REDDIE WP4
Analysis plans, Implementation and Results
Swedish group
Outline
- Statistical Analysis Plans and changes
- Implementation
- Results
Statistical Analysis Plans
| Inspired by | Comparison | Outcome | zenodo.org |
|---|---|---|---|
| LEADER | GLP1 vs DPP4 | Cardiovascular | https://doi.org/10.5281/zenodo.15311401 |
| EMPA-REG | SGLT2 vs DPP4 | Cardiovascular | https://doi.org/10.5281/zenodo.15320146 |
| LEAD2-3 | GLP1 vs DPP4 | Metabolic (HbA1c & Weight) | https://doi.org/10.5281/zenodo.15347461 |
Linked registries:
Swedish National Diabetes Register (NDR)
Swedish National Prescribed Drug Register (NPDR)
Swedish National Patient Register (NPR)
Swedish Cause of Death Register (CODR)
Swedish Longitudinal Integrated Database of Health Insurance and Labour Market Studies (LISA)
Study design / Methodology:
New user design - Persons in NDR with type 2 diabetes are included if/when they redeem relevant prescriptions within a predefined time window, and satisfy eligibility criteria emulating/inspired by the corresponding trial.
Follow-up until earliest occurrence of an outcome, death, emigration or end of trial, split into 6-month intervals.
Baseline covariates determined at inclusion looking back a predetermined period of time (any event within 180 days before inclusion for comorbidities, redemption of prescriptions within 180 days before inclusion for medications).
Time dependent covariates and outcome variables are summarized per interval (any event or prescription within interval, or, mean for continuous variables).
Efficacy is defined through absolute risk differences and risk ratios with bootstrap confidence intervals at prespecified follow-up time using the LTMLE framework with penalized outcome- and propensity score models, with penalty parameter determined by cross validation and without truncation. All-cause mortality is treated as competing event and emigration/end-of-trial as censoring event.
LEADER
EMPA-REG
LEAD 2-3
Primary Outcome:
MACE at 3.5 years
Primary Outcome:
MACE at 4 years
Primary Outcome(s):
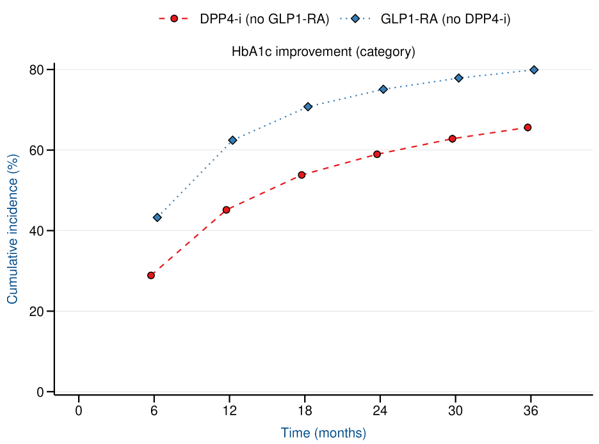
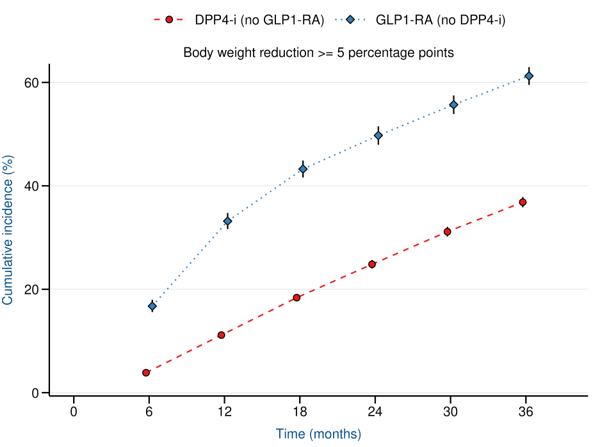
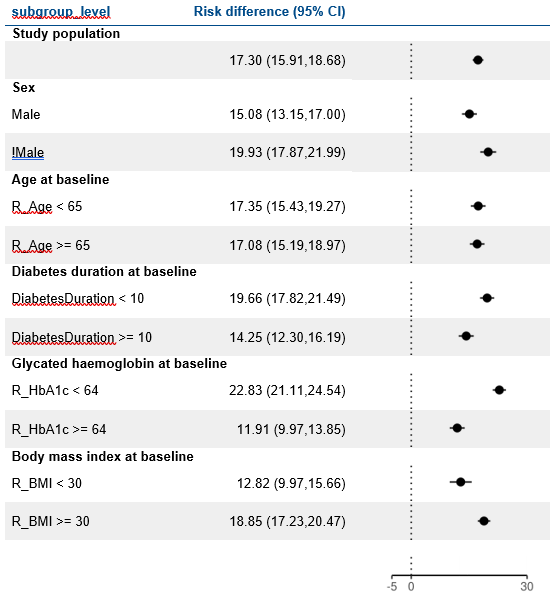
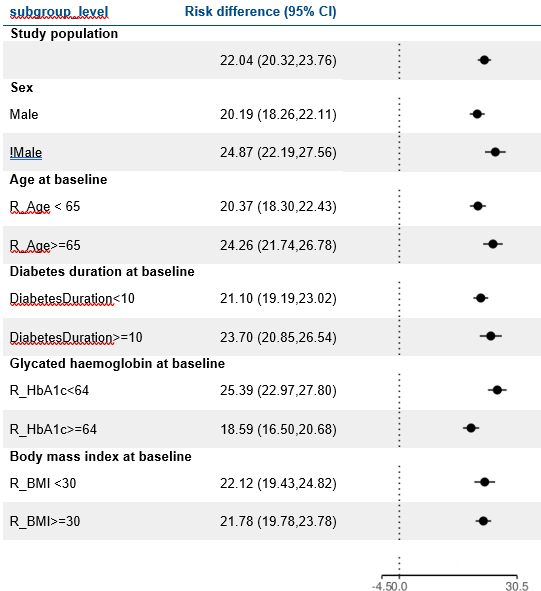
Reduction in HbA1c
Reduction in Weight
at 1 year
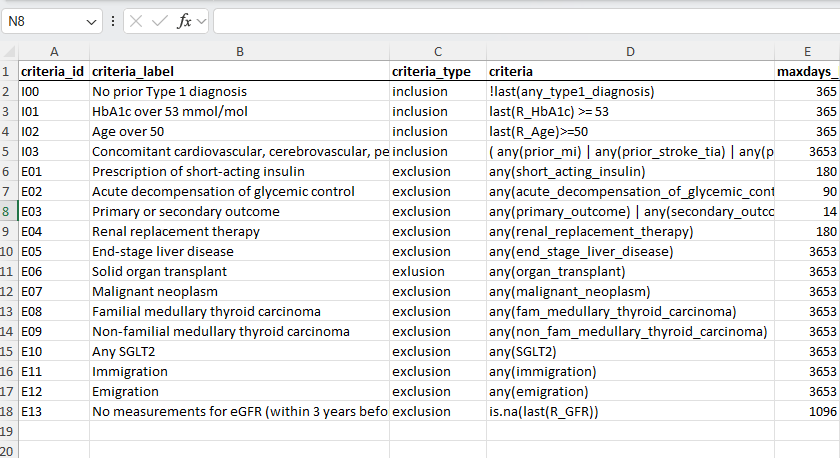
| Criteria | within time before BL |
|---|---|
| ✅No prior type 1 diagnosis | |
| ✅HbA1c over 53 mmol/mol | 1 year |
| ✅Age over 50 | 1 year |
| ✅Concomitant cardiovascular, cerebrovascular, peripheral vascular disease or renal disease or chronic heart failure OR age over 60 and other specified risk factors of vascular disease | 10 years |
