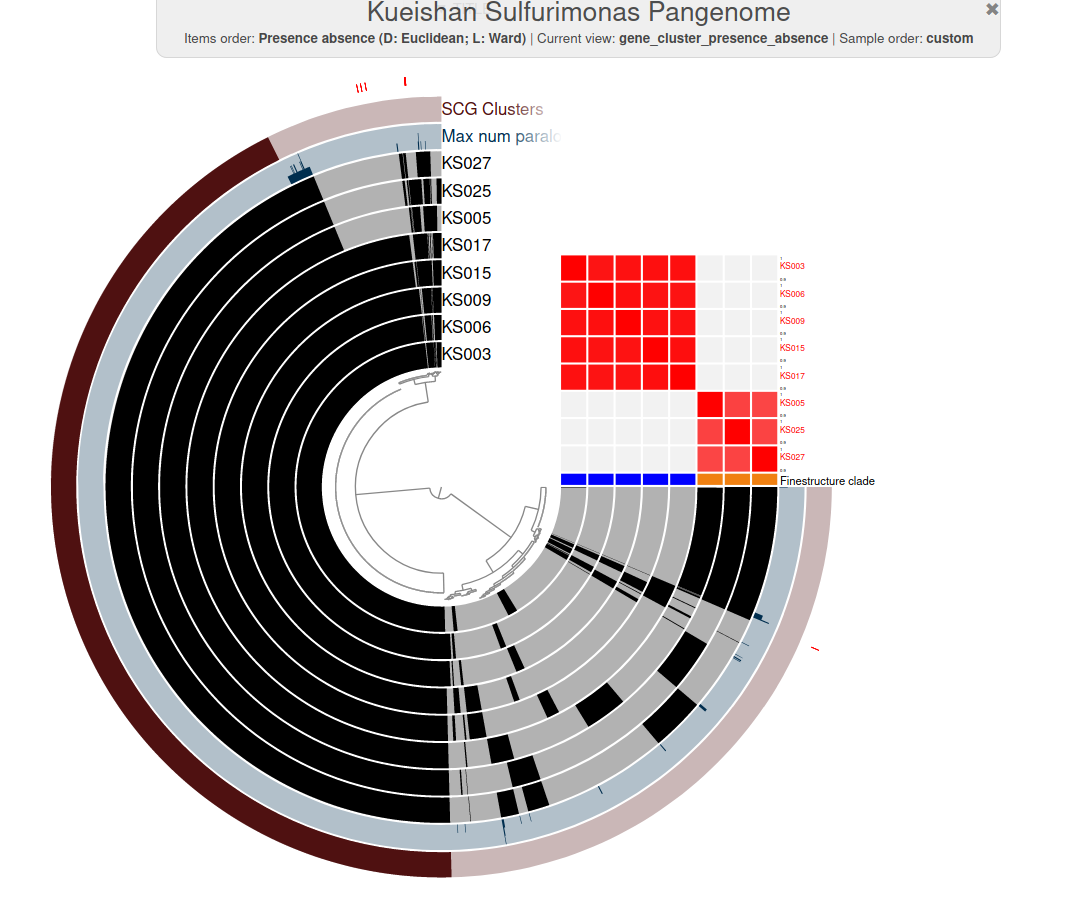
Genomics to biogeochemical insights
Plot made with anvi'o
Clade 1:
+ denit, + N2'ase
N source & sink
Clade 2:
- denit, + N2'ase
N source

Diagram = Li et al., 2018.

N2
- What characterizes a new genus of chemoautotrophs isolated from shallow-water vents in Taiwan?
Advantages for mentees:
- Fun, hands-on introduction to microbiology
- Impactful publication
- Could be extended with lab experiments, metagenomics, field work
- Many similar projects possible


Example student project #2
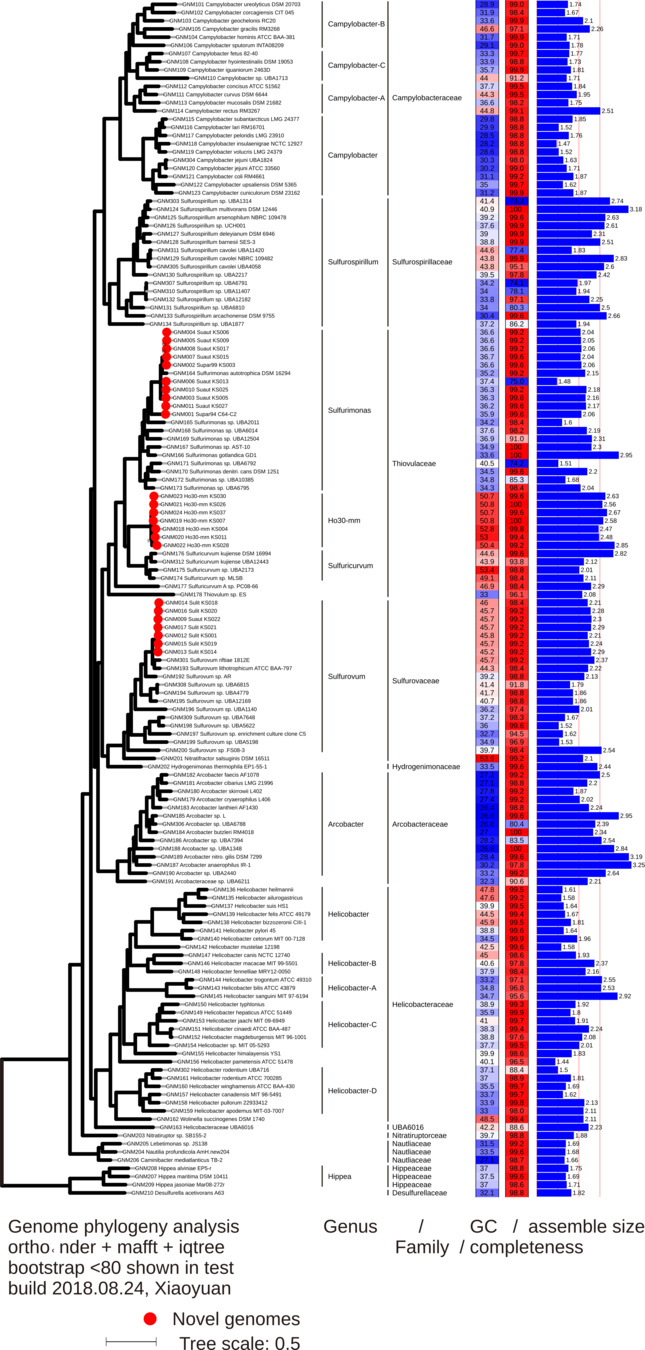
New genus
Xiaoyuan Feng, CUHK


1. Current work

Global-scale metabarcoding & StFX
Ecology & environmental science
Bioinformatics
Research questions
An in silico global phytoplankton model (DARWIN project, MIT)
-
How robust are model predictions?
- Need "ground truth" biogeographic distribution data
- What is the function of microbial "black box"?
- Can models and data help predict consequences of climate change?
- Goal: measure abundance of microbial life across space / time in global oceans using 3-domain metabarcoding
- Comprehensive (primers target all cellular life)
- Sensitive with deep sequencing
- Specific with "denoising" algorithms




Microbe art: @claudia_traboni
Assembling "ground truth" data
- Unique global dataset from 2003 - present
- Part of international, interdisciplinary collaboration
- Potential further funding from Simons Foundation
- Potential to access sample material from collaborators





Global-scale metabarcoding & StFX
- How well do imaging-based methods (flow cytometry, imaging flow cytobot) align with 3-domain metabarcoding?
Advantages for mentees:
- Simple but impactful
- Simons CMAP database makes possible
- Extend by mining global distribution data


Kalmbach et al. (2017), arXiv:1703.07309v1

Example student project #3
A 5-year plan to build a lab at StFX
|
|
Years 1-2 |
|---|---|
|
|
Years 3-4 |
|
|
Year 5 onwards |


Long term vision: A lab that links microbial diversity & biogeochemical function in the context of global ecosystem change with modern 'omics-enabled techniques
Y1-2: Equipment needs
|
|
Years 1-2 |
|---|---|
|
|
Years 3-4 |
|
|
Year 5 onwards |

Epifluorescence microscope

Microoxic cultivation equipment for non-pathogenic bacteria
CL2
A small computer lab for open-source bioinformatics software (qiime2, anvi'o) and databases (PR2, SILVA, GTDB)
|
|
Years 1-2 |
|---|---|
|
|
Years 3-4 |
|
|
Year 5 onwards |


- Begin "shovel-ready" projects
- Explore local systems
A 5-year plan to build a lab at StFX
Integrating disciplinary pillars = holistic research
Holistic, process studies require a "model system"
Ecology & environmental science

Microbiology

Bioinformatics




Open ocean systems
Whycocomagh Bay (WB)
- Diversity of metabolisms within ~1 hour drive
- Create freezer fossil record: DNA / RNA, cells for cultivation, FISH
- Small projects that build a bigger story, provide basis for process studies
Developing a local "model system" - rationale

- Explore collaboration with Bruce Hatcher (Bras D’Or Institute for Ecosystem Research, CBU)
-
Use 3-domain metabarcoding to publish on:
- Phytoplankton bloom time-series in WB
- Depth profiles of WB = influence of geo- chemistry on community composition

Needham & Fuhrman (2016), 10.1038/nmicrobiol.2016.5

Developing a local "model system" - how?






