A Study of the ISOMAP Algorithm and Its Applications in Machine Learning
Lucas Oliveira David
Universidade Federal de São Carlos
December 2015
Introduction
Introduction
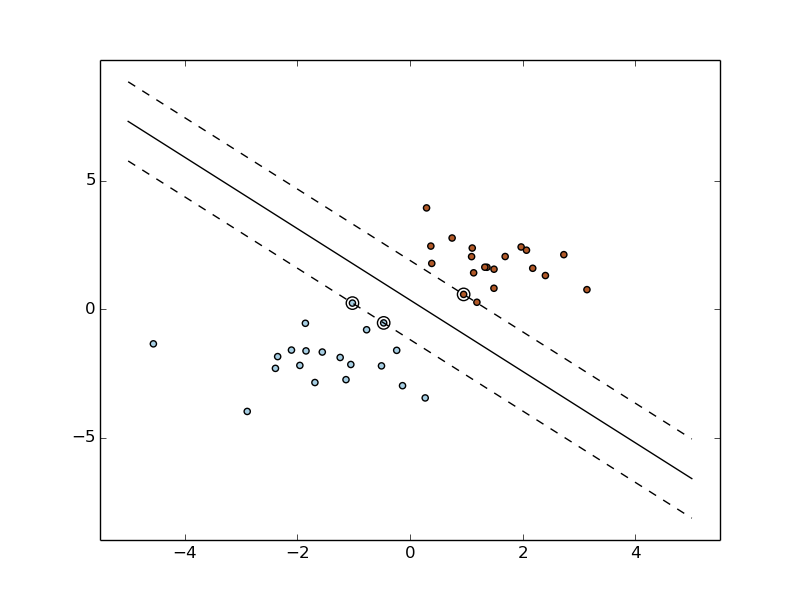
Machine Learning can help us with many tasks:
classification, estimation, data analysis and decision taking.
Data is an important piece of the learning process.
Visualization
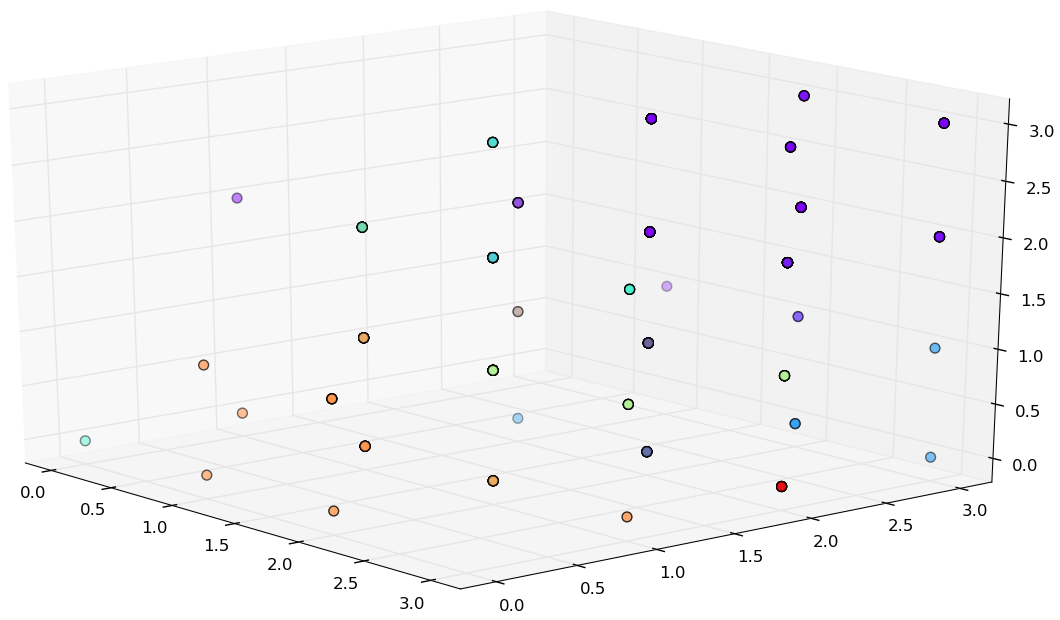

Original Dermatology data set
Reduced Dermatology data set
Memory and Processing Time Reduction
Original Spam data set
2048.88 KB
Reduced Spam data set
107.84 KB
Data Preprocessing
Original R data set
(linearly separable).
Reduced R data set,
(still linearly separable).