| ❌Prescription of short-acting insulin | 180 days |
| ❌Acute decompensation of glycemic control | 90 days |
| ❌Primary or secondary outcome | 14 days |
| ❌Renal replacement therapy | 180 days |
| ❌End-stage liver disease | 10 years |
| ❌Solid organ transplant | 10years |
| ❌Malignant neoplasm | 10 years |
| ❌Familial medullary thyroid carcinoma | 10 years |
| ❌Non-familial medullary thyroid carcinoma | 10 years |
| ❌Any SGLT2 | 10 years |
| ❌Immigration | 10 years |
| ❌Emigration | 10 years |
| ❌No measurements for eGFR, |
|
| ❌ |
| Criteria | within time before BL |
|---|---|
| ✅No prior Type 1 diagnosis | |
| ✅HbA1c between 53-83 mmol/mol | 1 year |
| ✅Age over 18 | 1 year |
| ✅Established cardiovascular disease | 10 years |
| ✅eGFR over 30 | 2 years |
| ❌Liver disease | 10 years |
| ❌eGFR under 30 | 2 years |
| ❌Bariatric surgery | 10 years |
| ❌Blood dyscrasias | 20 years |
| ❌Cancer (excluding non-melanoma skin cancer) | 10 years |
| ❌Any systemic steroids | 90 days |
| ❌Uncontrolled endocrine disorder | 10 years |
| ❌Familial medullary thyroid carcinoma | 10 years |
| ❌Occurrence of primary or secondary outcomes | 60 days |
| ❌Any GLP1 | 90 days |
| ❌Immigration | 10 years |
| ❌Emigration | 10 years |
| ❌No measurements for eGFR, |
|
| Criteria | within time before BL |
|---|---|
| ✅No prior type 1 diagnosis | |
| ✅Existing measurement for HbA1c (or weight) | 28 days |
| ✅Age 18-90 years | 1 year |
| ✅BMI less than 42 kg./m2 | 90 days |
| ✅HbA1c between 53 and 97 mmol/mol | 90 days |
| ❌Insulin prescription redeemed | 90 days |
| ❌Prescription of systemic corticosteroids | 180 days |
| ❌Liver-disease diagnosis | 10 years |
| ❌Prescription of excluded GLP1 | 180 days |
| ❌Pregnancy | 1 year |
| ❌Any SGLT2 | 10 years |
| ❌Immigration | 10 years |
| ❌Emigration | 10 years |
| ❌No measurements for eGFR or Albuminuria | 2 years |
List of changes
-
Confidence intervals:
Influence Function
→ Bootstrap -
"Non-fatal" (alive after 30 days) MI and Stroke
→ Any MI and Stroke from NPR
(results also for non-fatal and including events from CoDR) -
Require measurements for HbA1c,
BMI(within 1 year) and eGFR,Albuminuria(within 2 years) for baseline values.
→ Only HbA1c and eGFR at baseline for larger cohorts and power. -
HbA1c, BMI, eGFR, Albuminuria, Smoking as time dependent covariates using LOCF to impute missing values in intervals.
→ Only comedication and comorbidities as time dependent covaritates -
LEAD2-3: Continuous, time dependent HbA1c outcome
→ Categorical improvement, cumulative incidence
Implementation
Trial meta data
Targets - pipeline
Report / Diagnostics
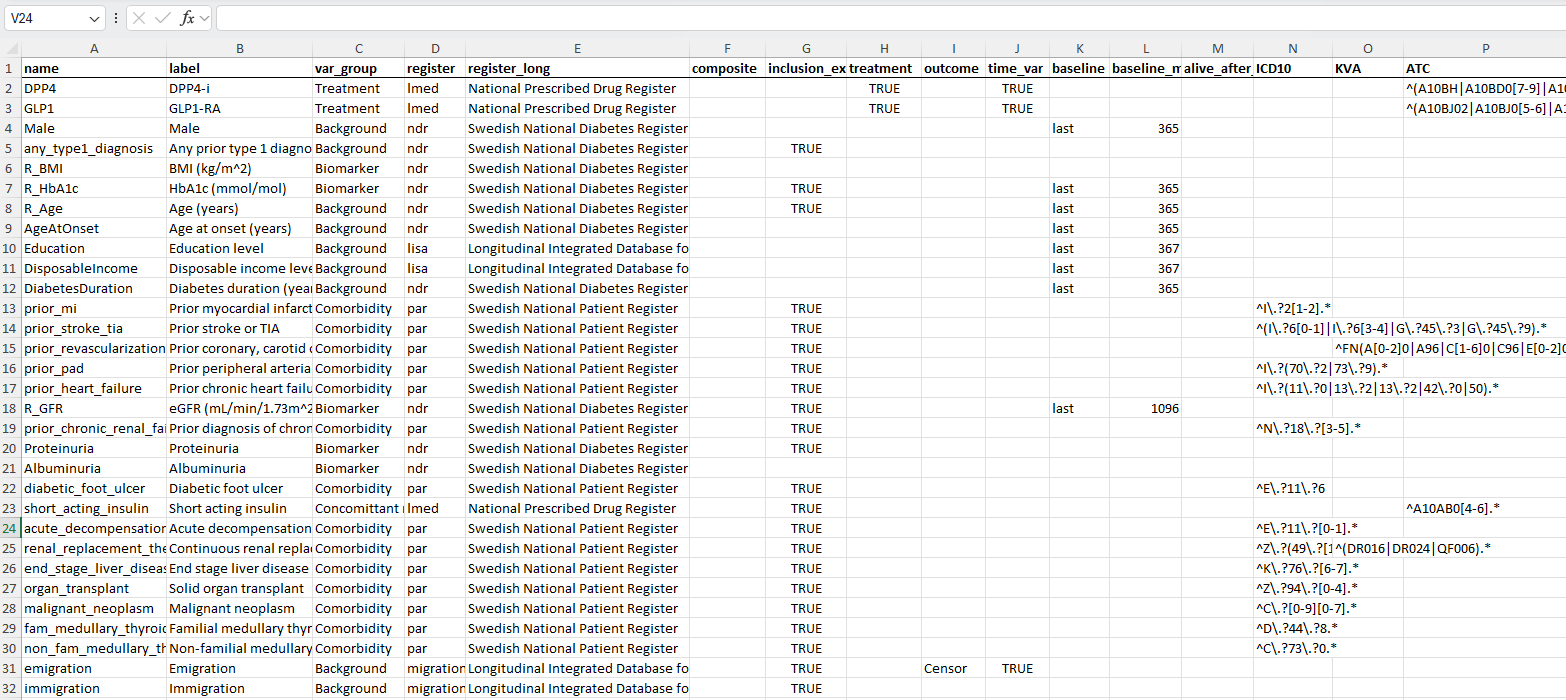
- Defines variables (register, ICD-codes, etc.)