3-domain metabarcoding a foundation for holistic

Identity + Activity
ecological research


Y 3-4
|
|
Years 1-2 |
|---|---|
|
|
Years 3-4 |
|
|
Year 5 onwards |


- Start local sampling, publish results
- Build infrastructure for process studies

- Begin "shovel-ready" projects
- Explore local systems

Y3-4: Equipment needs
|
|
Years 1-2 |
|---|---|
|
|
Years 3-4 |
|
|
Year 5 onwards |




Robotic automation for high-throughput DNA prep

Automatic cell sorter to measure in situ activity, sort cells for sequencing
Optional, could collaborate or buy second-hand
~$20,000 USD for full NGS automation
Long term vision: A lab that links microbial diversity & biogeochemical function in the context of global ecosystem change with modern 'omics-enabled techniques

Model system
In silico
Laboratory
Field work
Holistic ecosystem knowledge
Reductionist (multiple entry points)
Interdisciplinary opportunities
Diverse entry points for students
Mentoring philosophy & DEI
Instrumental
-
Real data, relevant skills, important questions, diverse collaborations
- Flexible, adaptable, part of "big picture"
Psychosocial
Evidence-based approaches
-
Mentoring to foster emotional belonging and research success
- Realize full potential
- Foster collaboration
- Modern pedagogy, research project scaffolding, culturally-aware mentoring
Ecosystem-driven research

Motivated by desire to understand the Earth System

We anticipate that as trait-based biogeography continues to evolve, micro- and macroorganisms will be studied in concert, establishing a science that is informed by and relevant to all domains of life.” -Green, Bohannan, and Whitaker, Science (2008)
Acknowledgements
WHOI / UFZ: Stefan Sievert, Jeff Seewald, François Thomas, Niculina Musat, Craig Taylor
CUHK / Academia Sinica: Haiwei Luo, Annie Wing-Yi Lo, Benny Chan
USC/MIT/UCSB/Dalhousie/Carnegie: Jed Fuhrman, Yubin Raut, Enrico Ser-Giacomi, Paul Berube, Steven Biller, Mick Follows, Stephanie Dutkiewicz, Craig Carlson, Zoe Finkel, Emily Zakem









Microbe art: @claudia_traboni
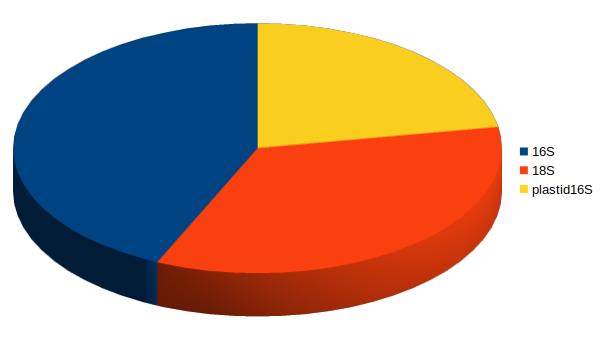
p16S
e16S
18S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
3-domain metabarcoding quantifies the whole ecosystem

Microbe art: @claudia_traboni
18S
p16S
e16S
-
Comprehensive community data from single PCR assay:
- p(rokaryotic)16S
- e(ukaryotic)16S
- Eukaryotic 18S
3-domain metabarcoding quantifies the whole ecosystem
Long-term scientific questions
Local work (Whycocomagh or elsewhere):
- What can spatiotemporal abundance patterns and interaction networks from 3-domain metabarcoding contribute to models?
- What biogeochemical functions do the prokaryotes catalyze?
- What are the biogeochemical consequences persistent stratification in W. Whycocomagh bay?
Global/local work:
- Can targeted mini-metagenomes improve databases & models?
- Can we cultivate microbial dark matter?
Vent work:
- What proportion of organisms are active?
- How abundant are oxygen-sensitive / tolerant types in situ?

Models
Microbe art: @claudia_traboni
Ecosystem
Kalmbach et al. (2017), arXiv:1703.07309v1



Physics
Biology
Chemistry

Long term: holistic vision
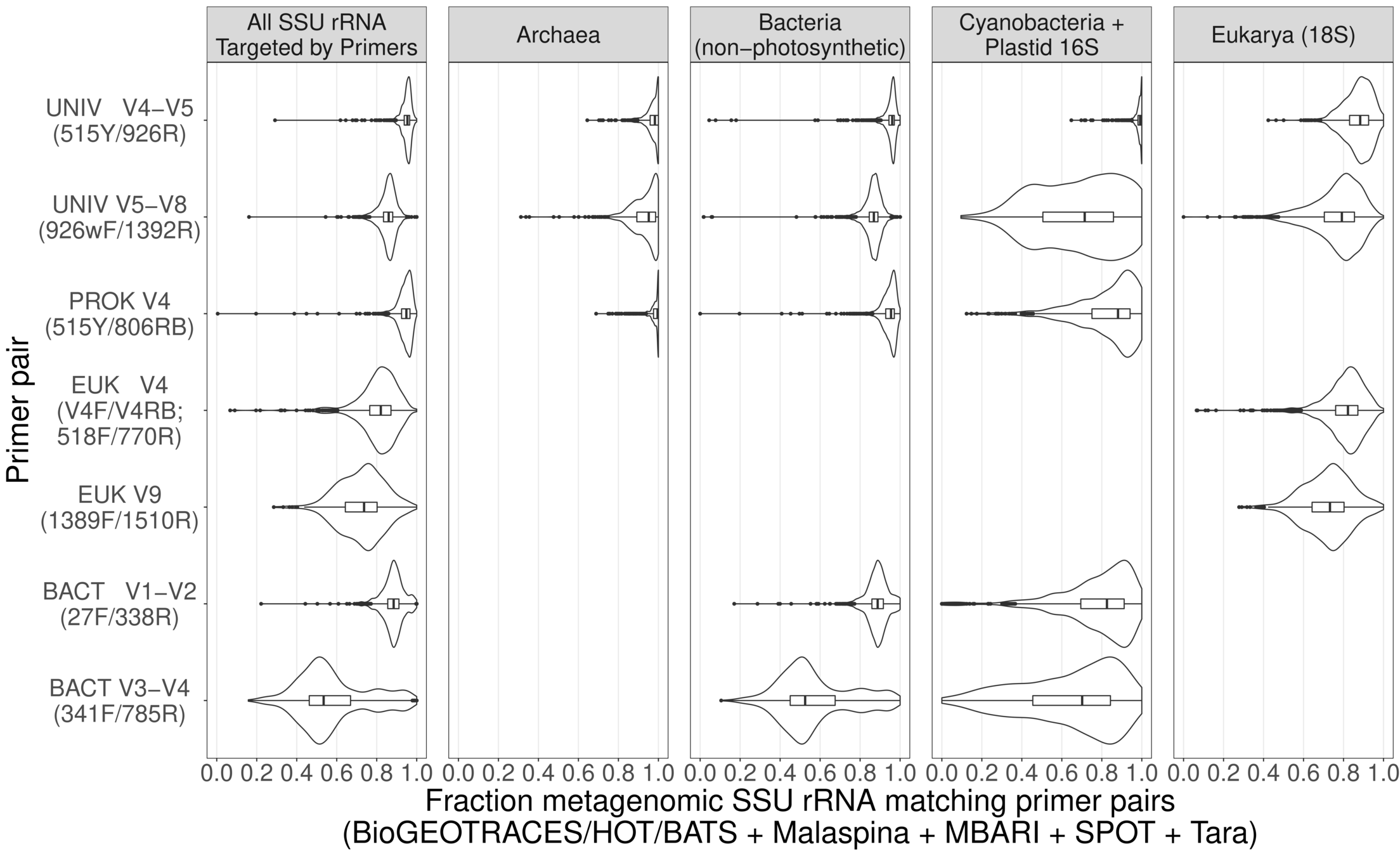
- How can we infer changes in microbial community composition change across decadal time-scales using barcoding data from different regions of SSU rRNA?
Advantages for mentees:
- "Rosetta stone" allows intercomparisons, FISH probe design from barcodes
- Address previously impossible questions
- "Data science" project experience


Dueholm et al. (2020) mBio, e01557-20
Database of full-length 16S rRNA

Example student project #4
Techniques for working at the microbial scale


Sebastián & Gasol (2019), 10.1098/rstb.2019.0083 ; McNichol et al (2018) 10.1073/pnas.1804351115 ; Zehr (2015) 10.1126/science.aac9752; Bramucci et al (2021) 10.1038/s43705-021-00079-z

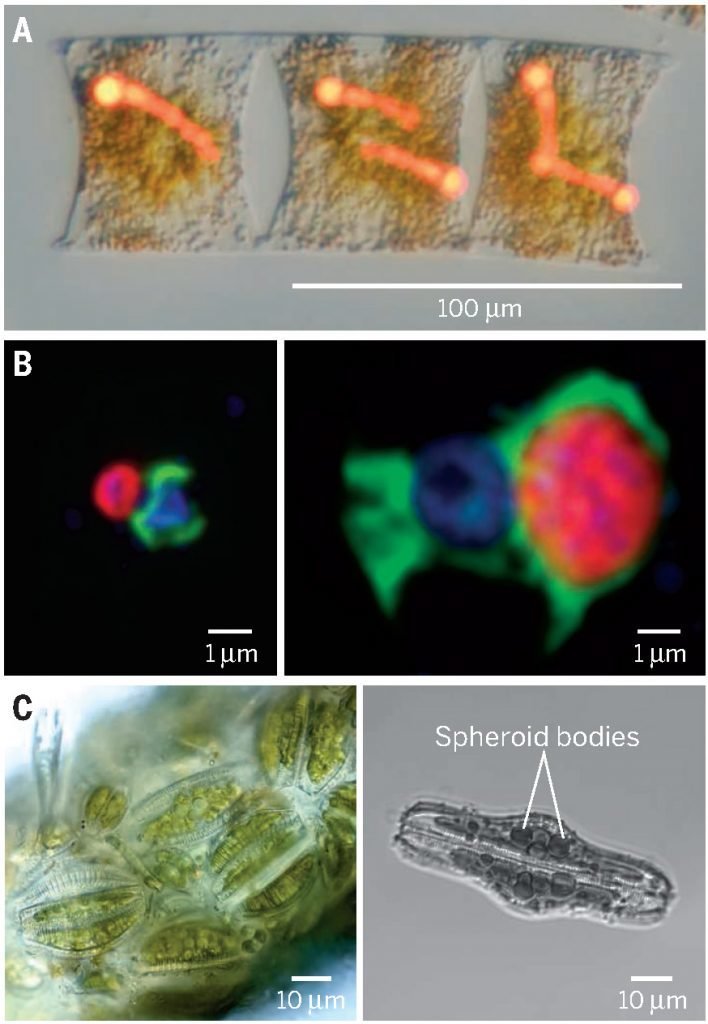

Long term: single-cell activity methods

Experience with single-cell activity methods
Specific FISH labeling + radiocarbon incubations = Carbon fixation rates were measured at the genus level and across different conditions
- Taxonomy (CARD/HCR-FISH) / transcription (FISH-TAMB)
- Respiration, cell viability (RSG); Polyphosphate content (DAPI)
Automatic cell-sorting + environmental databases = taxon-specific activity or genetic potential measurements
Many uses for sorted cells:
- Taxon-specific isotope uptake
- Mini-metagenomes with few cells
- Cell isolation for cultivation
Pjevac et al (2019) 10.1111/1462-2920.14739
Multiple sorting axes now possible

Long term: single-cell activity methods
Research impact

Subsurface C production > 10 - 10,000x photic zone export
Subsurface C production rivals chemosynthetic symbioses



Hydrothermal vent research - summary
-
Address basic questions about oxygen and carbon dioxide metabolism
- Biogeochemical, Earth Systems History implications
- Constrained projects with samples, data, and clear goals
- Bioinformatics, genomics a critical component
- International collaborations
- Similar organisms present in any sulfidic system


?