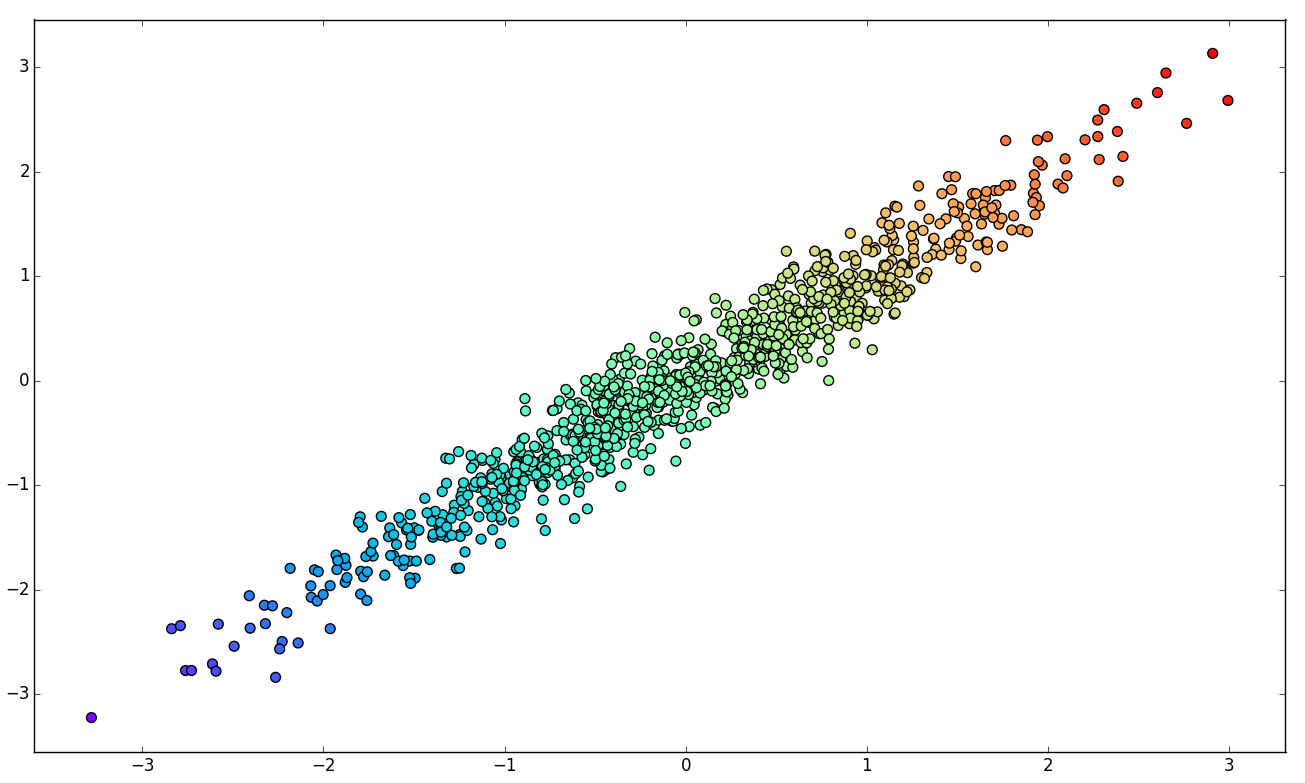
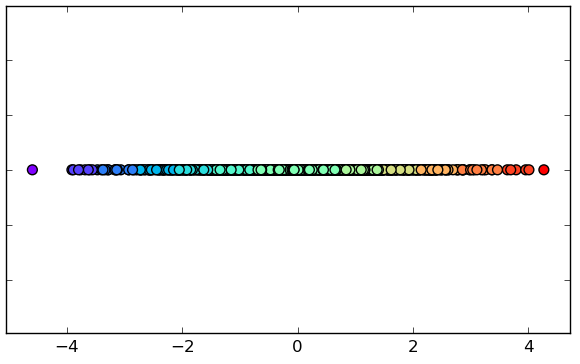
Linear Dimensionality Reduction
Linear DR
- Assumes linear distribution of data.
- Combines features to create new components that point out the direction in which the data varies.
- Sorts components by the variance.
- Removes components with "small" variance.