- Specifies eligibility criteria
- Specifies analyses
- Flowchart
- Cohort characteristics
- Overview of results
- Diagnostic plots
- Reads in trial meta data
- Constructs cohorts and applies eligibility criteria
- Performs specified analyses
- Outputs a report and log
# Created by use_targets().
#
# LEADER trial emulation
# Jens Michelsen 2024-12-09
Sys.setenv(OMP_THREAD_LIMIT=2,R_DATATABLE_NUM_PROC_PERCENT=5)
# Load packages required to define the pipeline:
library(targets)
library(tarchetypes) # Load other packages as needed.
library(crew)
# Set target options:
tar_option_set(
# Packages that your targets need for their tasks.
packages = c("data.table","dtplyr","tidyverse","purrr","qs",
"qs2","officer","flextable","survival",
"rtmle"),
# default storage format. qs is fast.
format = "qs",
# set number of workers
controller = crew_controller_local(
workers = future::availableCores(),
options_local = crew_options_local(log_directory="Logs/crew/"),
# #options_metrics = crew_options_metrics(path = "/dev/stdout", seconds_interval = 1),
garbage_collection = TRUE
),
storage = "worker",
retrieval = "worker"
)
# if (tar_active()) {
# log_start(
# path = "main_process.txt",
# seconds = 1
# )
# }
# Run the R scripts in the R/ folder with your custom functions:
tar_source("R")
# Target list
list(
#-----------------------------------------------------------------------------
# Define variables for study start and study end
tar_target(
name = study_start,
command = lubridate::as_date("2012-01-01")
),
tar_target(
name = study_end,
command = lubridate::as_date("2022-12-31")
),
#-----------------------------------------------------------------------------
# Define data sources to be used.
tar_target(
name = ndr_file, # National Diabetes Register
command = "/safe/data/REDDIE/Data/Clean/ndr_plus.qs",
format = "file"
),
tar_target(
name = lmed_file, # Prescribed Drug register
command = "/safe/data/REDDIE/Data/Import/lmed.qs",
format = "file"
),
tar_target(
name = par_file, # Patient in-/out care
command = "/safe/data/REDDIE/Data/Clean/par_plus.qs",
format = "file"
),
tar_target(
name = dors_file, # Cause of death register
command = "/safe/data/REDDIE/Data/Import/dors.qs",
format = "file"
),
tar_target(
name = lev_base_file, # Basic population data (Sex/Birth date/etc)
command = "/safe/data/REDDIE/Data/Import/lev_base.qs",
format = "file"
),
tar_target(
name = migrations_file, # Patient in-/out care
command = "/safe/data/REDDIE/Data/Import/migrations.qs",
format = "file"
),
tar_target(
name = lisa_file, # Patient in-/out care
command = "/safe/data/REDDIE/Data/Clean/lisa_plus.qs",
format = "file"
),
#-----------------------------------------------------------------------------
# load xlsx-file with variable meta data (incl. ICD, KVA and ATC codes)
tar_target(
name = variable_meta_data_xlsx,
command = "variable_meta_data.xlsx",
format = "file",
cue = tar_cue(mode = "thorough")
),
tar_target(
name = variable_lookup,
command = openxlsx::read.xlsx(variable_meta_data_xlsx,
sheet = "lookup",
rowNames=TRUE),
cue = tar_cue(mode = "thorough")
),
#-----------------------------------------------------------------------------
# Extract a data set containing id's of people with type-2 diabetes
tar_target(
name = type2_cohort,
command = get_type2_cohort(ndr_file, lev_base_file)
),
#-----------------------------------------------------------------------------
# collect data on GLP1/DPP4, define initial treatment arms,
# reduce data set to treatment cohort,
#
tar_target(
name = treatment_cohort,
command = get_treatment_cohort(variable_lookup = variable_lookup,
type2_cohort = type2_cohort,
lmed_file = lmed_file,
study_start = study_start,
study_end = study_end)
),
#-----------------------------------------------------------------------------
#### Now store reduced, cohort-specific, versions of ndr, lmed, dors, lev
#### for speed and memory purposes
tar_target(
name = ndr,
command = left_join(treatment_cohort,qread(ndr_file),by="LopNr")
),
tar_target(
name = lmed,
command = left_join(treatment_cohort,qread(lmed_file),by="LopNr")
),
tar_target(
name = par,
command = left_join(treatment_cohort,qread(par_file),by="LopNr")
),
tar_target(
name = dors,
command = left_join(treatment_cohort,qread(dors_file),by="LopNr")
),
tar_target(
name = migrations,
command = left_join(treatment_cohort,qread(migrations_file),by="LopNr")
),
tar_target(
name = lisa,
command = left_join(treatment_cohort,qread(lisa_file),by="LopNr")
),
#-----------------------------------------------------------------------------
#### get inclusion and exclusion criteria
tar_target(
name = incl_excl,
command = openxlsx::read.xlsx(variable_meta_data_xlsx,
sheet = "inclusion_exclusion"),
cue = tar_cue(mode = "thorough")
),
#-----------------------------------------------------------------------------
#### and create study cohort
tar_target(
name = study_cohort_with_criteria_values, # can be used later for flow chart
command = get_study_cohort(treatment_cohort,
variable_lookup, incl_excl,
ndr=ndr, lmed=lmed, par=par, dors=dors,
migrations=migrations, lisa=lisa),
cue = tar_cue("thorough")
),
tar_target(
name = study_cohort,
command = filter(study_cohort_with_criteria_values,included) |>
select(LopNr,index,treat,study_start,study_end),
cue = tar_cue("thorough")
),
#-----------------------------------------------------------------------------
# Collect baseline- and timedependent data for study cohort
tar_target(
name = baseline_data,
command = get_baseline_data(study_cohort,
variable_lookup,
ndr=ndr, par=par, lmed=lmed, lisa=lisa)
),
tar_target(
name = timevar_data,
command = get_timevar_data(study_cohort,
variable_lookup,
ndr=ndr, par=par, lmed=lmed, dors, migrations)
),
#-----------------------------------------------------------------------------
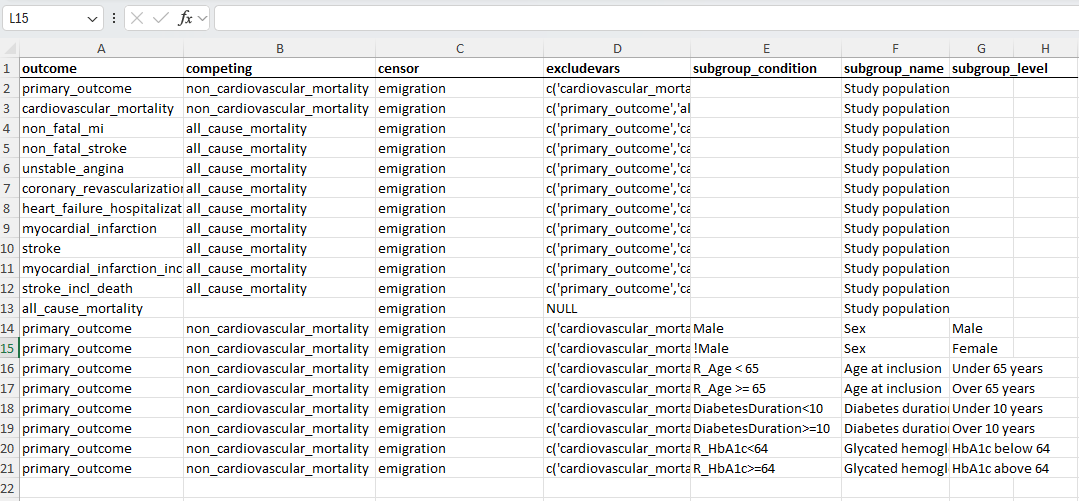
# Define specifications for the different analyses (from metadata_file)
tar_target(
name = analysis_specs,
command = openxlsx::read.xlsx(variable_meta_data_xlsx,
sheet = "outcomes"),
cue = tar_cue(mode = "thorough")
),
tar_target(
name = args,
command = split(analysis_specs,seq(nrow(analysis_specs))),
iteration = "list"
),
#-----------------------------------------------------------------------------
# Compute LTMLE estimates
tar_target(
name = estimate,
command = fit_rtmle_args(args,
study_cohort, variable_lookup,
baseline_data, timevar_data,
bootstrap = TRUE,
M_boot = 200,
rnd_seed = 321),
pattern = map(args),
iteration = "list"#,
#error = "null"
),
tar_target(
name = estimates,
command = dplyr::bind_rows(estimate)
),
#-----------------------------------------------------------------------------
# Create report
tar_target(
name = report,
command = get_report(estimates, study_cohort,
study_cohort_with_criteria_values,
variable_lookup, incl_excl,
baseline_data, timevar_data),
cue = tar_cue(mode="always")
),
# Create log
tar_target(
name = log,
command = print_log(),
cue = tar_cue(mode="always")
)
)
Trial meta data



Targets pipeline
# Created by use_targets().
#
# LEADER trial emulation
# Jens Michelsen 2024-12-09
Sys.setenv(OMP_THREAD_LIMIT=2,R_DATATABLE_NUM_PROC_PERCENT=5)
# Load packages required to define the pipeline:
library(targets)
library(tarchetypes) # Load other packages as needed.