Accomplishments & novelty
1. Led in silico method optimization
Universal primers for barcoding work almost* perfectly across global oceans
2. Developed collaborations and generated data




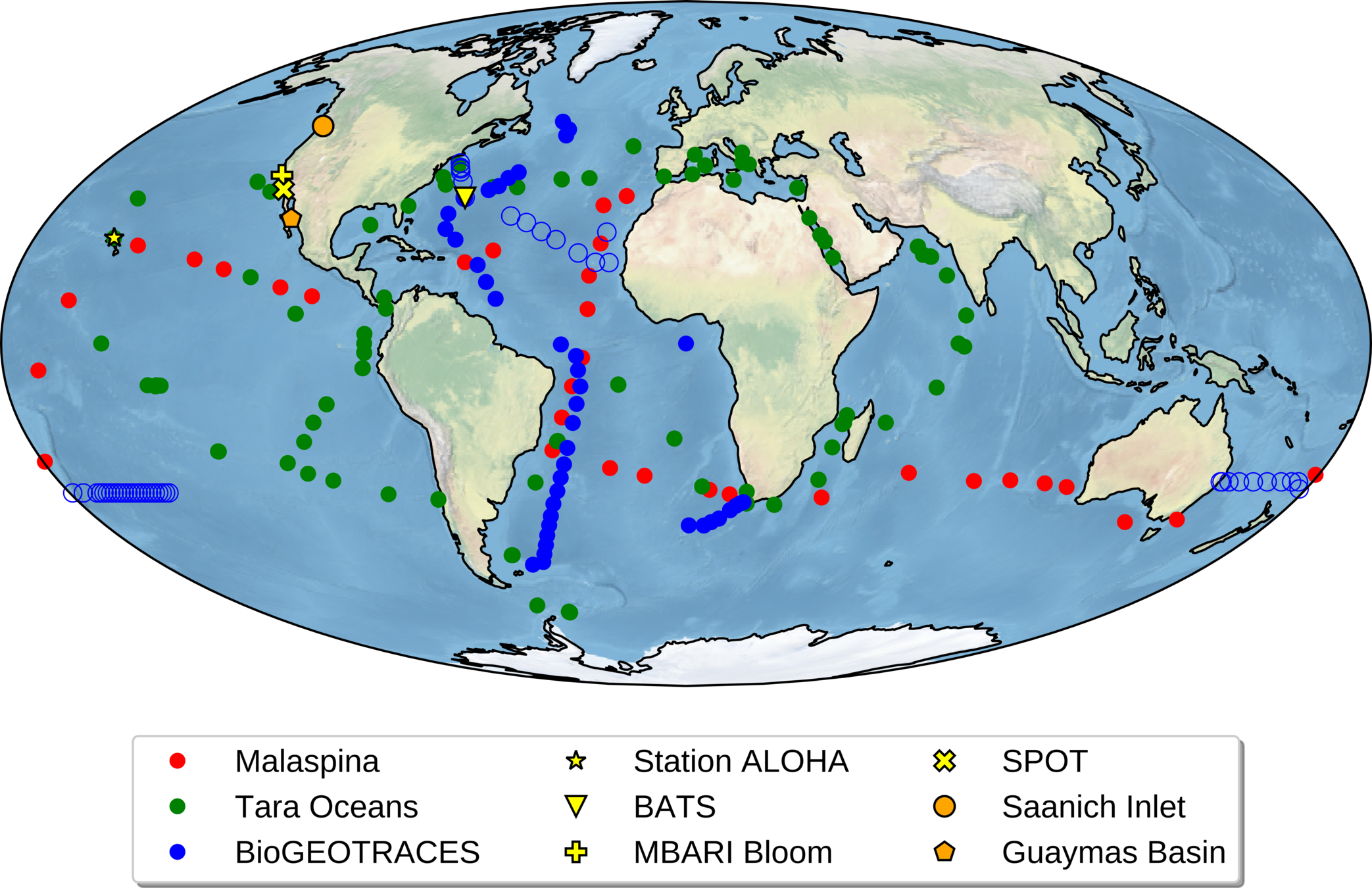
Global metagenomes
Over 800 globally-distributed barcode samples allow model-data intercomparison


Craig Carlson, UCSB
Potential for training HQP
-
Data Science
- Genomics, amplicon sequencing, FAIR principles
- conda, snakemake, github, bash/python
-
Microbiology
- High-throughput cultivation approaches
-
Environmental science
- Oceanography, water quality
- Holistic ecosystem measurements
Why microbial ecology at StFX?
- Opportunity to focus on fundamental biological research
- Build model (eco)systems, interdepartment collaborations
- Diverse questions / techniques fits with liberal arts education
- Flexible, student-centred projects; CURE
- Maritimes offers a wealth of natural marine environments
- Collaborations in oceanography, ecosystem monitoring



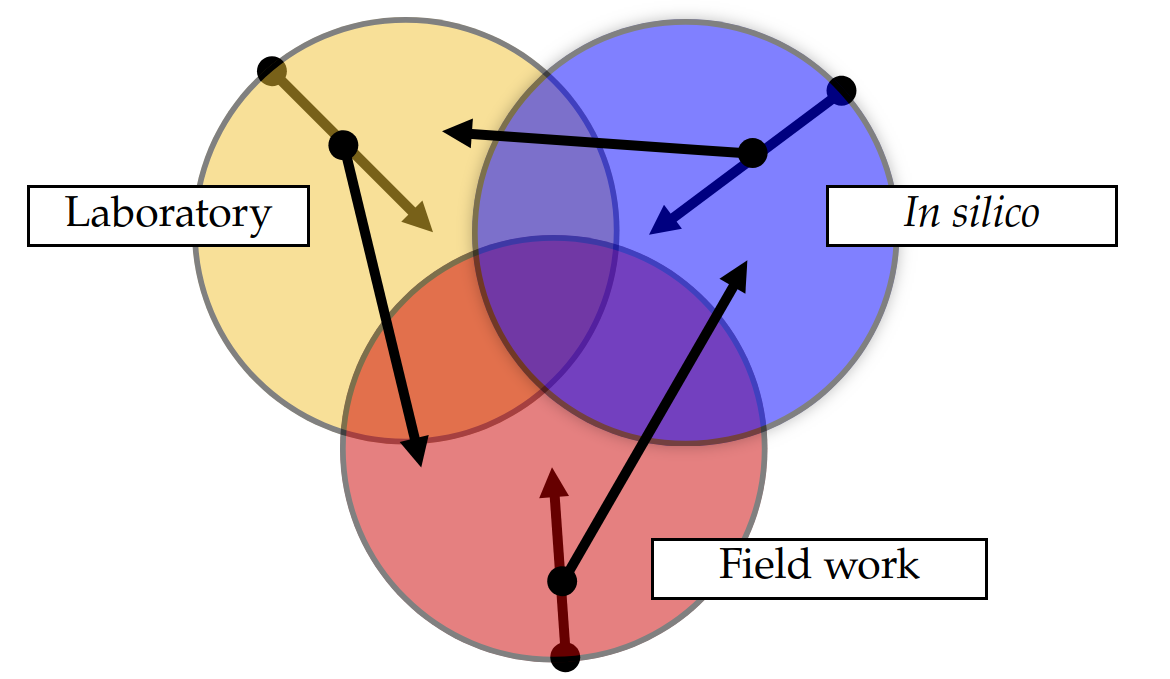
- Methods interact and inform one another
- Points to "microbial natural history"

Holistic picture






Holistic techniques "at sea"
-
"Rosetta stone" => FISH probes:
- Cell enrichment for mini-metagenomes
- Visualize predicted symbioses / other ecological interactions
- Measure cell-specific traits such as biovolume
-
Attempt cultivation of "microbial dark matter":
- Flow cytometry with "live stains"
- Enrichment experiments


Native Microbiota CURE*
- Based on undergraduate botany experience (Native Flora)
- Students collect microbial "herbaria":
- Characterize with barcoding
- Visualize with FISH
- Attempt to cultivate
- Goal: Develop "microbial natural history"
- Bring new scientists into the fold
- X-Oceans Outreach
- Discover new taxa, study sites


*CURE = Course-based undergraduate research experience
Outside the box
Departmental / interdepartmental collaborations:
- Identify microbial symbioses / interactions with plants & animals with single-cell methods
- Use 3-domain metabarcoding during incubation studies / field sampling
- Collaboration with Earth Science for ongoing biogeochemical work
Access work:
- How to make microbiome profiling more accessible, affordable, reliable?
- How can diverse students be nurtured?
Natural history:
- What unique projects can students of Native Microbiota devise?
- A "SEA-PHAGES"-like project?
- Intercalibration, intercomparison
- Methods development
- Interdisciplinary by nature
- Lots of room for young scientists!