(e.g., PCA, MDS)
PCA
Principal Component Analysis
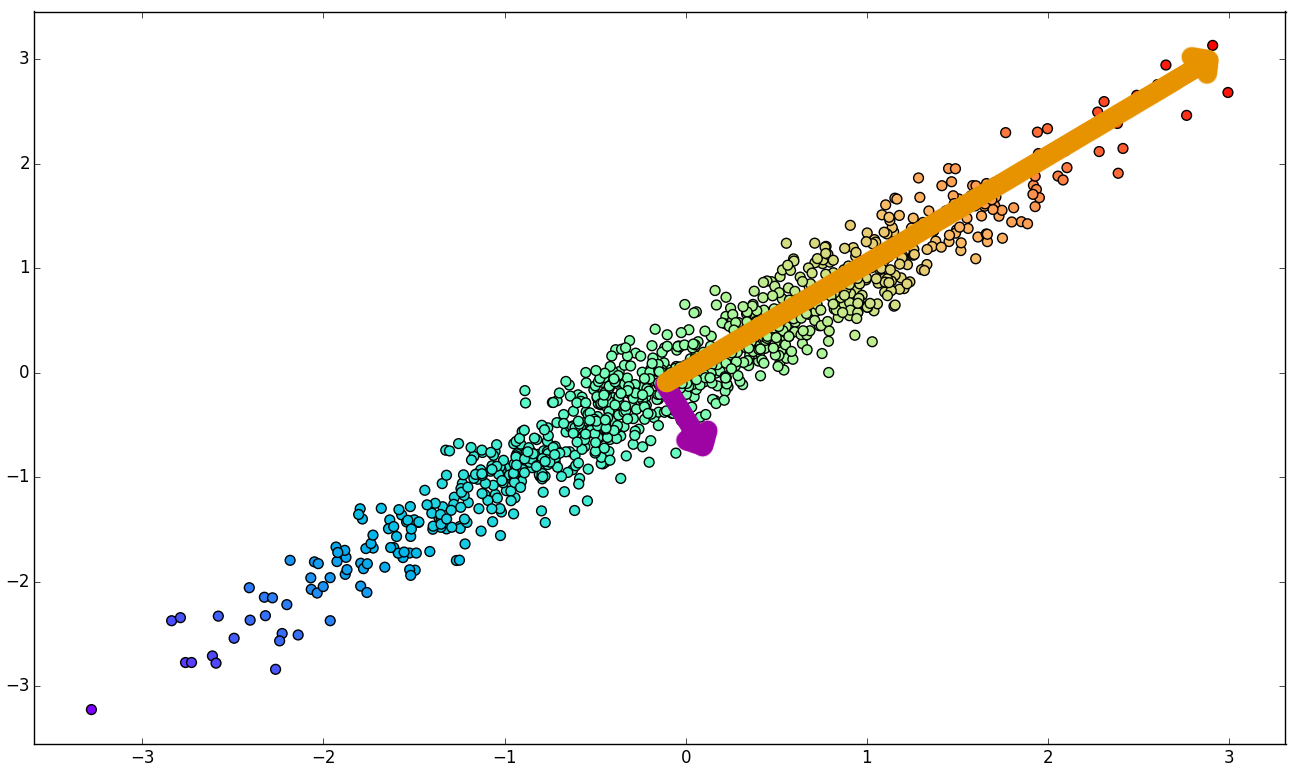

1. Original data set.
2. Principal Components of K.
3. Basis change.
4. Component elimination.
PCA
Feature Covariance Matrix.
V is a change of basis orthonormal matrix.
Use V' to transform samples from X to Y.
Find V', the base formed by the most important principal components.
SVD
MDS
Multidimensional Scaling
MDS
Pairwise distances.
Full space reconstruction.
Sort eigenvalues and eigenvectors by
the value of the eigenvalues.
Embed it!
Nonlinear Dimensionality Reduction
Nonlinear DR
(e.g., ISOMAP, Kernel PCA)

Q: what happens when data that follows a nonlinear distribution is reduced with linear methods?
A: very dissimilar samples become mixed as they are crushed onto lower dimensions.
Nonlinear DR