library(crew)
# Set target options:
tar_option_set(
# Packages that your targets need for their tasks.
packages = c("data.table","dtplyr","tidyverse","purrr","qs",
"qs2","officer","flextable","survival",
"rtmle"),
# default storage format. qs is fast.
format = "qs",
# set number of workers
controller = crew_controller_local(
workers = future::availableCores(),
options_local = crew_options_local(log_directory="Logs/crew/"),
# #options_metrics = crew_options_metrics(path = "/dev/stdout", seconds_interval = 1),
garbage_collection = TRUE
),
storage = "worker",
retrieval = "worker"
)
# if (tar_active()) {
# log_start(
# path = "main_process.txt",
# seconds = 1
# )
# }
# Run the R scripts in the R/ folder with your custom functions:
tar_source("R")
# Target list
list(
#-----------------------------------------------------------------------------
# Define variables for study start and study end
tar_target(
name = study_start,
command = lubridate::as_date("2012-01-01")
),
tar_target(
name = study_end,
command = lubridate::as_date("2022-12-31")
),
#-----------------------------------------------------------------------------
# Define data sources to be used.
tar_target(
name = ndr_file, # National Diabetes Register
command = "/safe/data/REDDIE/Data/Clean/ndr_plus.qs",
format = "file"
),
tar_target(
name = lmed_file, # Prescribed Drug register
command = "/safe/data/REDDIE/Data/Import/lmed.qs",
format = "file"