BioGeoSCAPES*
*Depending on funding, availability of reference material

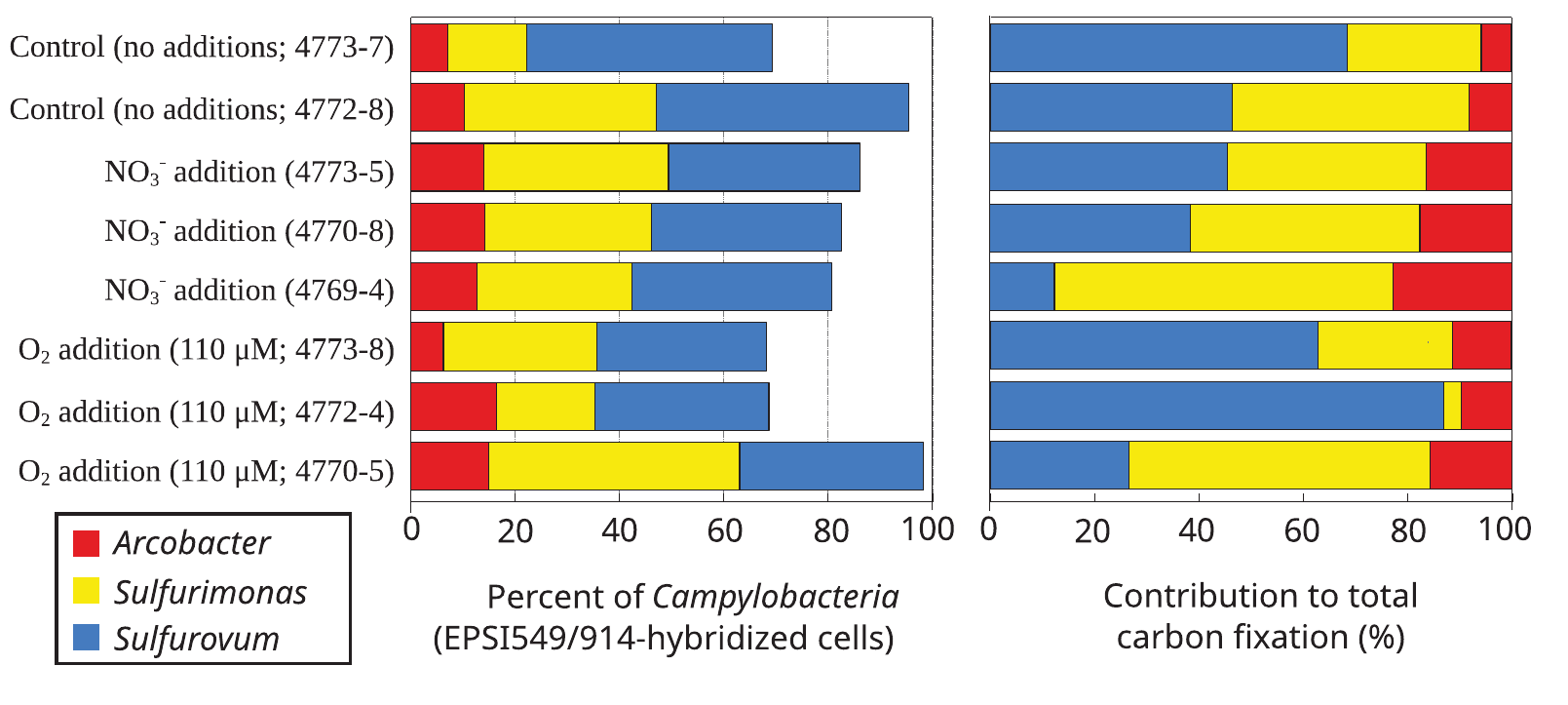
- Developed theoretical metabolic model of chemoautotrophic Campylobacteria during PhD
Y 3-4: Campylobacteria, physiology, and evolution

Targets for physiological / evolutionary studies
- Campylobacteria are oxygen-sensitive and capnophiles (CO2-loving), existing in an "intermediate evolutionary state"
-
How does the emergence of genomic traits/clades relate to the development of a more oxic, lower CO2 atmosphere?
- Method: Molecular dating using genome phylogeny
- Japanese collaborator has agreed to contribute genomes








Campylobacteria, physiology, and evolution
Model-data intercomparison (Yubin Raut, USC)



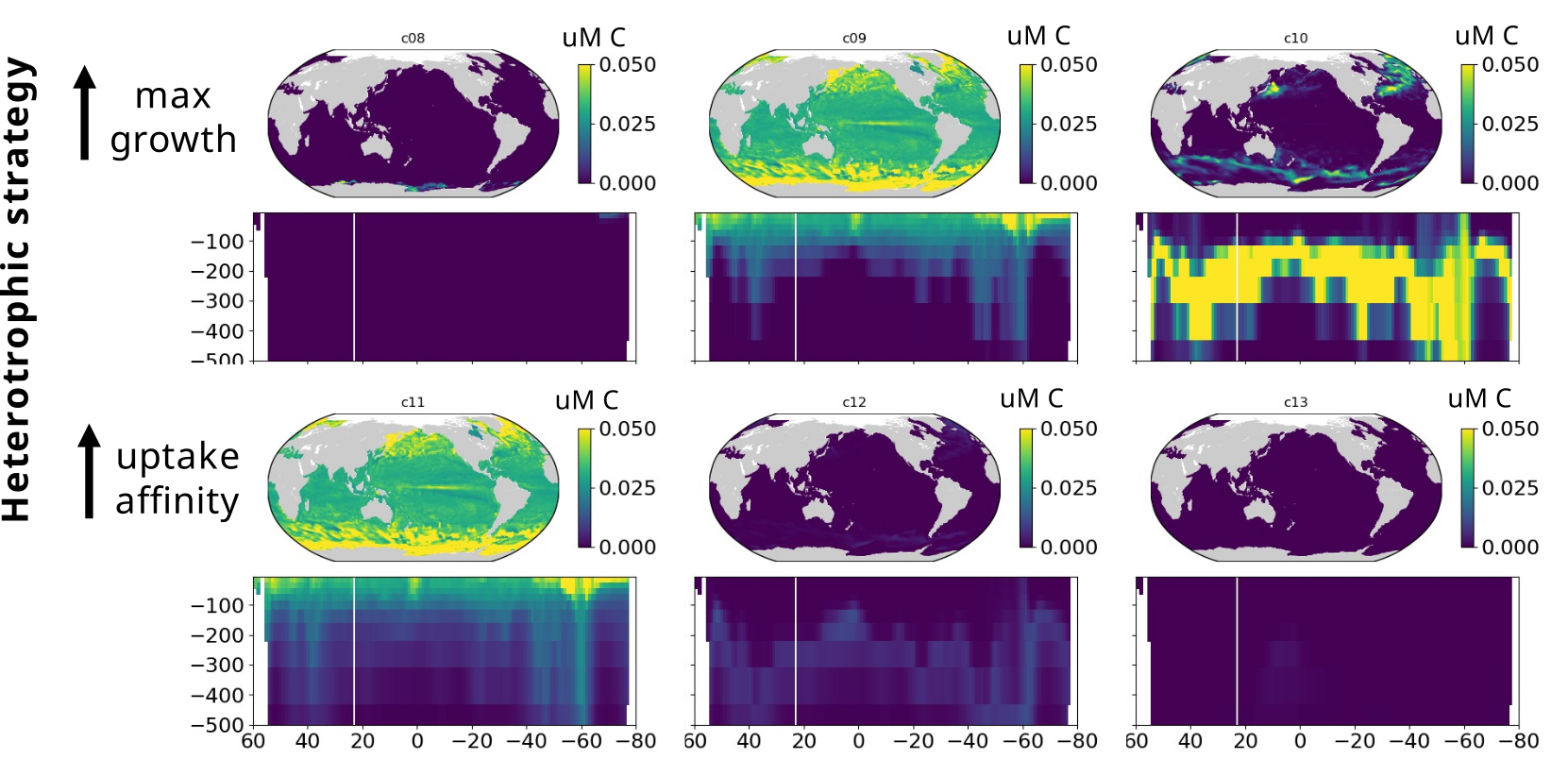
Modelling marine heterotrophs (Emily Zakem, Carnegie Inst.)
- Play key intermediate role between data and modelling worlds
- Basis of application for Simons Foundation "Early Career Investigator in Marine Microbial Ecology and Evolution Awards"
- Contingent on successful NSERC application or other federal award
Y1-2: Biogeography, modelling, and funding

Y5 - onwards: Other potential collaborators
- Niculina Musat, Leipzig, Germany
- ProVis: Single-cell techniques including NanoSIMS
- Jeff Seewald, WHOI, USA
- Hydrocarbon chemistry
- Irene Wagner-Döbler / Meinhard Simon, Germany
- Plymouth Marine Laboratory, UK
- Camila Signori, Brazil
- Shengwei Hou, SUSTech, China
- Grad student community
Clade 1:
+ denit, + N2'ase
N source & sink
Clade 2:
- denit, + N2'ase
N source



Diagram = Li et al., 2018.

