Using the tools at GEDmatch:
the Basics for Finding Relatives and more
By Kitty Cooper, blogging at blog.kittycooper.com -
slides always at slides.com/kittycooper
This is the landing page for GEDmatch


CIick the link above for a video on how to register for GEDmatch
Go to the GEDmatch education page for many videos, no log in needed gedmatch.com/education/

GEDmatch was founded to let you compare GEDCOMs and autosomal DNA results from different companies with powerful analysis tools



A problem is that the various DNA testing companies test different SNP sets which are not the same although there is much overlapping
So GEDmatch developed a special template for comparing them !



My Home page at GEDmatch
Your dashboard (aka home) page has much help for new users plus messages about what's new on GEDmatch



When you log in your information and uploaded resources are listed on the left side of the page below the day's messages and the welcome
we call this the Dashboard Page
The bottom of your Dashboard Page
notice the CONTACT US button!

The free tools are listed in a panel on the right of the Dashboard


And they are listed with other tools in the top drop down menu

If you have just uploaded a kit there are a few things you can do while you wait for it to be ready to use (aka "tokenized")


See these slides of mine for how to use the admix tools: https://slides.com/kittycooper/gedmatch
You can play with the admixture tools, many have not been updated in years but they are still fun and different from your testing company


I am often asked about slimming
for the purpose of quick comparisons the heterozygous SNPs are removed because with one of each base, those SNPs match everyone

Next check use the "Are your parents related" tool as this can make it harder to figure out new relatives
Unless you are from an endogamous population or know your parents are cousins this will likely be negative


sample negative result
sample negative result

people from endogamous populations often find that their parents are distantly related

At the bottom right of your dashboard are the tools for family trees stored as GEDCOMs
Next Upload a GEDCOM of your Family Tree
You can download your family tree as a GEDCOM from Ancestry, MyHeritage, GENI, WIKItree or any genealogy program
you can also add your DNA kit to your profile at WIKItree
What is a GEDCOM -https://en.wikipedia.org/wiki/GEDCOM

I recommend uploading a privatized GEDCOM with about 10 generations of ancestors for your kit

Since a GEDCOM is a text file, you can edit it in a plain text editor like the notepad on a PC
I recommend changing the first names of anyone living to just LIVING
and removing birth month and day leaving the birth year


Another great free tool lets you look at the GEDcoms of your DNA matches who have uploaded them
GEDCOM + DNA matches


There can be a lot of information on the first page, the individual, in a gedcom

But typically we just click to see the pedigree

Top half of my Aunt's pedigree, the clickable little tree shows where a match to another GEDCOM has been confirmed
2 GEDCOMs comparison




The next thing to do is set up your tag groups
You need to know the kit numbers of your known relatives (easier once you can do a one to many)
The next page has a set of tabbed headings; click on Tag Group Management

I recommend setting up a group for each great grandparent line using the same colors you used at Ancestry or on your Leeds Chart
Also perhaps a group for close family and for localities see
https://blog.kittycooper.com/2017/03/gedmatch-tag-groups-plus-new-one-to-many/

Click on the green button Display/Edit tag group to get to this page
Once you have created a tag group. add the kit numbers of your known relatives at GEDmatch via the Manually Add Kits

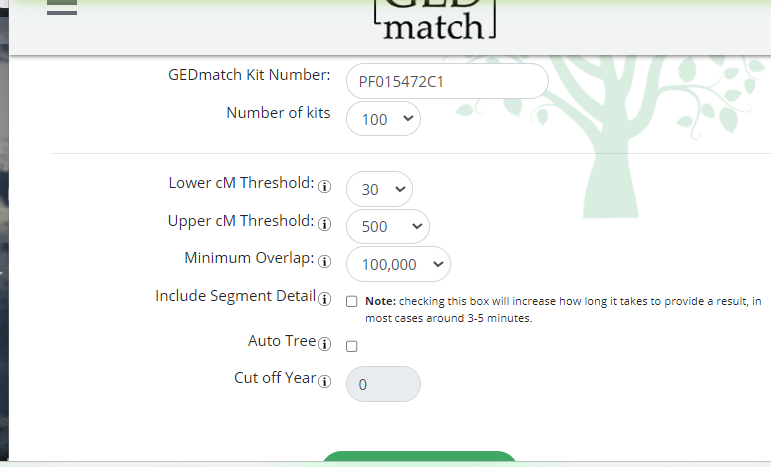
Once you have a GEDCOM uploaded and tag groups set up, it is time to use the most important tool for finding relatives: the One-To-Many