),
tar_target(
name = par_file, # Patient in-/out care
command = "/safe/data/REDDIE/Data/Clean/par_plus.qs",
format = "file"
),
tar_target(
name = dors_file, # Cause of death register
command = "/safe/data/REDDIE/Data/Import/dors.qs",
format = "file"
),
tar_target(
name = lev_base_file, # Basic population data (Sex/Birth date/etc)
command = "/safe/data/REDDIE/Data/Import/lev_base.qs",
format = "file"
),
tar_target(
name = migrations_file, # Patient in-/out care
command = "/safe/data/REDDIE/Data/Import/migrations.qs",
format = "file"
),
tar_target(
name = lisa_file, # Patient in-/out care
command = "/safe/data/REDDIE/Data/Clean/lisa_plus.qs",
format = "file"
),
#-----------------------------------------------------------------------------
# load xlsx-file with variable meta data (incl. ICD, KVA and ATC codes)
tar_target(
name = variable_meta_data_xlsx,
command = "variable_meta_data.xlsx",
format = "file",
cue = tar_cue(mode = "thorough")
),
tar_target(
name = variable_lookup,
command = openxlsx::read.xlsx(variable_meta_data_xlsx,
sheet = "lookup",
rowNames=TRUE),
cue = tar_cue(mode = "thorough")
),
#-----------------------------------------------------------------------------
# Extract a data set containing id's of people with type-2 diabetes
tar_target(
name = type2_cohort,
command = get_type2_cohort(ndr_file, lev_base_file)
),
#-----------------------------------------------------------------------------