N2
Campylobacteria and biogeochemistry
- Use metagenomics to assess impact in natural system
Craig Carlson, Elisa Halewood, UCSB
P16S/N
Fraction of 18S amplicon sequences

16S
plastid 16S
18S




Jan-Feb 2005
Feb-Mar 2006
With deep sequencing, good coverage for all 3 domains
Looking for interactions
plastid
16S
mito 16S
nuclear 18S
Space / time
Abundance
A eukaryotic phytoplankter
Complementary to existing techniques
"eDNA"




Jan-Feb 2005
Feb-Mar 2006
Craig Carlson, Elisa Halewood, UCSB
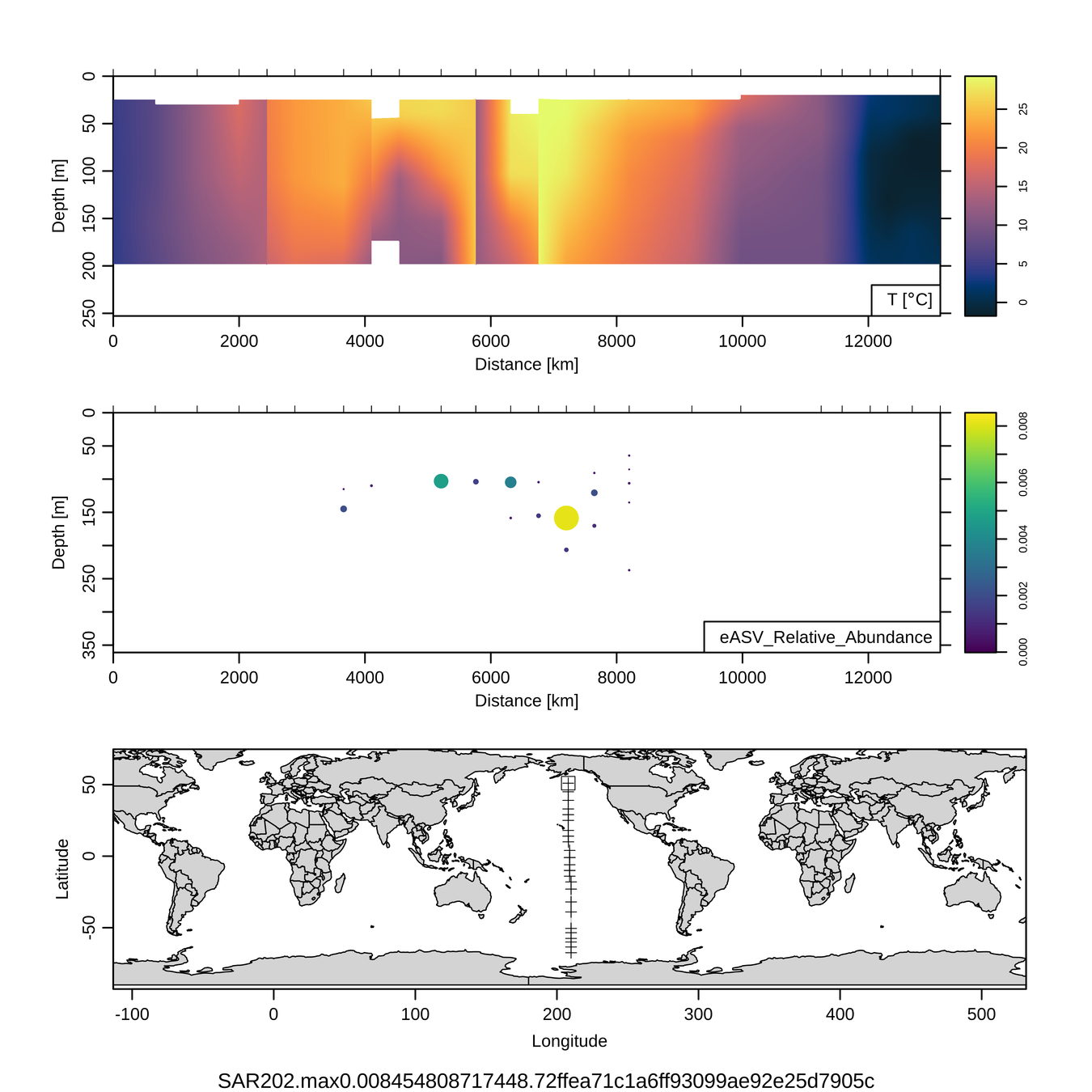
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
Craig Carlson, Elisa Halewood, UCSB
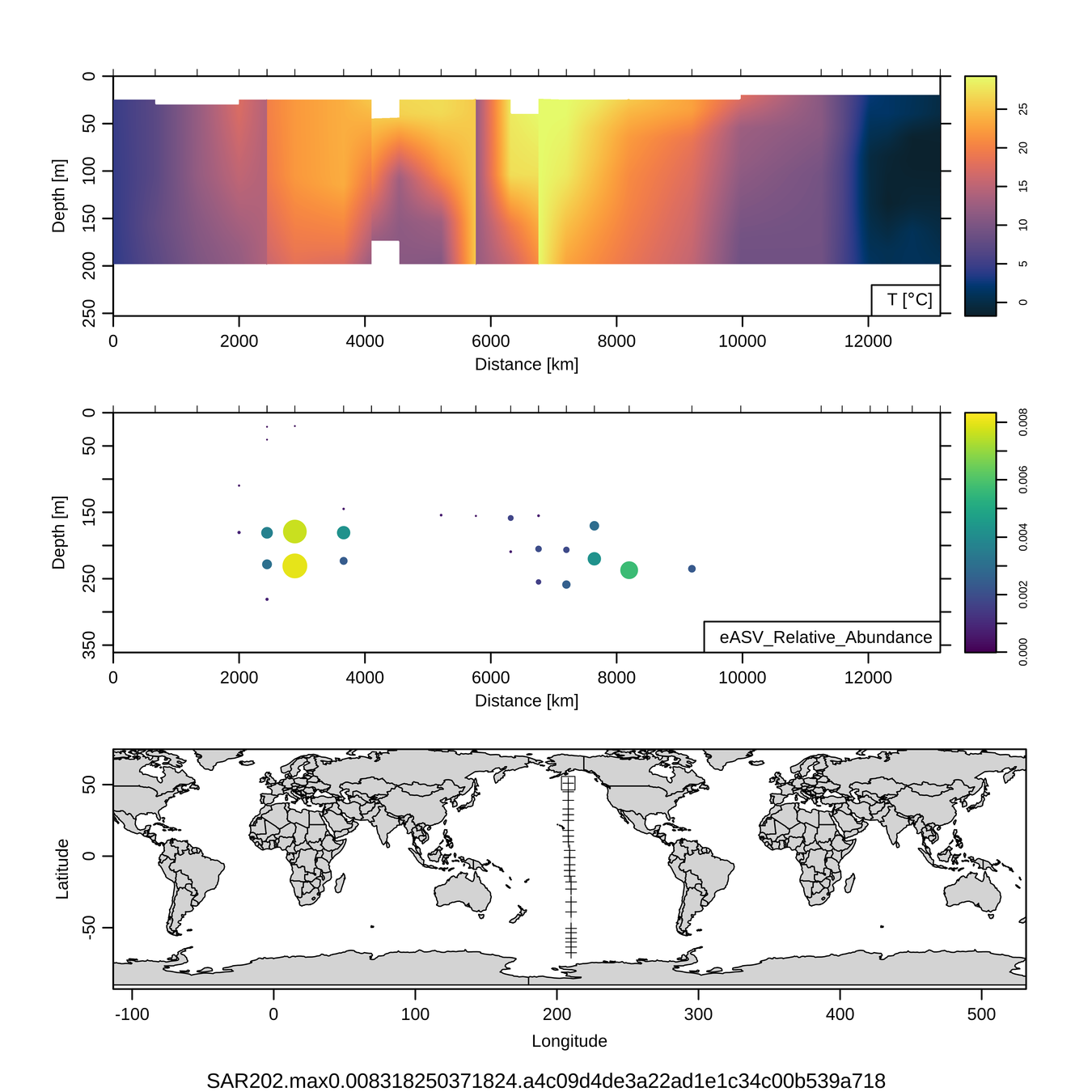