- Manifold Assumption.
- Explores manifolds' properties such as local linearity and neighborhood.
- Unfolding before reducing.
(e.g., ISOMAP, Kernel PCA)

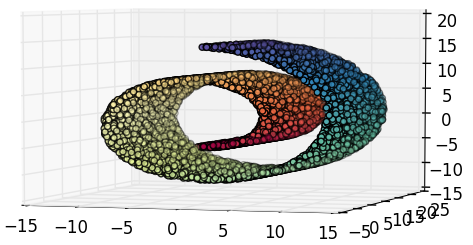
ISOMAP
Isometric Feature Mapping
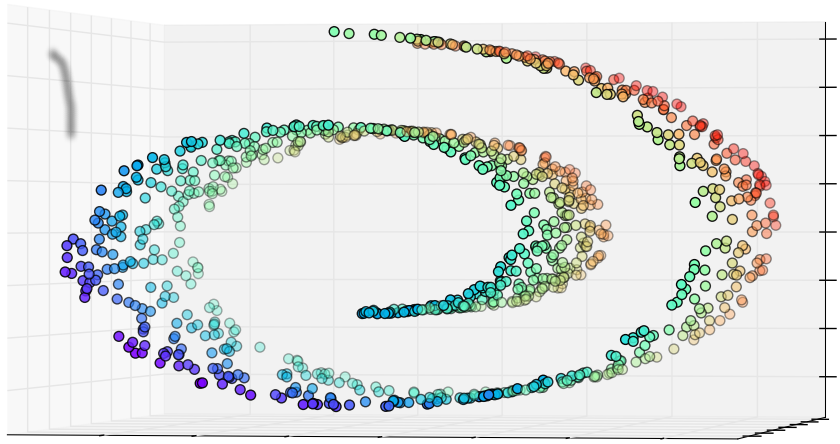
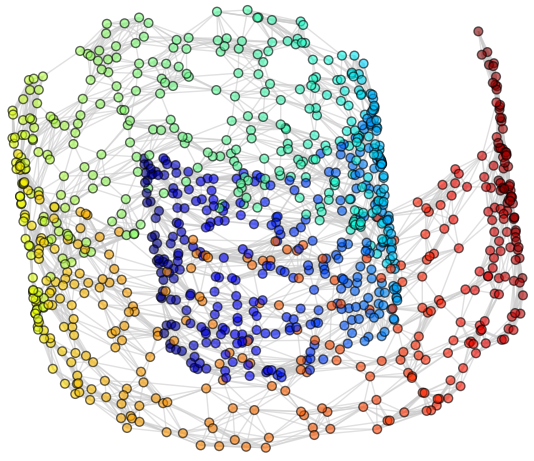
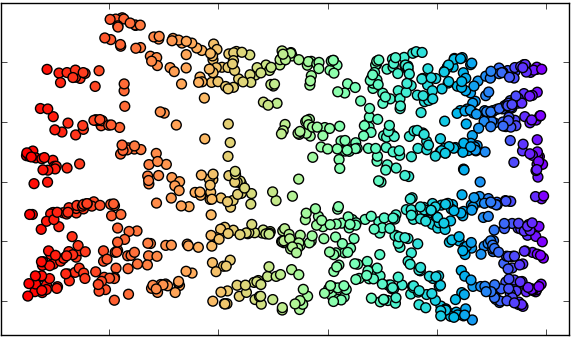
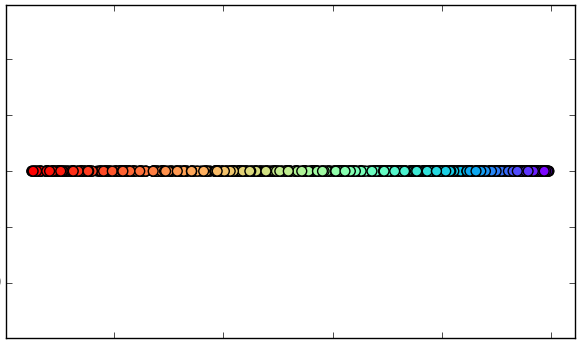
1. Original data set.
2. Compute neighborhood graph.
4.A. Reduction with MDS
(2 dimensions).
4.B. Reduction with MDS
(1 dimension).
ISOMAP
def isomap(data_set, n_components=2,
k=10, epsilon=1.0, use_k=True,
path_method='dijkstra'):
# Compute neighborhood graph.
delta = nearest_neighbors(delta, k if use_k else epsilon)
# Compute geodesic distances.
if path_method == 'dijkstra':
delta = all_pairs_dijkstra(delta)
else:
delta = floyd_warshall(delta)
# Embed the data.
embedding = mds(delta, n_components)
return embeddingISOMAP
In Practice
ISOMAP In Practice
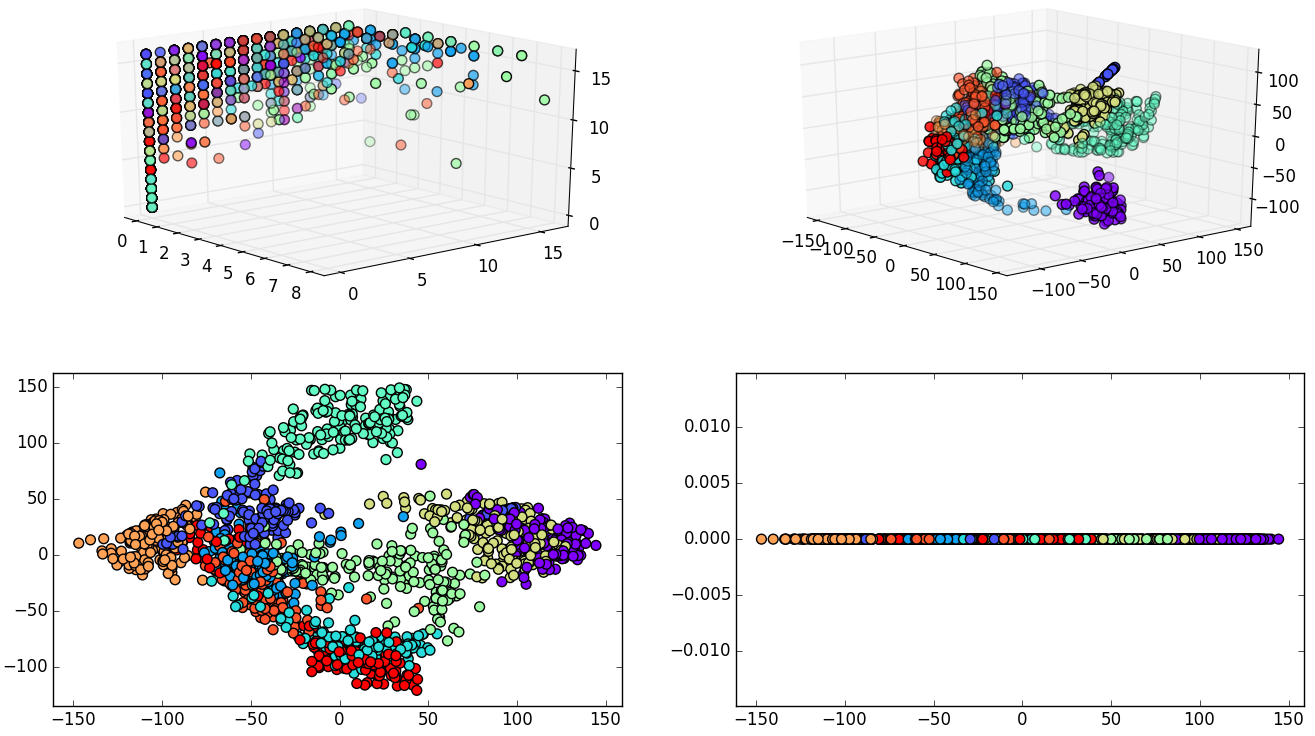
Experiment 1: Digits, 1797 samples and 64 dimensions.
Grid search was performed using a Support Vector Classifier.
| 64 dimensions | 10 dimensions | |
|---|---|---|
| Accuracy | 98% | 96% |
| Data size | 898.5 KB | 140.39 KB |
| Grid time | 11.12 sec | 61.66 sec |
| Best parameters | 'kernel': 'rbf', 'gamma': 0.001, 'C': 10 | 'kernel': 'linear', 'C': 1 |
- 54 dimensions eliminated with only 2% of accuracy loss.
- It is also important to mention that grid search time has increased when the data dimensionality was reduced.
ISOMAP In Practice
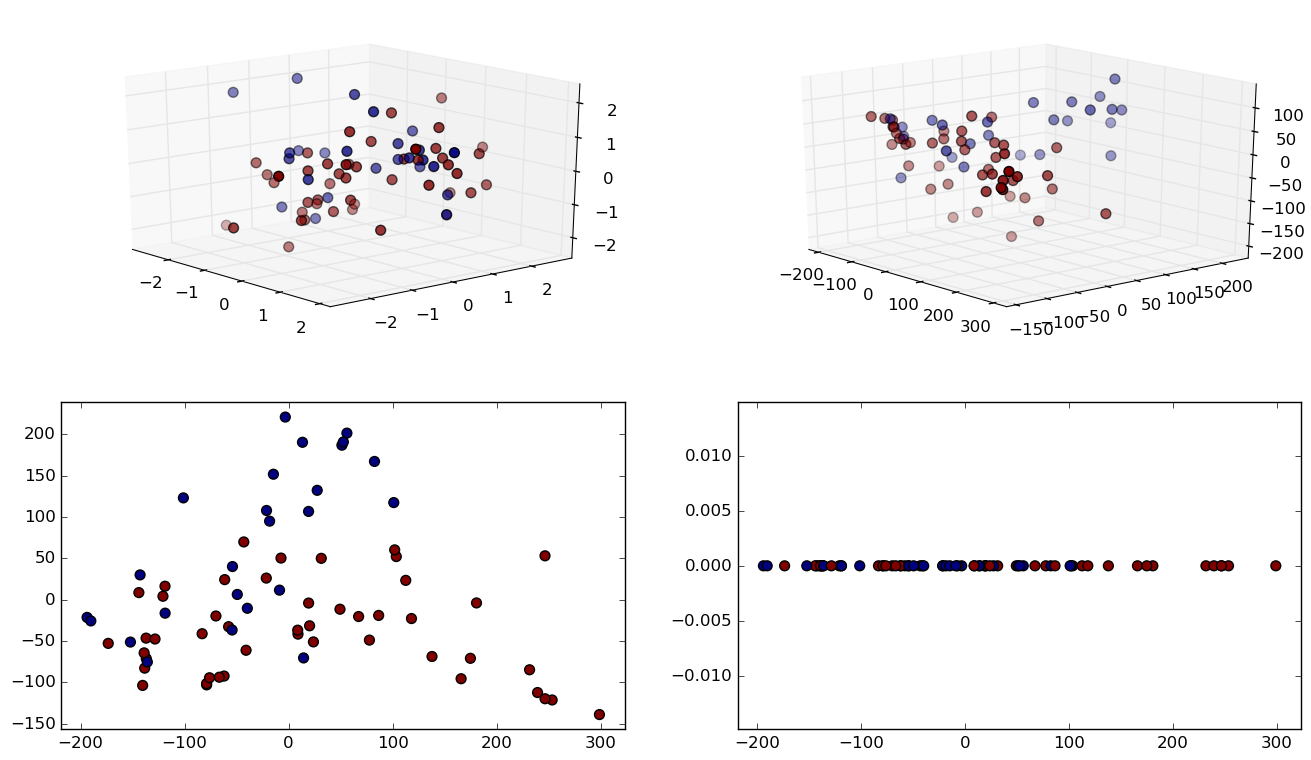
Experiment 2: Leukemia, 72 samples and 7130 features.