# collect data on GLP1/DPP4, define initial treatment arms,
# reduce data set to treatment cohort,
#
tar_target(
name = treatment_cohort,
command = get_treatment_cohort(variable_lookup = variable_lookup,
type2_cohort = type2_cohort,
lmed_file = lmed_file,
study_start = study_start,
study_end = study_end)
),
#-----------------------------------------------------------------------------
#### Now store reduced, cohort-specific, versions of ndr, lmed, dors, lev
#### for speed and memory purposes
tar_target(
name = ndr,
command = left_join(treatment_cohort,qread(ndr_file),by="LopNr")
),
tar_target(
name = lmed,
command = left_join(treatment_cohort,qread(lmed_file),by="LopNr")
),
tar_target(
name = par,
command = left_join(treatment_cohort,qread(par_file),by="LopNr")
),
tar_target(
name = dors,
command = left_join(treatment_cohort,qread(dors_file),by="LopNr")
),
tar_target(
name = migrations,
command = left_join(treatment_cohort,qread(migrations_file),by="LopNr")
),
tar_target(
name = lisa,
command = left_join(treatment_cohort,qread(lisa_file),by="LopNr")
),
#-----------------------------------------------------------------------------
#### get inclusion and exclusion criteria
tar_target(
name = incl_excl,
command = openxlsx::read.xlsx(variable_meta_data_xlsx,
sheet = "inclusion_exclusion"),
cue = tar_cue(mode = "thorough")
),
#-----------------------------------------------------------------------------