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
Craig Carlson, Elisa Halewood, UCSB
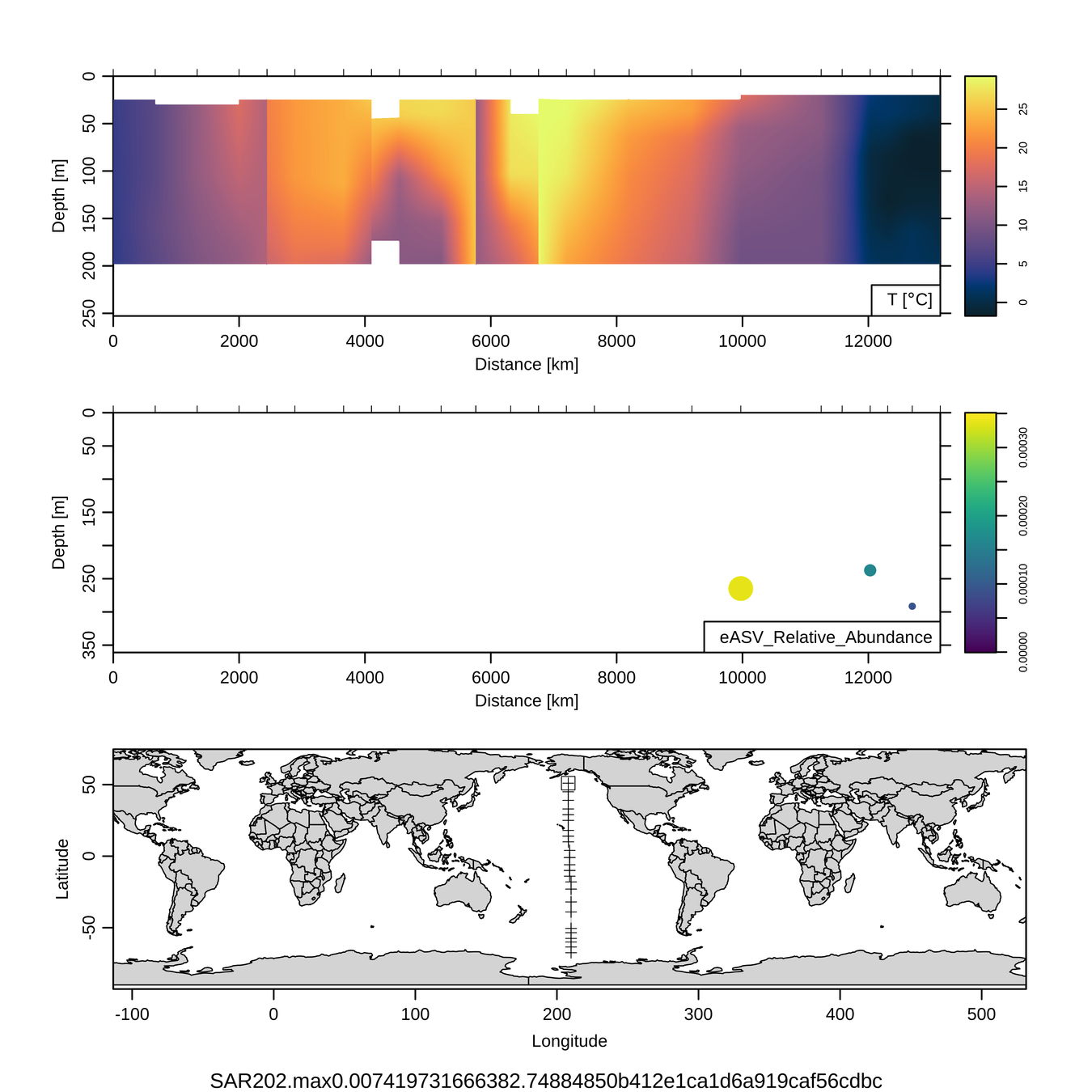
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
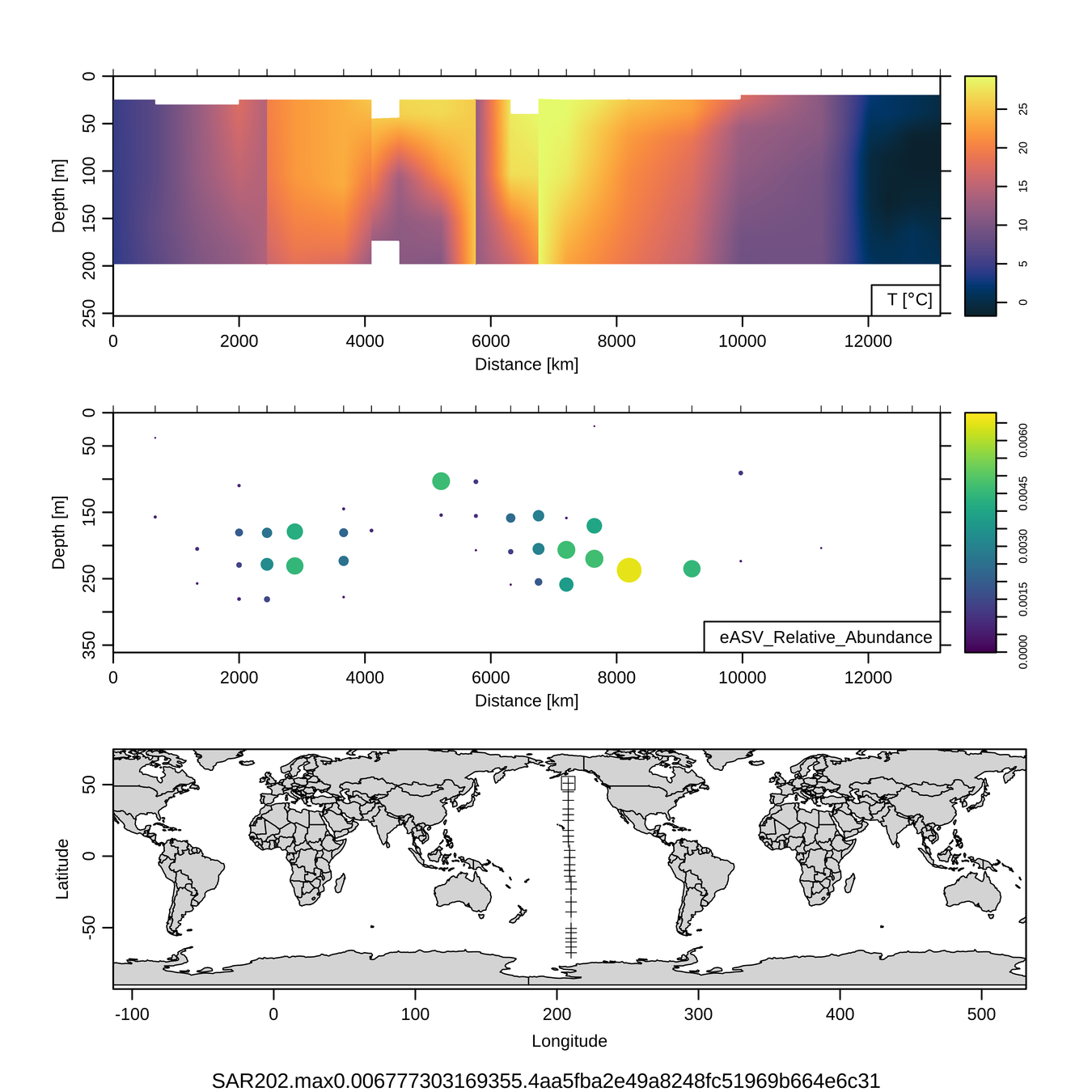
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
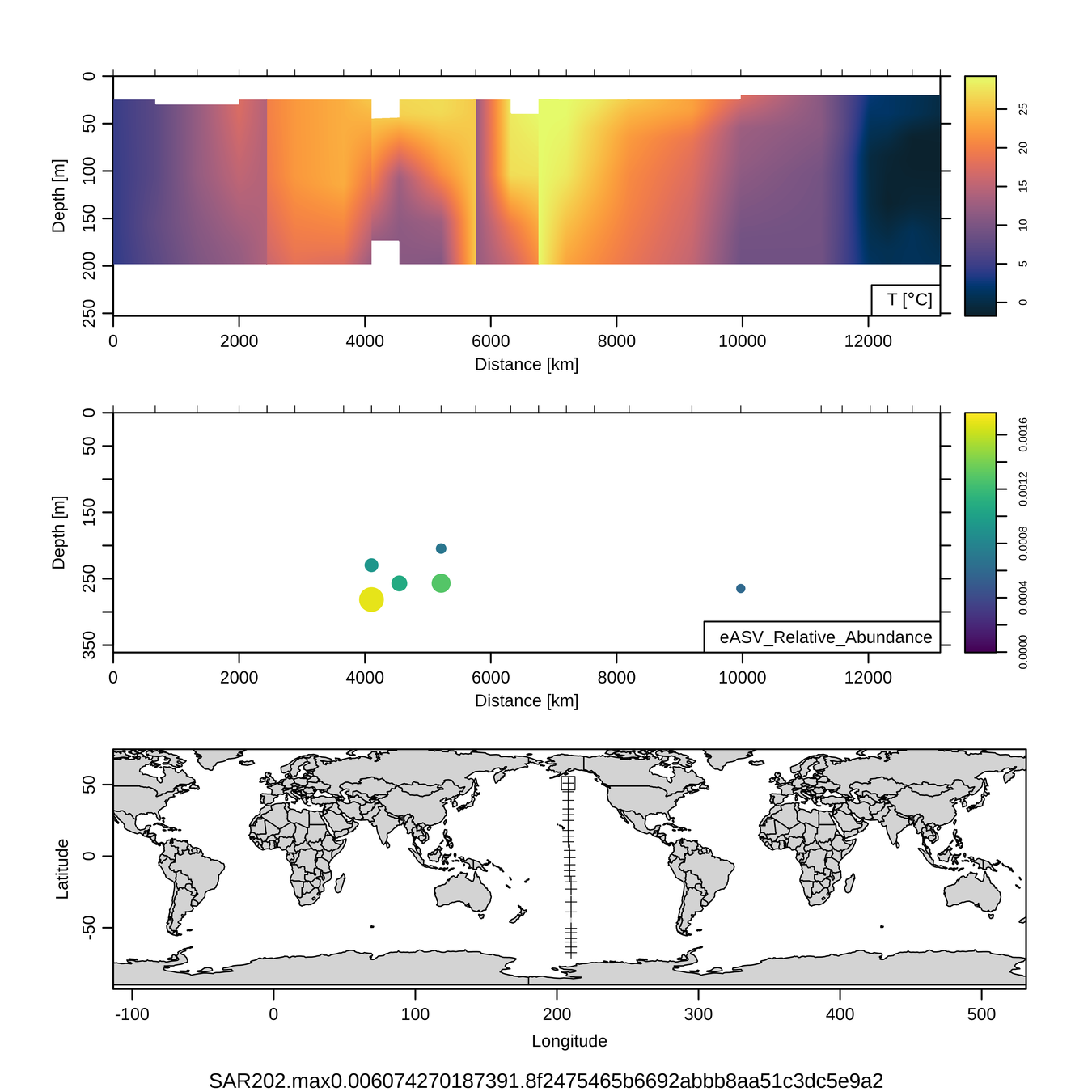
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
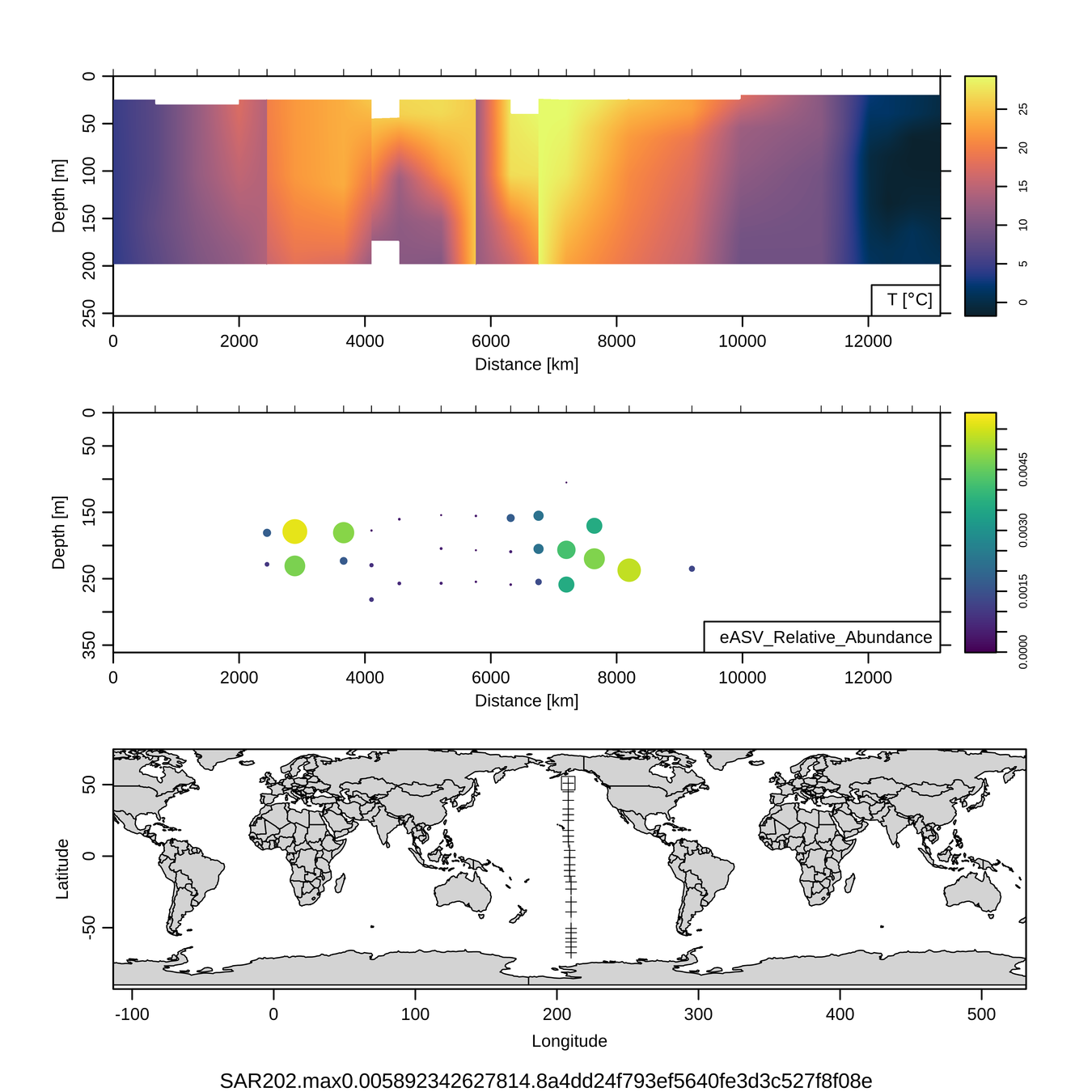
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
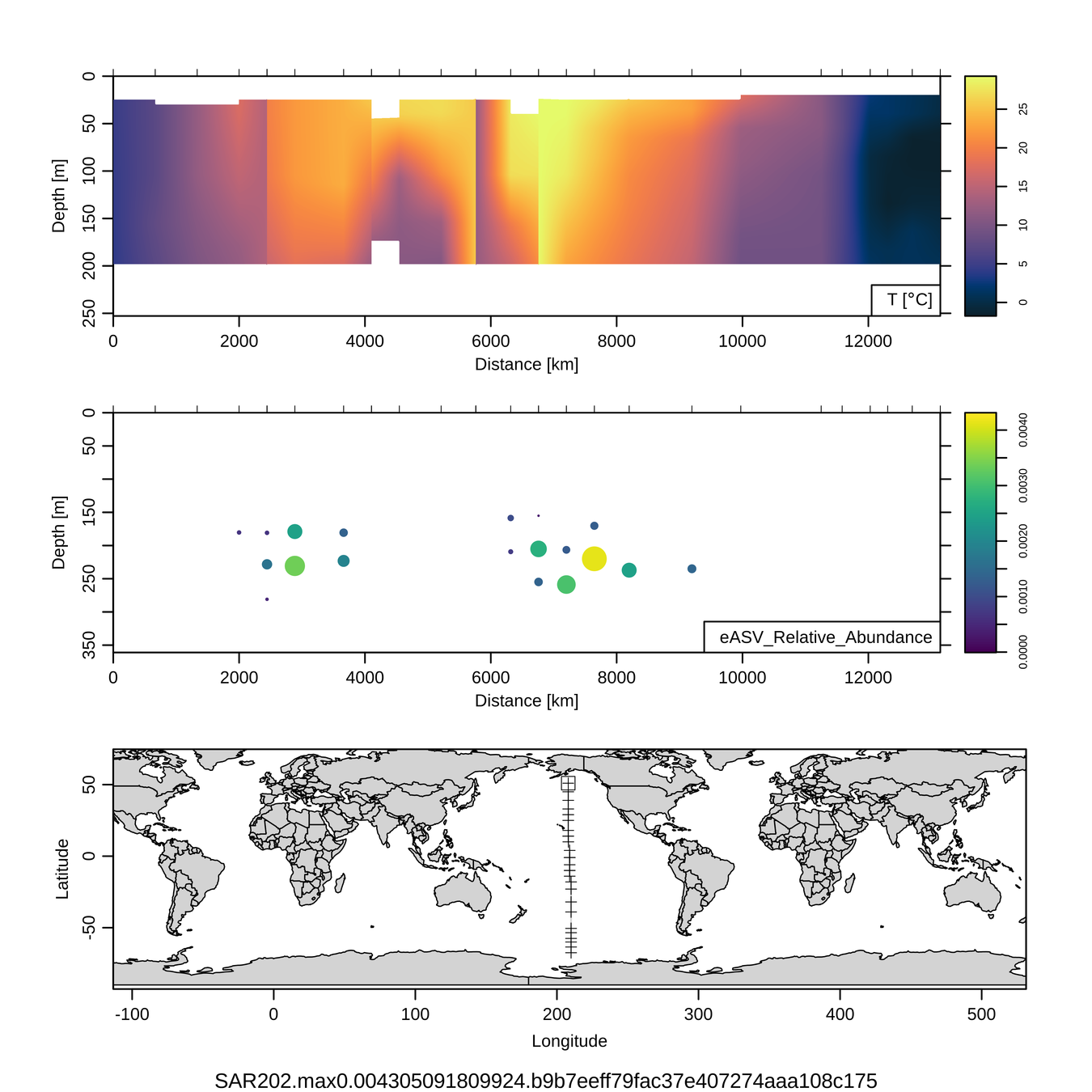
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
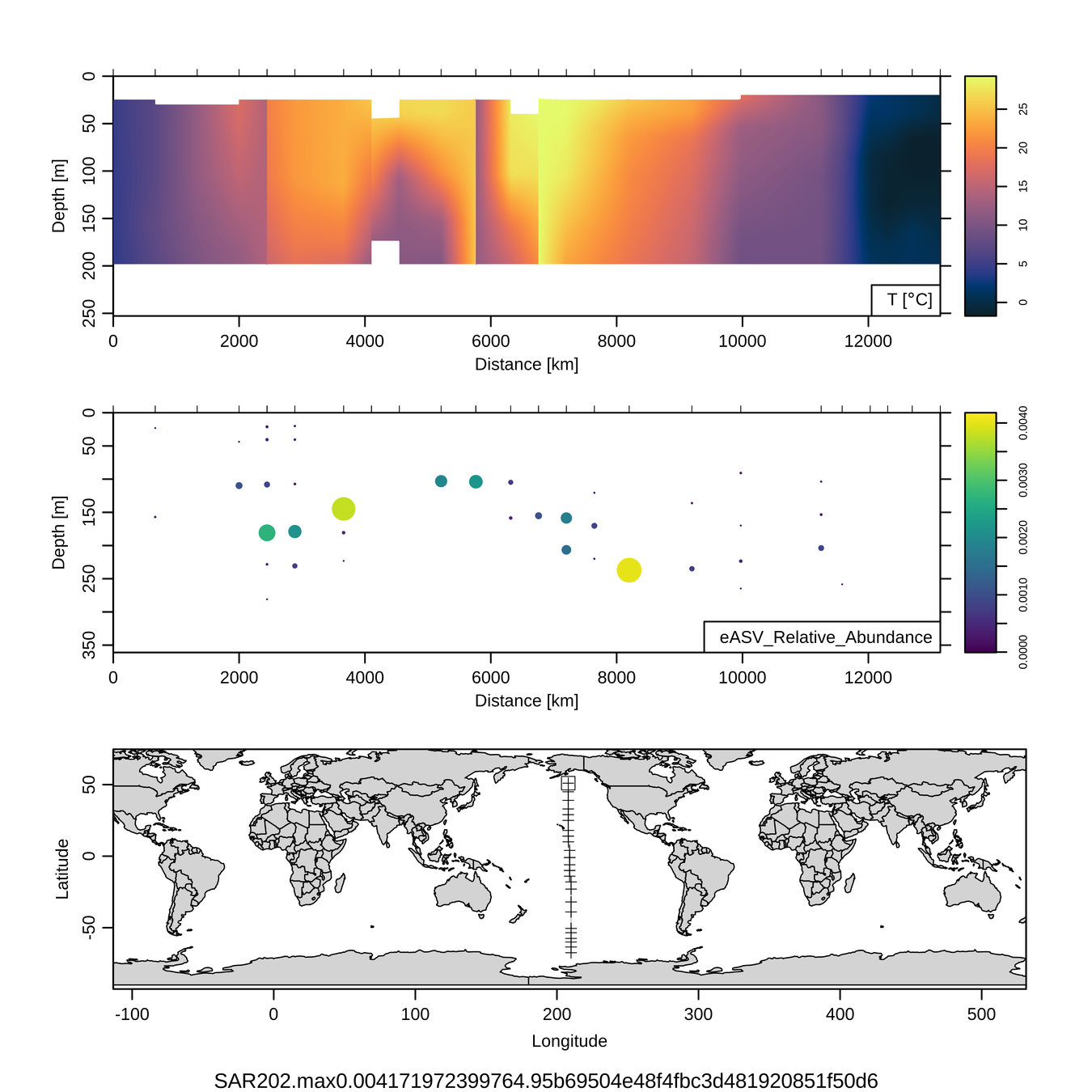
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"