ISOMAP In Practice
Grid search was performed using a Support Vector Classifier.
- We observe massive memory requirement reduction, while loosing 11% of prediction accuracy.
ISOMAP In Practice
| 7130 dimensions | 10 dimensions | |
|---|---|---|
| Accuracy | 99% | 88% |
| Data size | 4010.06 KB | 16.88 KB |
| Grid time | 2.61 sec | .36 sec |
| Best parameters | 'degree': 2, 'coef0': 10, 'kernel': 'poly' | 'C': 1, 'kernel': 'linear' |
ISOMAP's
Complexity
Complexity
def isomap(data_set, n_components=2,
k=10, epsilon=1.0, use_k=True,
path_method='dijkstra'):
# 1. Compute neighborhood graph.
delta = nearest_neighbors(delta, k if use_k else epsilon)
# 2. Compute geodesic distances.
if path_method == 'dijkstra':
delta = all_pairs_dijkstra(delta) # 2.A
else:
delta = floyd_warshall(delta) # 2.B
# 3. Embed the data.
embedding = mds(delta, n_components)
return embeddingComplexity
Reducing it to 3 dimensions took...
- 29.9 minutes, with my implementation.
- 10.76 seconds, with scikit-learn's!
Experiment 3: Spam, 4601 samples and 57 features.
Complexity
Obviously, we can do do better!
Studying scikit-learn's implementation:
- Ball Tree is used for efficient neighbor search.
- Shortest-path searches are implemented in C.
- ISOMAP can be interpreted as the precomputed kernel for KernelPCA.
- ARPACK is used for the eigen-decomposition.
- Many of the steps can be parallelized.
Extensions
Extensions
ISOMAP as Kernel PCA
"Kernel Trick"
Sample Covariance Matrix.
Extensions
L-ISOMAP
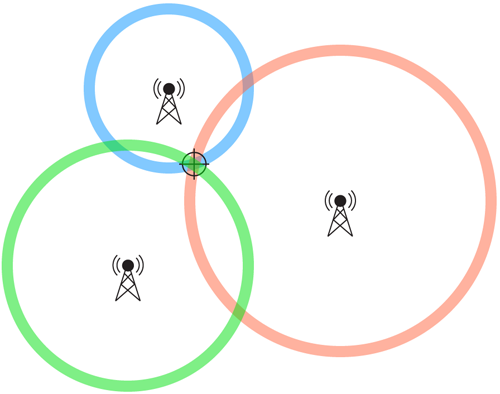
- In a n-dimensional space with N samples, a point's position can be found from its distance to n+1 landmarks and their respective position.
- Calculates the dissimilarity matrix
- Apply MDS to the sub-matrix
- Embed the rest of the data using the eigenvectors found.
Limitations
Limitations
Manifold Assumption
Disconnected graphs.
Incorrect reductions.
Possible work-around: increasing the parameters k or epsilon.
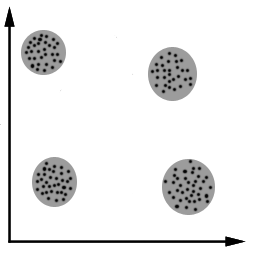
Limitations
Manifold Convexity
Deformation around the "hole".