#### and create study cohort
tar_target(
name = study_cohort_with_criteria_values, # can be used later for flow chart
command = get_study_cohort(treatment_cohort,
variable_lookup, incl_excl,
ndr=ndr, lmed=lmed, par=par, dors=dors,
migrations=migrations, lisa=lisa),
cue = tar_cue("thorough")
),
tar_target(
name = study_cohort,
command = filter(study_cohort_with_criteria_values,included) |>
select(LopNr,index,treat,study_start,study_end),
cue = tar_cue("thorough")
),
#-----------------------------------------------------------------------------
# Collect baseline- and timedependent data for study cohort
tar_target(
name = baseline_data,
command = get_baseline_data(study_cohort,
variable_lookup,
ndr=ndr, par=par, lmed=lmed, lisa=lisa)
),
tar_target(
name = timevar_data,
command = get_timevar_data(study_cohort,
variable_lookup,
ndr=ndr, par=par, lmed=lmed, dors, migrations)
),
#-----------------------------------------------------------------------------
# Define specifications for the different analyses (from metadata_file)
tar_target(
name = analysis_specs,
command = openxlsx::read.xlsx(variable_meta_data_xlsx,
sheet = "outcomes"),
cue = tar_cue(mode = "thorough")
),
tar_target(
name = args,
command = split(analysis_specs,seq(nrow(analysis_specs))),
iteration = "list"
),
#-----------------------------------------------------------------------------
# Compute LTMLE estimates
tar_target(