Jan-Feb 2005
Feb-Mar 2006
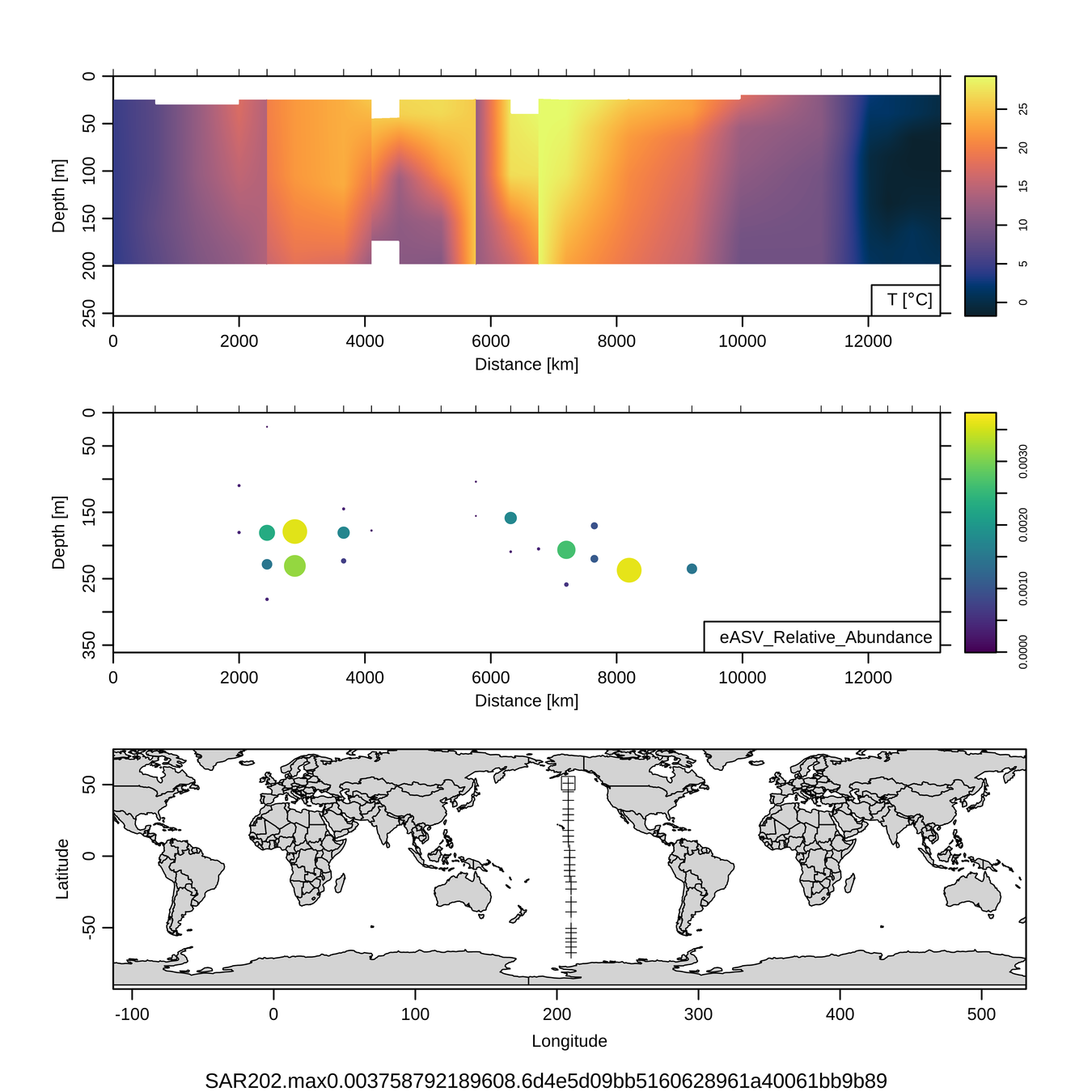
This is only the top 10 from one bacterial taxon!
Similar patterns for phytoplankton (and even some Metazoa)
Craig Carlson, Elisa Halewood, UCSB
P16S/N - Biogeography of top 10 SAR202 "species"
Sources
| Images used in presentation were adapted from: |
|---|
|
Line diagram (slide 9): J. A. Fuhrman, J. A. Cram, D. M. Needham, Marine microbial community dynamics and their ecological interpretation. Nature Reviews Microbiology 13, 133–146 (2015). Metagenome image (slide 19): V.A. Iverson et al., Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science, 335(6068): 587-590 (2012). Anvi'o used to make image on slide 20: https://merenlab.org/software/anvio/; https://peerj.com/articles/1319/ Shallow-water vent image (slides 20, 22): Y. Li et al., Coupled Carbon, Sulfur, and Nitrogen Cycles Mediated by Microorganisms in the Water Column of a Shallow-Water Hydrothermal Ecosystem. Frontiers in Microbiology. 9:2718 (2018). Speciation image (slides 20, 22): R. Stepanauskas et al., Gene exchange networks define species-like units in marine prokaryotes. bioRxiv, 2020.09.10.291518 (2020). Oxygen diagram (slide 25): L.R. Kump, The Rise of Atmospheric Oxygen. Nature, 451(7176): 277-8 (2008). Water column image on slide 30 adapted from: M. Hügler, S. M. Sievert, Beyond the Calvin Cycle: Autotrophic Carbon Fixation in the Ocean. Annu. Rev. Marine. Sci. 3, 261–289 (2011). |
Unless otherwise noted, other images are either my own (un)published work, ⓒWHOI, or public domain images from Wikimedia Commons
Notes
- Center for Global Change Science
- Explain how techniques could apply to soil, host-associated microbiomes, etc
In silico
- Collaboration with modellers
Laboratory
- Method optimization / intercalibration
Field work
- New data collection
Pathways for mentees