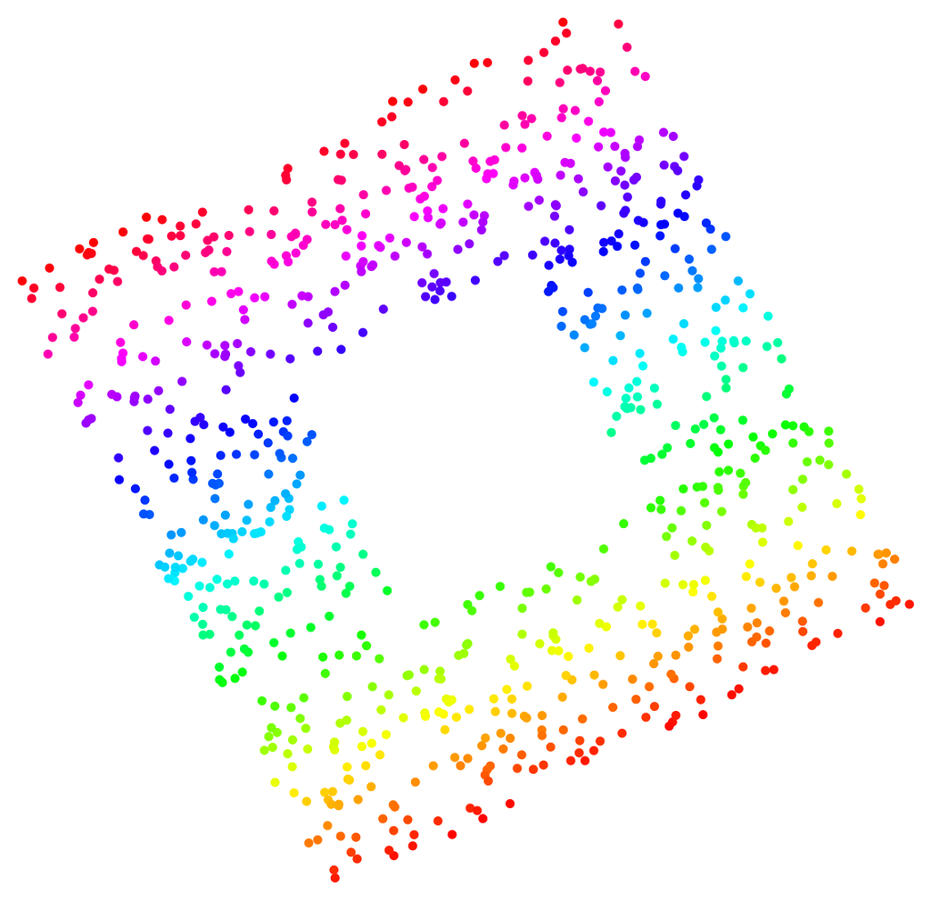
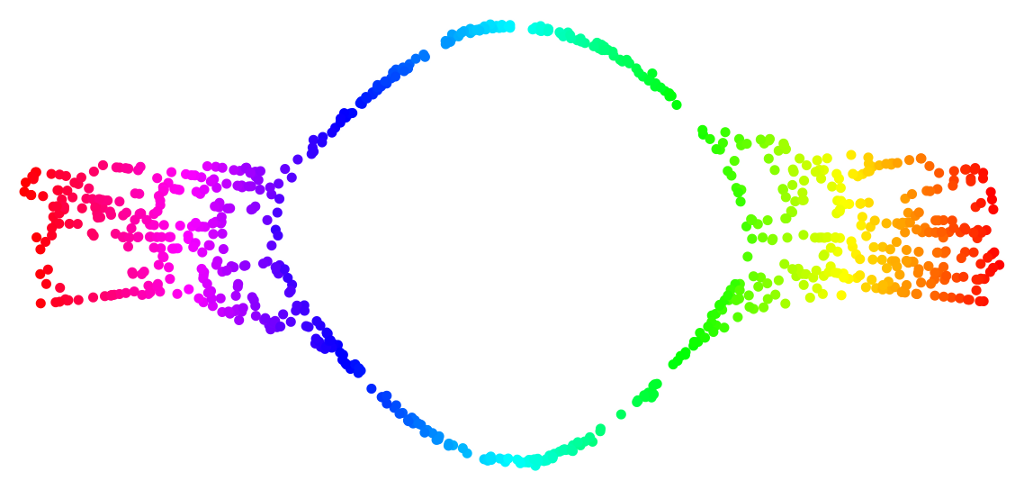
Limitations
Noise
Incorrect unfolding.
Possible work-around: reducing the parameters k or epsilon.