First make your selections on the the One-To-Many form. the defaults do not include tag groups so check that

I also prefer to raise the minimum cM to at least 10 and for endogamous people to 20 cM
The basic one-to-many shows the kits that match the kit number you give it with the overlap indicating how many SNPs are in the comparison, red means too few, and it shades down from there

Notice that the kit number is colored with its tag group
https://blog.kittycooper.com/2017/04/using-gedmatch-tag-groups/

If the kit is connected to a GEDCOM or to a tree at WIKItree that shows here
The haplogroup information is entered by the user, notice that it can be connected to the mitoYDNA web site ....
This old post explains the column headings like Gen
Every column is sortable, I often sort by largest segment, see
https://blog.kittycooper.com/2015/08/size-matters-for-matching-dna-segments/

Recently I had a close match with a name I did not recognize, so I clicked on her kit number to look at her one to many

I could tell immediately from the tag group colors of her matches that she was a Munson
If tag groups don't solve where a match is related try
the ICW function:
"People who match
one or both kits"

Comparing my Dad and my new Munson cousin

Some of the FREE tools that I find invaluable are highlighted

Checking my new cousin in the relationship probability tool by entering the shared cM

SEGMENT DATA
A matching segment of DNA is where a long run of SNPs are the same in two different kits. The problem is that the testing process cannot tell which side they are from. Thus the match can even be from a mix of the two sides, maternal and paternal.


Clicking the largest segment on the one to many brings you to this comparison form with the kits filled out
One to one with numbers and graphics (partial page)
My double third AJ cousin


One to one without the graphics, position only, min of 10 cM
My double third AJ cousin

One to one just graphics with my double third AJ cousin at min = 7cM


Full siblings always have some fully identical regions (FIRs) - where both strands of DNA got the same SNPs from their parents - those are the green bars
this is the graphic only, one-to-one, for my brother and myself

FIR only version of my comparison of my brother and my Ancestry kits

One to one on the X chromosome with my double third AJ cousin


New exciting tools typically come to Tier 1
The first thing to do is make a combined kit if you have tested at multiple companies



If you have tested at more than one company, since they may have tested some different SNPs, best to upload both and combine them into a super kit (then mark the base kits research) see
https://blog.kittycooper.com/2019/04/make-a-combined-dna-kit-for-yourself/
Visual comparison of my 23andme kit to my Ancestry kit

Tier 1 One to Many
You can check boxes next to multiple kits and then use the visulize button to use the multikit analysis tools


Clicking on the total shared brings up the relationship estimate

MULTI-KIT ANALYSIS (Tier 1)


You can do a matching segment search for just the selected kits
Alternately you can do a triangulation for just the selected kits




My Compact Chromosome Browser

The 3D chromosome browser includes multiple matrices, autosomal. X, and number of segments

FInd common ancestors with your DNA matches











Can software figure out the family tree from the shared DNA?
AUTO Kinship
The result has to be downloaded

After you download it and unzip it click on the folder Autokinship then on the HTML file autokinship to see a long page in your browser: first the clusters

My auto kinship clusters
one endogamous population grandparent, and another missing from the testing community


Since so many of my MUNSON cousins are tested ... what would autokinship came up with for that cluster 2 green box?

Lauritz + Josephine Monsen
Inga
Lawrence
Alfred
Christian
Pretty accurate. I have identified which child of my great great grandparents each match is descended from. Only the last one is not, he is actually descended from a sister of Josephine's
?
A closer look at my cousins with the one generational correction


?

Next is a matrix of how many cM they each share with each other

And finally there is a list of shared segments that met your criteria
Tim Janzen did a great Rootstech talk about these new features at

Shall we look at the auto tree for my endogamous cluster?

Here are the few trees created for cluster1 - the problem is not enough kits have linked trees on GEDmatch

Gugenheimer and Thannhauser are known ancestral lines, not Shadur
Phasing
- If you have at least one parent tested then you can separate what you got from each parent
- If the maternal or paternal phase can be determined the values will be assigned else the original unphased values will be kept


LAZARUS lets you create a kit for comparisons for a person who cannot be tested from their close relatives

There are many more fun Tier 1 tools you might try
Set the parameters for clustering, raising the low end will give clearer boxes