In silico
- Study evolution and explore new genomes
Laboratory
- Describe novel taxa, study their physiology
Field work
- Explore new ecosystems
Pathways for mentees





Annie Wing-Yi Lo, CUHK
Microbiology
Environmental science
Bioinformatics
Evolution, population genetics
Ecosystem modelling, global change biology
Microbial ecology, biogeochemistry
3 "pillars"
What I would bring to your department
Microbiology
Ecology & environmental science
Bioinformatics
- > 10 years experience in marine microbial ecology
- Can train mentees in lab, computational, field techniques
- Diverse interests: oceanography, modelling, analytical method development, natural history, evolution
- Passionate about integrating cutting-edge "microbiome" research techniques into my teaching (CURE)
- Track record of successful interdisciplinary collaborations, and excited to build connections at DPES / UofT
- Set up equipment for O2-sensitive microbes
- Retrieve cultures / samples / data
- Train students in cultivation, genomic analysis
- Medium term: Develop methods to identify essential genes (Tn-Seq)
- Long term: Conduct isolation campaigns in model systems, do biochemical assays


Cultivation and genomics at DPES





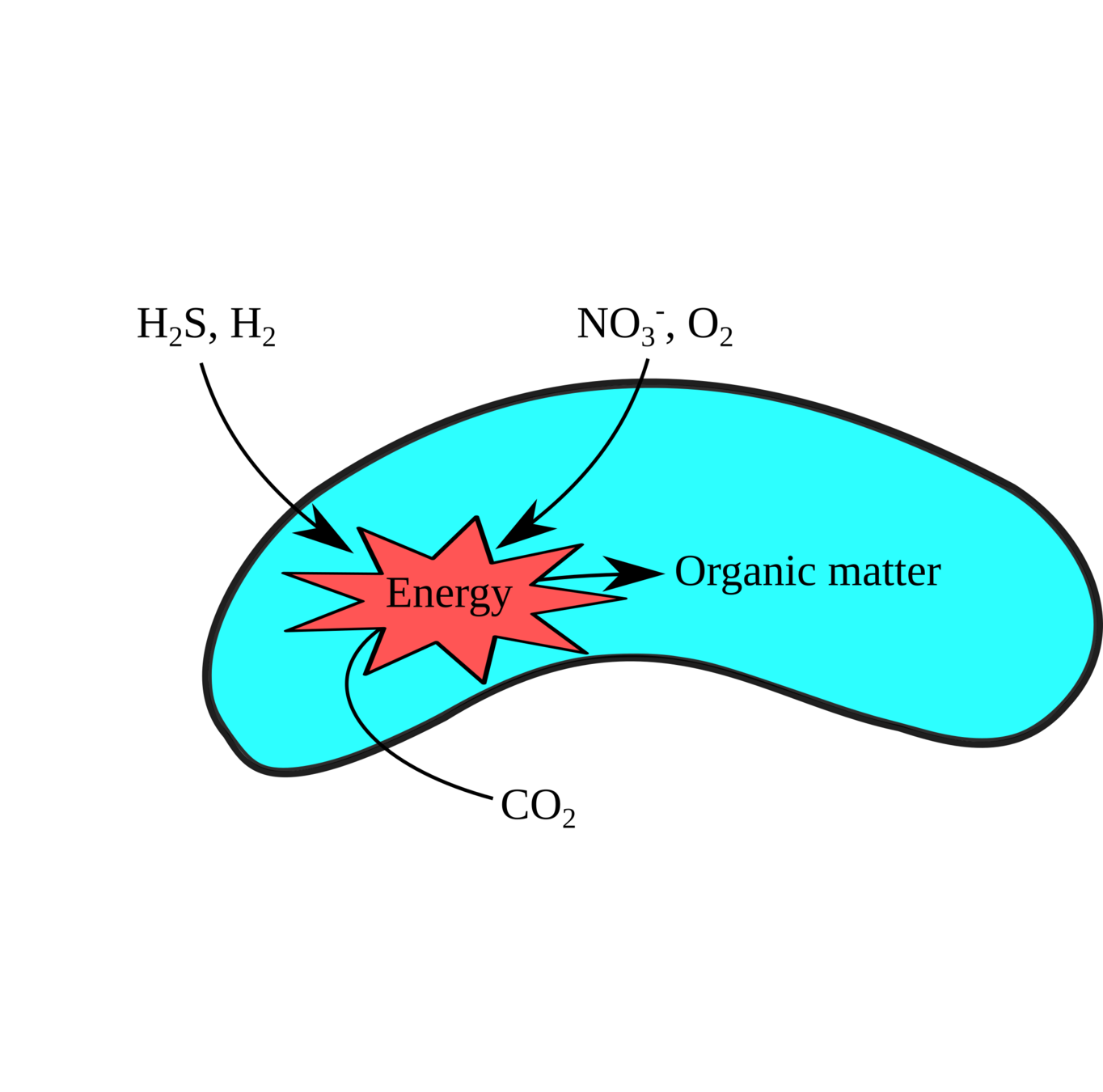
Pure culture physiology / genetics
Integrative techniques: A Microbe's Eye View


Metabolic enzyme function
Integrative techniques: A Microbe's Eye View



primary productivity, elemental cycling



primary productivity, elemental cycling

effect of anthropogenic pressure on ecosystems