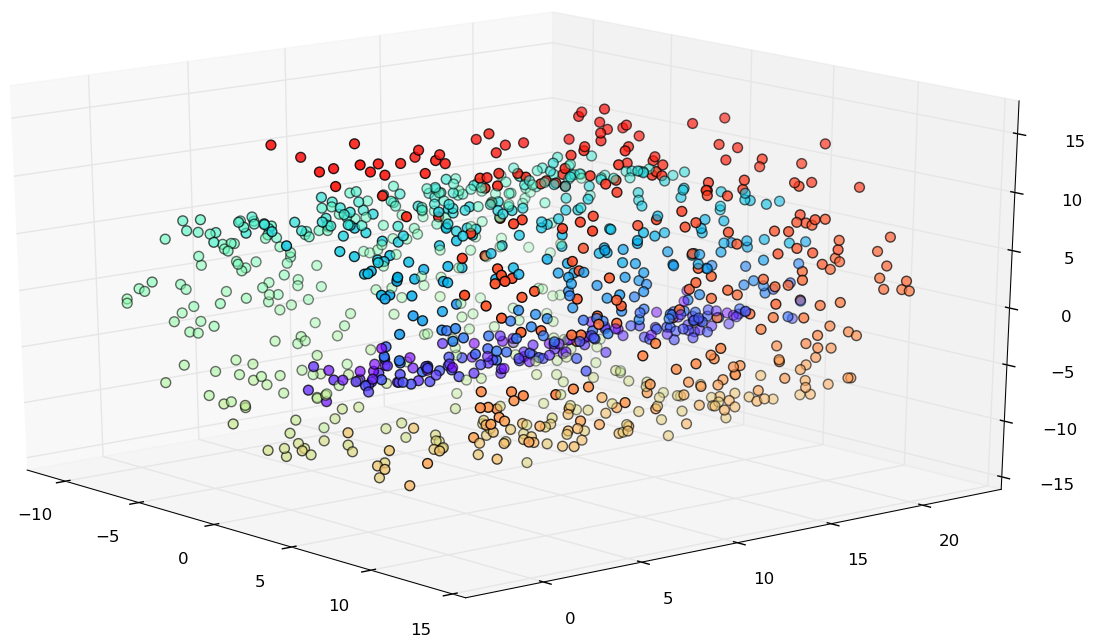
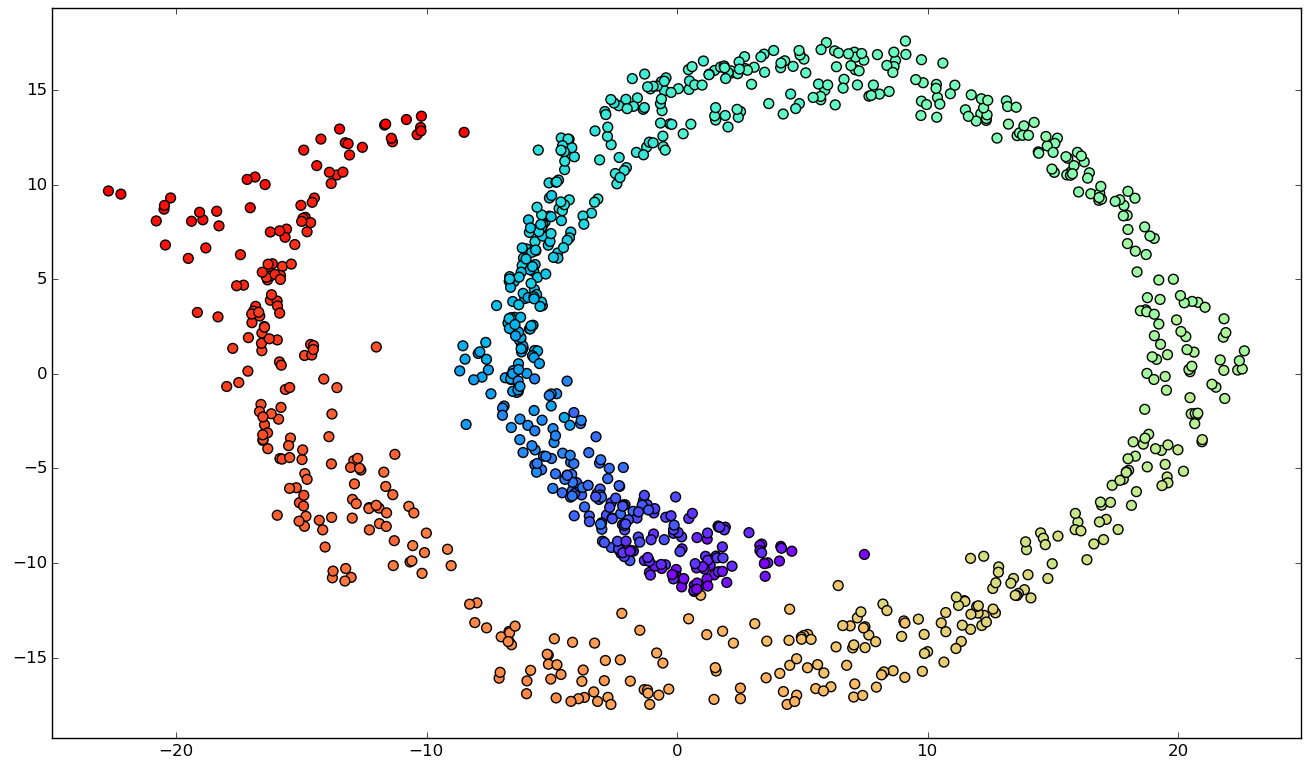
Final Considerations
- ISOMAP performed well on many experiments.
- However, it makes serious assumptions and it has its limitations. It is hard to say how it will behave on real-world data.
- ISOMAP is a landmark on Manifold Learning.
Thank you if you made it this far!
For ISOMAP implementation and experiments, check out my github: github.com/lucasdavid/manifold-learning.