name = estimate,
command = fit_rtmle_args(args,
study_cohort, variable_lookup,
baseline_data, timevar_data,
bootstrap = TRUE,
M_boot = 200,
rnd_seed = 321),
pattern = map(args),
iteration = "list"#,
#error = "null"
),
tar_target(
name = estimates,
command = dplyr::bind_rows(estimate)
),
#-----------------------------------------------------------------------------
# Create report
tar_target(
name = report,
command = get_report(estimates, study_cohort,
study_cohort_with_criteria_values,
variable_lookup, incl_excl,
baseline_data, timevar_data),
cue = tar_cue(mode="always")
),
# Create log
tar_target(
name = log,
command = print_log(),
cue = tar_cue(mode="always")
)
)
Report / Diagnostics
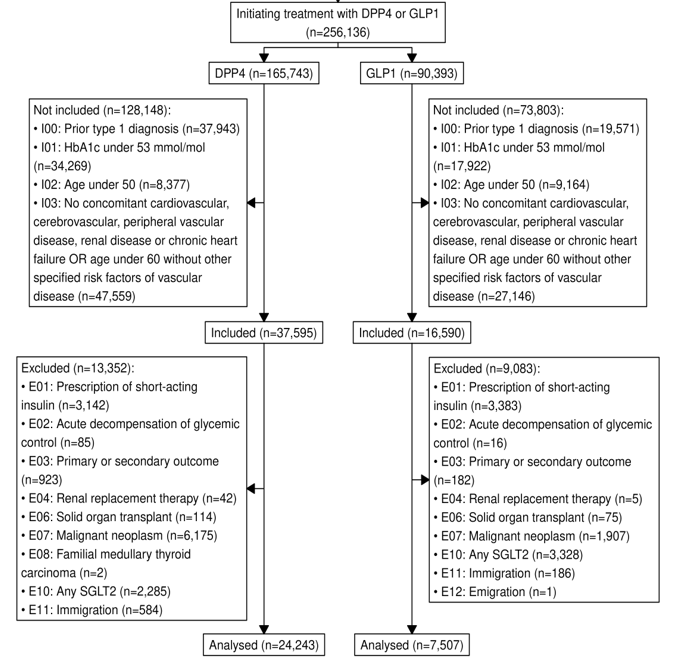
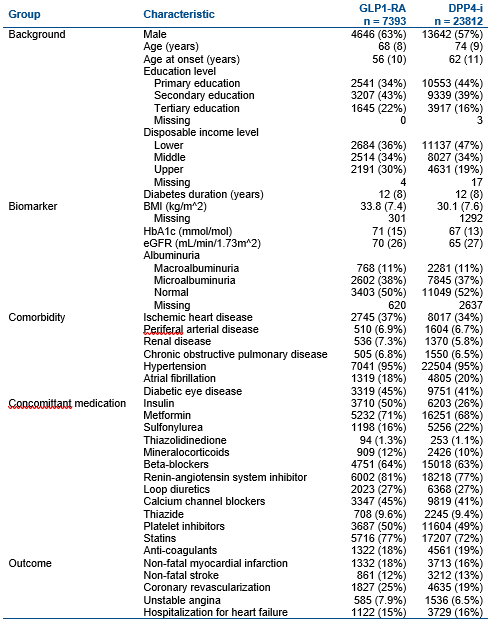
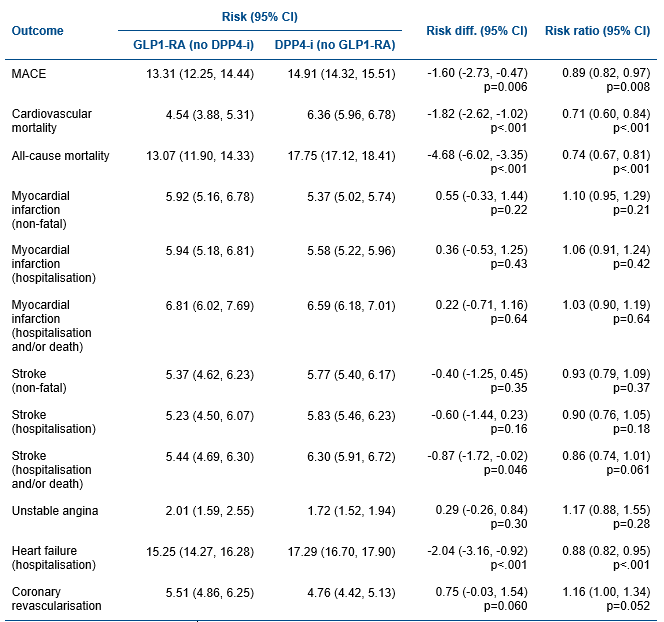
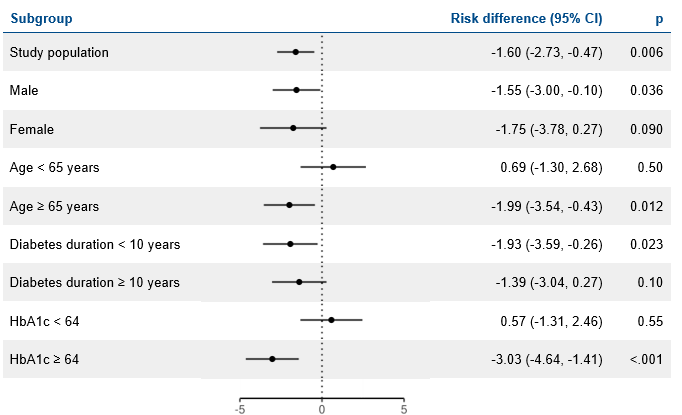
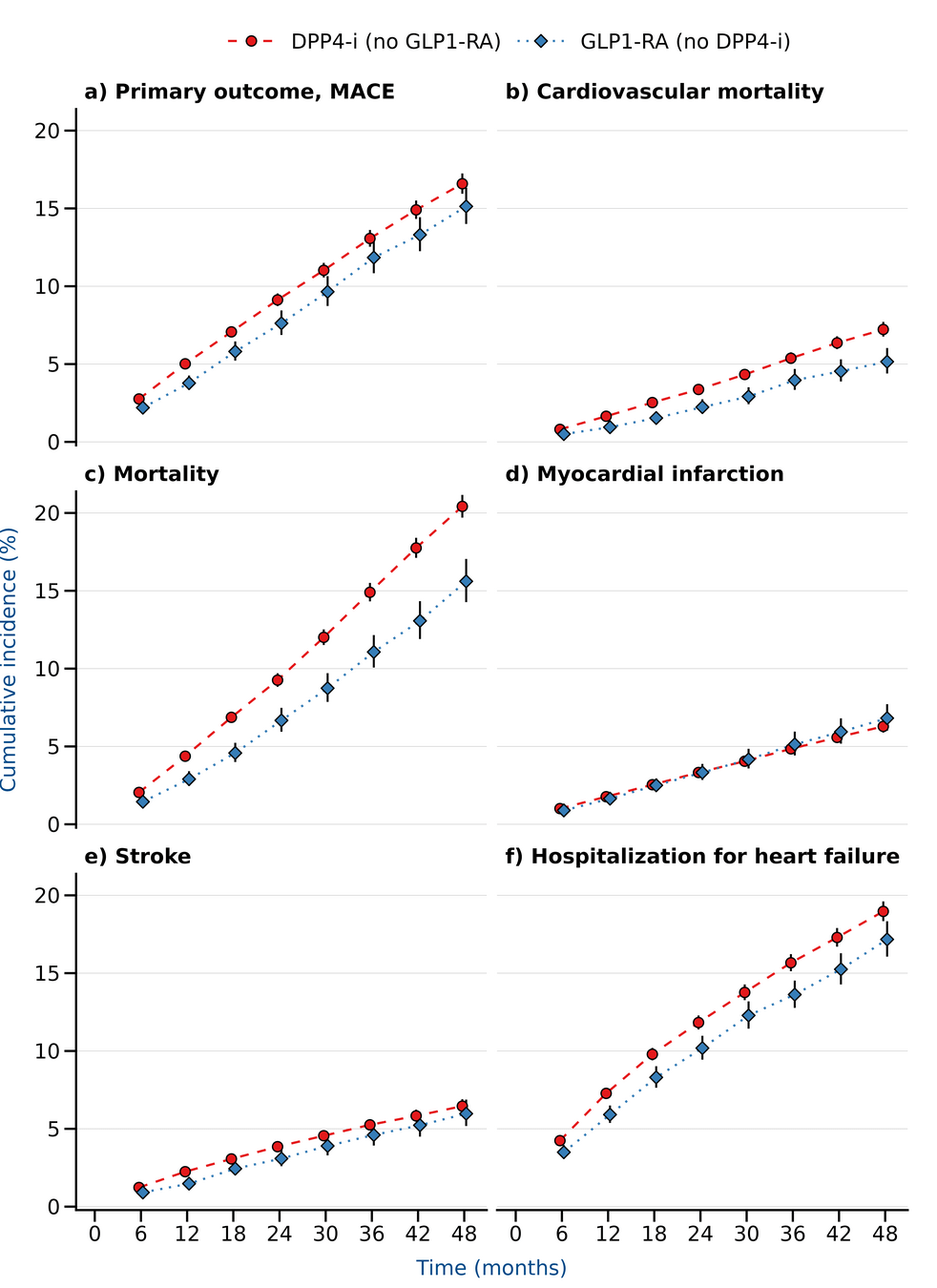
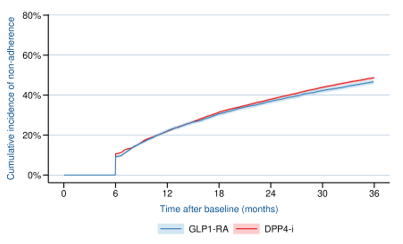
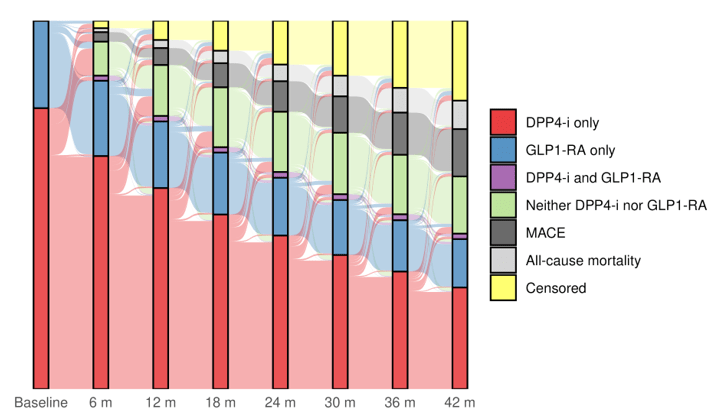
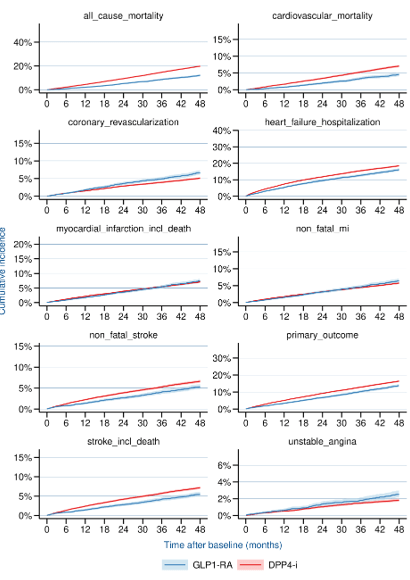
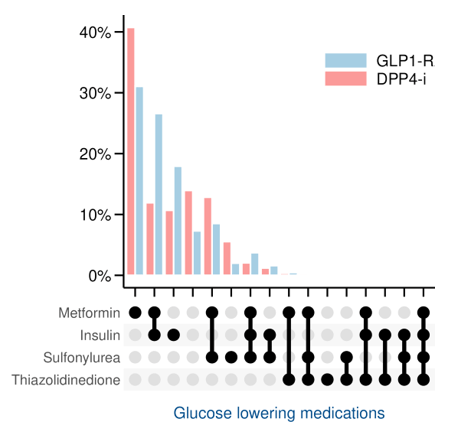
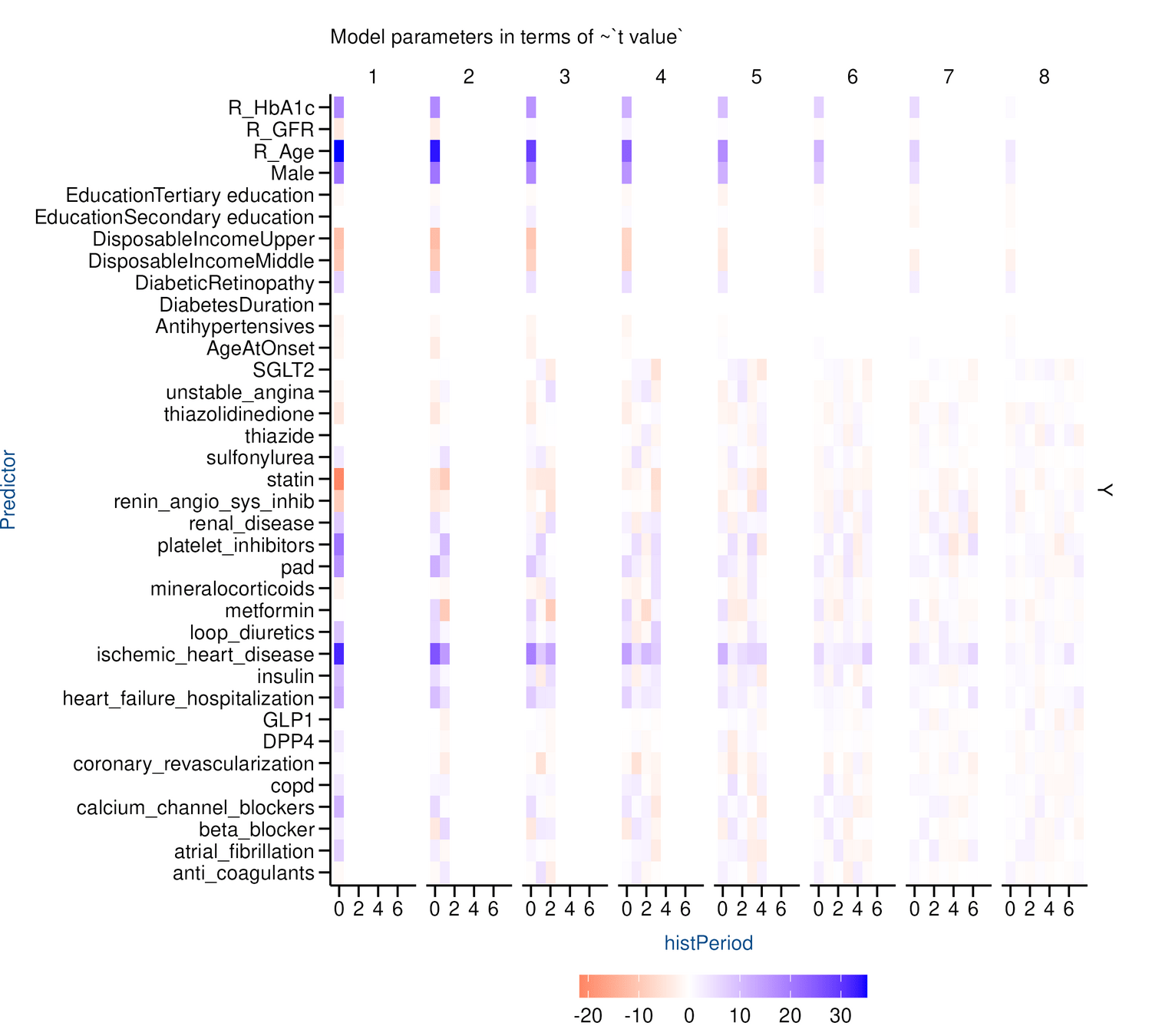

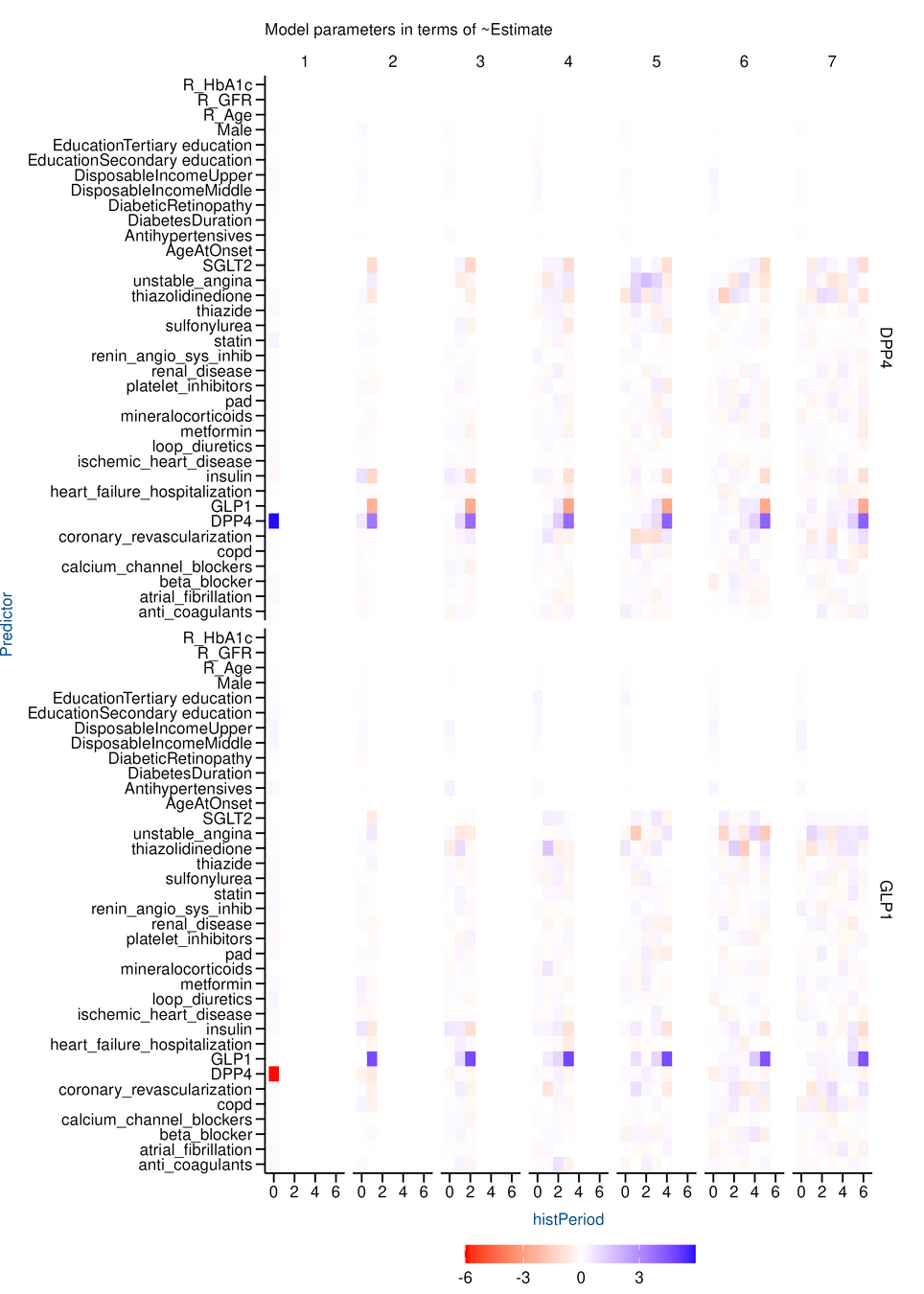
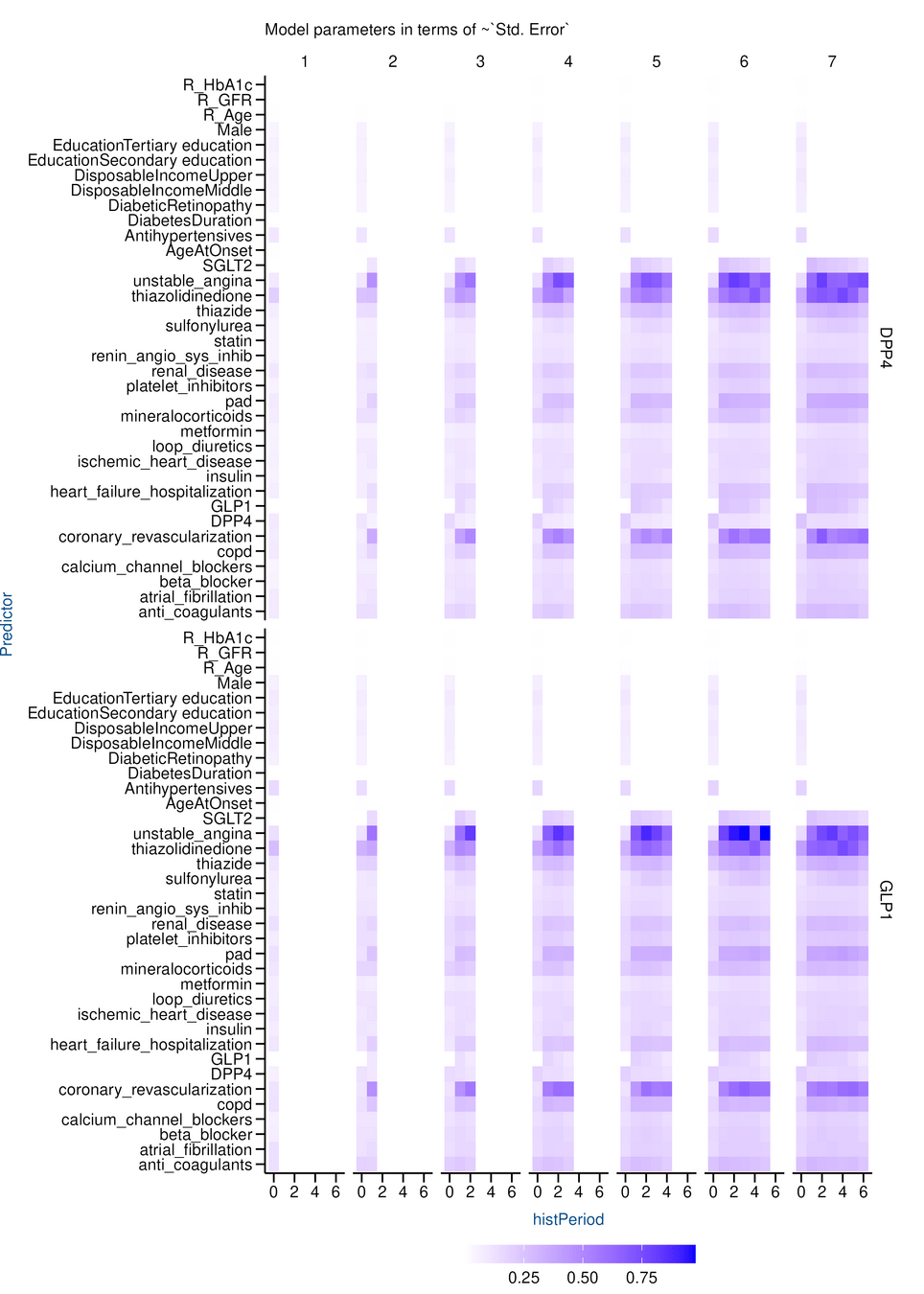
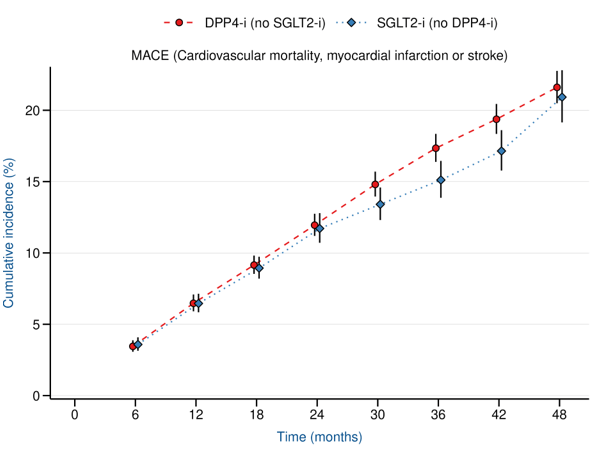
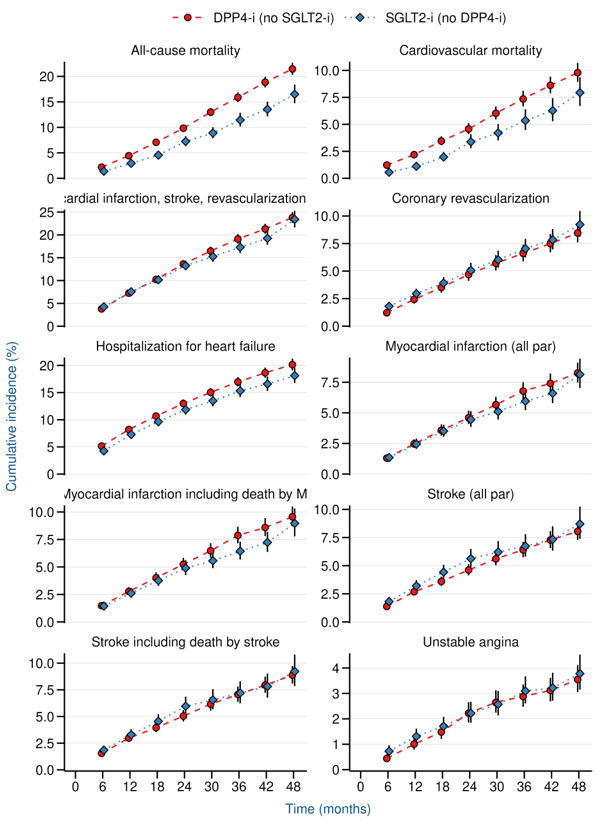
Results
LEADER


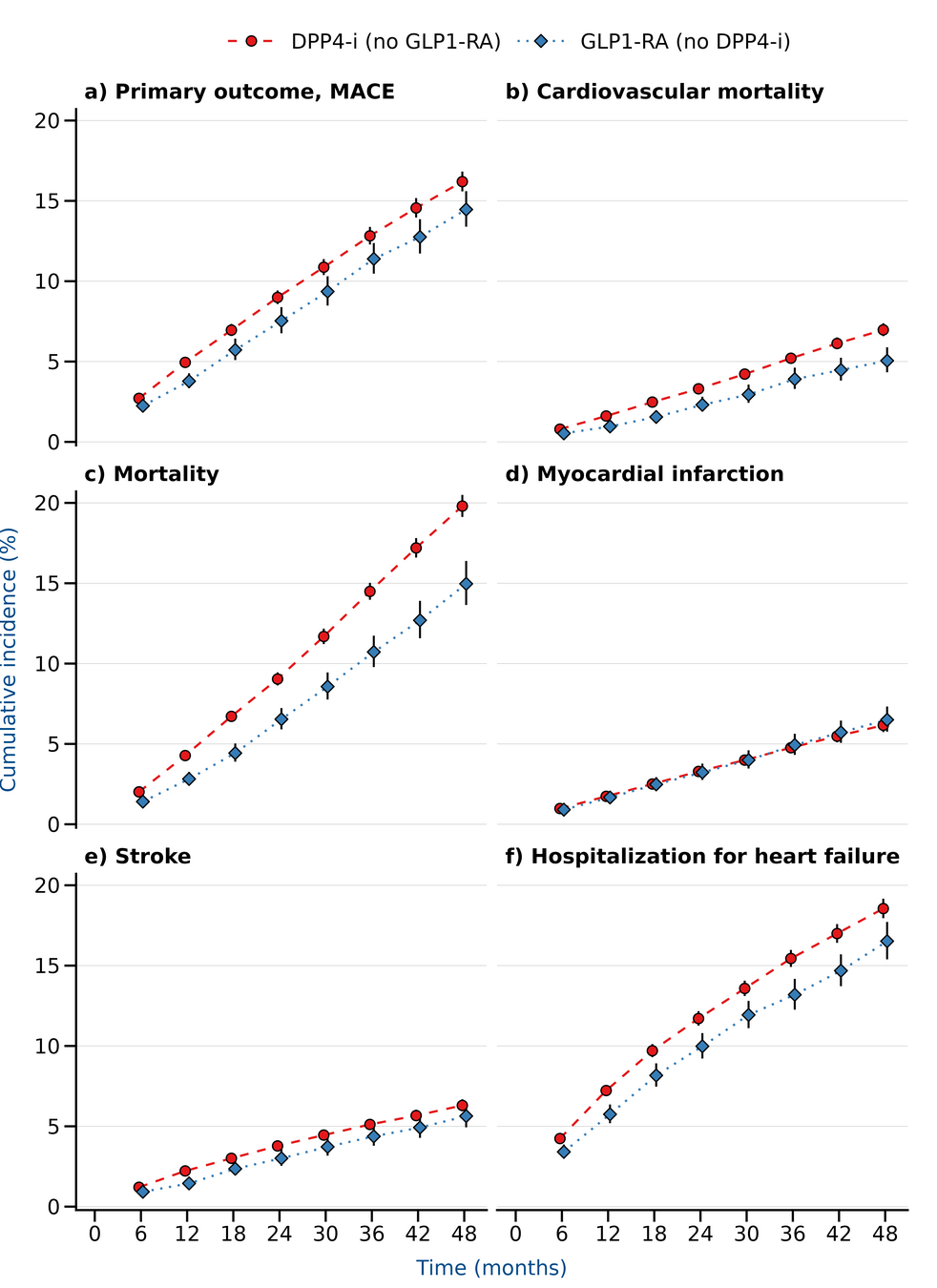

Only baseline HbA1c
Time dependent HbA1c
EMPA-REG
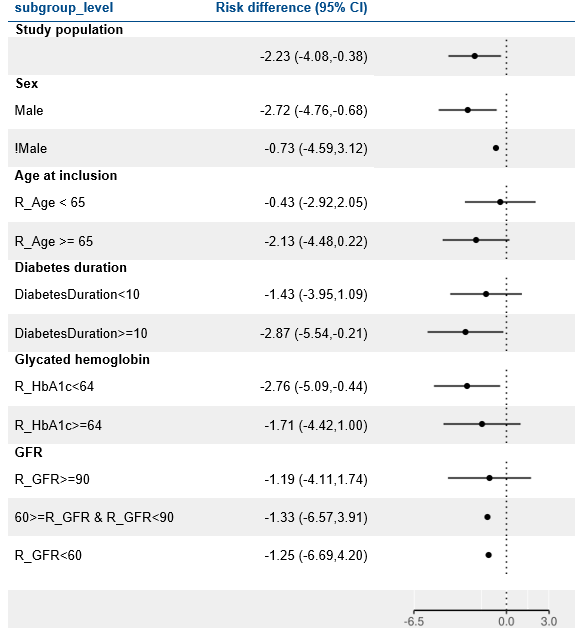
LEAD 2-3